Interferon-induced degradation of the persistent hepatitis B virus cccDNA form depends on ISG20 - PubMed (original) (raw)
. 2021 Jun 4;22(6):e49568.
doi: 10.15252/embr.201949568. Epub 2021 May 9.
Martin Kächele 1, Alisha N Jones 2 3, Julia Hess 4 5 6, Christian Urban 1, Jessica Schneider 1, Yuchen Xia 1 7, Andreas Oswald 1, Firat Nebioglu 8, Romina Bester 1, Felix Lasitschka 9, Marc Ringelhan 1 10, Chunkyu Ko 1, Wen-Min Chou 1, Arie Geerlof 2 3, Maarten A van de Klundert 1, Jochen M Wettengel 1, Peter Schirmacher 9 11, Mathias Heikenwälder 11 12, Sabrina Schreiner 1, Ralf Bartenschlager 8 11 13, Andreas Pichlmair 1 11, Michael Sattler 2 3, Kristian Unger 4 5 6, Ulrike Protzer 1 11
Affiliations
- PMID: 33969602
- PMCID: PMC8183418
- DOI: 10.15252/embr.201949568
Interferon-induced degradation of the persistent hepatitis B virus cccDNA form depends on ISG20
Daniela Stadler et al. EMBO Rep. 2021.
Abstract
Hepatitis B virus (HBV) persists by depositing a covalently closed circular DNA (cccDNA) in the nucleus of infected cells that cannot be targeted by available antivirals. Interferons can diminish HBV cccDNA via APOBEC3-mediated deamination. Here, we show that overexpression of APOBEC3A alone is not sufficient to reduce HBV cccDNA that requires additional treatment of cells with interferon indicating involvement of an interferon-stimulated gene (ISG) in cccDNA degradation. Transcriptome analyses identify ISG20 as the only type I and II interferon-induced, nuclear protein with annotated nuclease activity. ISG20 localizes to nucleoli of interferon-stimulated hepatocytes and is enriched on deoxyuridine-containing single-stranded DNA that mimics transcriptionally active, APOBEC3A-deaminated HBV DNA. ISG20 expression is detected in human livers in acute, self-limiting but not in chronic hepatitis B. ISG20 depletion mitigates the interferon-induced loss of cccDNA, and co-expression with APOBEC3A is sufficient to diminish cccDNA. In conclusion, non-cytolytic HBV cccDNA decline requires the concerted action of a deaminase and a nuclease. Our findings highlight that ISGs may cooperate in their antiviral activity that may be explored for therapeutic targeting.
Keywords: APOBEC3A; HBV; chronic hepatitis B; interferon alpha; interferon gamma.
© 2021 Helmholtz Zentrum Muenchen. Published under the terms of the CC BY NC ND 4.0 license.
Conflict of interest statement
The position of Daniela Stadler was in part financed by a research grant from ALIOS BioPharma. Ulrike Protzer is shareholder and board member of SCG Cell Therapy. The remaining authors declare no conflict of interest.
Figures
Figure EV1. IFNα induces APOBEC3 proteins, of which A3A is knocked down by siRNA
- A
dHepaRG cells were infected with HBV at a multiplicity of infection (MOI) of 100 virions/cell, transfected with siRNA against A3A (siA3A) or control siRNA (−) on day 6 postinfection (p.i.), and treated with 300 U/ml IFNα 1 day later for 6 h. Expression of indicated genes was analyzed by qRT–PCR relative to TATA‐box binding protein (TBP) mRNA (n = 3 biological replicates). mRNA from PBMCs was used as positive control in qRT–PCR for AICDA. - B–D
dHepaRG cells were infected with HBV at a multiplicity of infection (MOI) of 100 virions/cell and transfected with siRNA against A3A (siA3A) or control siRNA (−) on day 8 postinfection (p.i.). Cells were treated with 300 U/ml IFNα from days 9 to 15 p.i., and cell viability was determined thereafter by CellTiter‐Blue (CTB) assay (B), total intracellular HBV DNA by qPCR relative to the prion protein (Prnp) gene (C), and HBeAg by ELISA (D) (n = 4 biological replicates).
Data information: Data are represented as mean ± standard deviation (SD), nd: not detected. *P < 0.05, **P < 0.01, ***P < 0.001, and ns: not significant by Student’s unpaired _t_‐test with Welch’s correction.
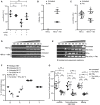
Figure 1. A3A deaminates HBV cccDNA dependent on transcription but is not sufficient to induce its degradation
- A
dHepaRG cells were infected with HBV at a multiplicity of infection (MOI) of 100 virions/cell and transfected with siRNA against A3A (siA3A) or control siRNA (−) on day 8 postinfection (p.i.). Cells were treated with 300 U/ml IFNα from days 9 to 15 p.i., and HBV cccDNA was determined thereafter by qPCR relative to the prion protein (Prnp) gene (n = 4 biological replicates). - B–D
dHepaRG‐TR‐X cells were infected with an HBx‐deficient HBV (HBVΔx) at an MOI of 200 virions/cell. Cells were treated with tetracycline to induce expression of HBx (+HBx) and/or 1,000 U/ml IFNα from days 10 to 17 p.i. HBeAg (B) was measured by ELISA (OD: optical density) and HBV cccDNA (C) by qPCR relative to Prnp (n = 3 biological replicates). cccDNA amplicons were analyzed by differential DNA denaturation PCR (3D‐PCR) (D) to visualize deamination. - E–G
dHepaRG‐TR‐A3A cells were infected with HBV at an MOI of 300 virions/cell. Cells were treated with tetracycline (Tet (A3A)) or/and 300 U/ml IFNα from days 9 to 18 p.i. cccDNA amplicons were analyzed by 3D‐PCR (E). Marked amplicons were sequenced and analyzed for nucleotide composition (F) in comparison with the wild‐type HBV sequence (n = 3–5 biological replicates). HBV infection level was determined by measuring cccDNA, intracellular HBV DNA by qPCR relative to Prnp, or HBeAg by ELISA (G) (n = 6 biological replicates from two independent experiments).
Data information: Data are represented as mean ± standard deviation (SD), nd: not detected. *P < 0.05, **P < 0.01, ***P < 0.001, and ns: not significant by Student’s unpaired _t_‐test with Welch’s correction.
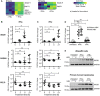
Figure 2. Differential gene expression analysis indicates ISG20 as candidate nuclease
- A
A3A‐expressing dHepaRG‐TR‐A3A cells were infected with HBV at an MOI of 300 virions/cell and treated 7 days after infection with 300 U/ml IFNα (+IFNα) or without (−IFNα). Microarray data were analyzed for upregulated nucleases after 6 or 72 h of treatment, respectively. - B, C
dHepaRG cells were infected with HBV at an MOI of 100 virions/cell and treated 9 days later with 300 U/ml IFNα (B) or 200 U/ml IFNγ (C) for indicated times (h: hours). Expression of indicated genes was analyzed by qRT–PCR relative to TATA‐box binding protein (TBP) mRNA (n = 6 biological replicates of two independent experiments). - D
dHepaRG cells were infected with HBV at an MOI of 100 virions/cell or not infected and treated 7 days later with 300 U/ml IFNα for 6 h or 200 U/ml IFNγ for 24 h. ISG20 mRNA levels were measured by qRT–PCR relative to TBP mRNA (n = 6 biological replicates of two independent experiments). - E
dHepaRG cells were infected with HBV at an MOI of 100 virions/cell and treated with 300 U/ml IFNα from days 7 to 9 p.i. or 200 U/ml IFNγ from days 7 to 10 p.i. Primary human hepatocytes were infected with HBV at an MOI of 100 virions/cell and treated with 500 U/ml IFNα or 200 U/ml IFNγ from days 4 to 6 p.i. Expression of ISG20 was detected by Western blot.
Data information: Data are represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, and ns: not significant by Student’s unpaired _t_‐test with Welch’s correction. Source data are available online for this figure.
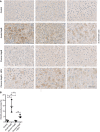
Figure 3. ISG20 is expressed in acute, self‐limiting HBV infection but not in chronic hepatitis B
- Liver tissue samples obtained from HBV‐negative patients undergoing metastasis resection (control) or patients with acute, self‐limiting HBV infection or chronic hepatitis B, or chronic coinfection with HBV and hepatitis D virus (HDV) were stained for ISG20 by immunohistochemistry. For each clinical entity, tissue sections from three different patients are shown; scale bar: 50 µm.
- ISG20‐positive area of each sample (n = 3 biological replicates) was determined by Tissue IA image analysis software and is given in % of total tissue area scanned.
Data information: Data are represented as mean ± SD. **P < 0.01 and ns: not significant by one‐way ANOVA.
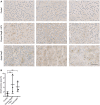
Figure EV2. ISG20 is expressed in HCV infection
- Liver tissue samples obtained from HBV‐negative patients undergoing metastasis resection (control), with chronic hepatitis B (HepB) and HCV coinfection or acute hepatitis C (HepC) were stained for ISG20 by immunohistochemistry. For each clinical entity, tissue sections from three different patients are shown; scale bar: 50 µm.
- ISG20‐positive area of each sample was determined by Tissue IA image analysis software (n = 3 biological replicates) and is given in % of total tissue area scanned.
Data information: Data are represented as mean ± SD. ns: not significant by one‐way ANOVA.
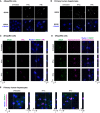
Figure 4. ISG20 localizes to nucleoli after interferon treatment
- A
dHepaRG cells were infected with HBV at an MOI of 100 virions/cell and treated with 600 U/ml IFNα or 400 U/ml IFNγ from days 10 to 14 p.i., and ISG20 was stained by immunofluorescence. - B
Primary human hepatocytes were infected with HBV at an MOI of 100 virions/cell and treated with 500 U/ml IFNα or 200 U/ml IFNγ from days 4 to 6 p.i., and ISG20 was stained by immunofluorescence. - C, D
dHepaRG cells were infected with HBV at an MOI of 500 virions/cell, treated with 600 U/ml IFNα or 400 U/ml IFNγ from days 7 to 9 p.i., and stained by immunofluorescence for ISG20 and PML (C) or ISG20 and nucleophosmin (D). - E
Primary human hepatocytes were infected with HBV at an MOI of 100 virions/cell, treated with 500 U/ml IFNα or 200 U/ml IFNγ from days 4 to 6 p.i., and stained for ISG20 and nucleophosmin. White arrowheads indicate double‐positive nuclei.
Data information: Scale bars: 10 µm.
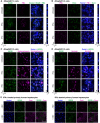
Figure EV3. ISG20 co‐localizes with nucleolar proteins and is expressed in HBV‐replicating cells
- A–D
dHepG2H1.3 cells were treated with 1,500 U/ml IFNα or 1,000 U/ml IFNγ from days 3 to 4 after differentiation start and stained by immunofluorescence for ISG20 and PML (A), PML and Daxx (B), ISG20 and nucleophosmin (C), or ISG20 and a nucleolar marker protein (D). Scale bar: 10 µm. - E
Primary human hepatocytes were infected with an MOI of 200 virions/cell, treated with 500 U/ml IFNα or 200 U/ml IFNγ from days 5 to 6 p.i., and stained for ISG20 and HBV core protein. Scale bar: 50 µm.
Figure EV4. ISG20 is knocked down by siRNA and binds and degrades ssDNA
- A–D
dHepaRG cells were infected with HBV at an MOI of 100 virions/cell and transfected with siRNA targeting ISG20 (siISG20) or control siRNA (−) at day 7 p.i. and treated with 300 U/ml IFNα from days 8 to 11 p.i. ISG20 mRNA (A) was measured by qRT–PCR relative to TBP mRNA (n = 3 biological replicates). cccDNA (B) and total intracellular HBV DNA (C) were determined by qPCR relative to Prnp, and HBeAg (D) was determined by ELISA (n = 5–6 biological replicates from two independent experiments). - E
ssDNA oligomers with and without dU modifications (left panel) or ssRNA oligomers (right panel) were digested in vitro for 4 h using 7 µM oligomer and 2.5 µM recombinant ISG20 each and separated by polyacrylamide gel electrophoresis. - F
NMR spectra of 15N‐labeled ISG20 bound to the (−) ssDNA and dU‐containing (−) ssDNA oligomers are shown. At a 1:1 ratio of oligomer to ISG20, both oligomers induce similar chemical shift perturbations in ISG20 amide resonances, as observed in the 1H,15N correlation NMR spectra.
Data information: Data are represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, and ns: not significant by Student’s unpaired _t_‐test with Welch’s correction.
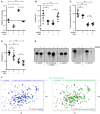
Figure 5. ISG20 knockdown mitigates interferon‐induced control of cccDNA
- A–D
dHepaRG cells were infected at an MOI of 100 virions/cell and transduced at day 7 p.i. with an adenoviral vector for expression of shRNA targeting ISG20 (AdV‐shISG20). After 2 days, cells were treated with 300 U/ml IFNα or 200 U/ml IFNγ. ISG20 expression (A) was analyzed by Western blot after 7 days of IFNα or 12 days of IFNγ treatment, respectively. The amount of ISG20/GAPDH was determined by signal density measurement. cccDNA (B) relative to Prnp (n = 4 biological replicates), total intracellular HBV DNA (C) relative to Prnp, and HBeAg (D) were measured 12 days after treatment (n = 8 biological replicates of two independent experiments). cccDNA was measured after T5 digestion of DNA.
Data information: Data are represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, and ns: not significant by Student’s unpaired _t_‐test with Welch’s correction. Source data are available online for this figure.
Figure 6. ISG20 degrades dsDNA and ssDNA and is enriched on deoxyuridine(dU)‐containing ssDNA
- A 3D‐PCR amplicon was digested in vitro with recombinant ISG20 and increasing concentration of manganese chloride.
- A subsequence of the 3D‐PCR amplicon was used as (−) ssDNA oligomer, complementary (+) ssDNA, and a dU‐containing (−) ssDNA, where all cytosines were replaced by uracils. These oligomers were digested in vitro with recombinant ISG20 with or without EDTA as indicated.
- Recombinant ISG20 and mA3A (mutant E72A, C72A) were mixed with indicated oligomers and analyzed in a single point binding mobility shift assay.
- dHepG2H1.3 cells were treated for 1 day with 1,000 U/ml IFNγ, and harvested proteins were subjected to an affinity purification–mass spectrometry assay (n = 4 technical replicates). Oligomers used for pulldown were single‐stranded or double‐stranded DNA with and without dU as indicated. Significantly enriched protein groups in the volcano plot were identified via two‐sided Welch’s _t_‐tests (S0 = 1) corrected for multiple hypothesis testing applying a permutation‐based FDR (FDR < 0.01, 250 randomizations). Dashed line and blue colored points indicate a significant enrichment according to the Welch’s _t_‐test cut‐offs. Statistical significance of log2 iBAQ intensities of ISG20 enrichment was determined using a two‐sided Welch’s _t_‐test.
Data information: Data are represented as mean ± SD. ***P ≤ 0.001 and n.s.: not significant (P > 0.05) by two‐sided Welch’s _t_‐test.
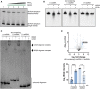
Figure 7. ISG20 overexpression together with A3A is sufficient for reduction of transcriptionally active cccDNA
- A
dHepaRG‐TR‐A3A cells were treated with tetracycline to induce A3A expression (Tet(A3A)) or transduced with an adenoviral vector for expression of ISG20 (AdV‐ISG20). After 4 days, expression of A3A and ISG20 was detected by Western blot. - B, C
dHepaRG‐TR‐A3A cells were infected with HBV at an MOI of 300 virions/cell and at day 7 p.i. transduced with AdV‐ISG20 and treated with tetracycline to induce A3A expression as indicated (± Tet (A3A)) for 7 days. Cell lysates were analyzed for cccDNA after T5 digest (B) and total intracellular HBV DNA (C) by qPCR relative to Prnp (n = 5–6 biological replicates from two independent experiments). - D
Primary human hepatocytes were infected with HBV at an MOI of 100 virions/cell and transfected on day 3 p.i. with mRNAs of A3A, ISG20, or catalytically inactive mutants thereof as indicated. Four days after transfection, cccDNA after T5 digest was measured by qPCR relative to Prnp (n = 4 biological replicates). - E, F
dHepaRG‐TR‐A3A cells were infected with wild‐type HBV (HBVwt) or HBVΔx at an MOI of 300 virions/cell and transduced on day 7 p.i. with AdV‐ISG20 and treated with tetracycline to induce A3A expression as indicated (± Tet (A3A)) for 7 days. HBeAg (E) was measured by ELISA and cccDNA after T5 digest (F) by qPCR relative to Prnp (n = 6 biological replicates from two independent experiments).
Data information: Data are represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, and ns: not significant by Student’s unpaired _t_‐test with Welch’s correction. Source data are available online for this figure.
Figure EV5. ISG20 overexpression together with A3A reduces cccDNA similar to interferon treatments
- A, B
dHepG2H1.3‐A3A cells constantly expressing A3A (A3A Ctrl) were transduced 10 days after differentiation with an empty adenoviral vector (AdV) or AdV‐ISG20. ISG20 levels (A) were determined by qRT–PCR relative to TBP mRNA 1 day after transduction (n = 4 biological replicates). cccDNA (B) was measured after T5 digest 7 days after transduction by qPCR relative to Prnp (n = 4 biological replicates). - C
dHepaRG‐TR‐A3A cells were infected with HBV at an MOI of 300 virions/cell and transduced on day 7 p.i. with AdV‐ISG20 and treated with tetracycline to induce A3A expression, 300 U/ml IFNα or 200 U/ml IFNγ. Seven days after transduction, cccDNA was measured after T5 digest by qPCR relative to Prnp (n = 5 biological replicates from two independent experiments). - D, E
dHepaRG‐TR‐A3A cells were infected with wild‐type HBV (HBVwt) or HBVΔx at an MOI of 300 virions/cell and transduced on day 7 p.i. with AdV‐ISG20 and treated with tetracycline to induce A3A expression as indicated (± Tet (A3A)) for 7 days. Cell viability (D) was measured by CTB assay and total intracellular HBV DNA (E) by qPCR relative to Prnp (n = 6 biological replicates from two independent experiments).
Data information: Data are represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, and ns: not significant by Student’s unpaired _t_‐test with Welch’s correction.
Similar articles
- Interferon-inducible MX2 is a host restriction factor of hepatitis B virus replication.
Wang YX, Niklasch M, Liu T, Wang Y, Shi B, Yuan W, Baumert TF, Yuan Z, Tong S, Nassal M, Wen YM. Wang YX, et al. J Hepatol. 2020 May;72(5):865-876. doi: 10.1016/j.jhep.2019.12.009. Epub 2019 Dec 18. J Hepatol. 2020. PMID: 31863794 - Intracellular interferon signalling pathways as potential regulators of covalently closed circular DNA in the treatment of chronic hepatitis B.
Goh ZY, Ren EC, Ko HL. Goh ZY, et al. World J Gastroenterol. 2021 Apr 14;27(14):1369-1391. doi: 10.3748/wjg.v27.i14.1369. World J Gastroenterol. 2021. PMID: 33911462 Free PMC article. Review. - Interferon-γ and Tumor Necrosis Factor-α Produced by T Cells Reduce the HBV Persistence Form, cccDNA, Without Cytolysis.
Xia Y, Stadler D, Lucifora J, Reisinger F, Webb D, Hösel M, Michler T, Wisskirchen K, Cheng X, Zhang K, Chou WM, Wettengel JM, Malo A, Bohne F, Hoffmann D, Eyer F, Thimme R, Falk CS, Thasler WE, Heikenwalder M, Protzer U. Xia Y, et al. Gastroenterology. 2016 Jan;150(1):194-205. doi: 10.1053/j.gastro.2015.09.026. Epub 2015 Sep 28. Gastroenterology. 2016. PMID: 26416327 - Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA.
Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Hüser N, Durantel D, Liang TJ, Münk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. Lucifora J, et al. Science. 2014 Mar 14;343(6176):1221-8. doi: 10.1126/science.1243462. Epub 2014 Feb 20. Science. 2014. PMID: 24557838 Free PMC article. - Hepatitis B virus cccDNA: Formation, regulation and therapeutic potential.
Xia Y, Guo H. Xia Y, et al. Antiviral Res. 2020 Aug;180:104824. doi: 10.1016/j.antiviral.2020.104824. Epub 2020 May 22. Antiviral Res. 2020. PMID: 32450266 Free PMC article. Review.
Cited by
- Role of epigenetic modification in interferon treatment of hepatitis B virus infection.
Yang Z, Sun B, Xiang J, Wu H, Kan S, Hao M, Chang L, Liu H, Wang D, Liu W. Yang Z, et al. Front Immunol. 2022 Oct 17;13:1018053. doi: 10.3389/fimmu.2022.1018053. eCollection 2022. Front Immunol. 2022. PMID: 36325353 Free PMC article. Review. - Stable structures or PABP1 loading protects cellular and viral RNAs against ISG20-mediated decay.
Louvat C, Deymier S, Nguyen XN, Labaronne E, Noy K, Cariou M, Corbin A, Mateo M, Ricci EP, Fiorini F, Cimarelli A. Louvat C, et al. Life Sci Alliance. 2024 Feb 28;7(5):e202302233. doi: 10.26508/lsa.202302233. Print 2024 May. Life Sci Alliance. 2024. PMID: 38418089 Free PMC article. - Identification of disulfidptosis-related subtypes and development of a prognosis model based on stacking framework in renal clear cell carcinoma.
Peng K, Wang N, Liu Q, Wang L, Duan X, Xie G, Li J, Ding D. Peng K, et al. J Cancer Res Clin Oncol. 2023 Nov;149(15):13793-13810. doi: 10.1007/s00432-023-05201-3. Epub 2023 Aug 2. J Cancer Res Clin Oncol. 2023. PMID: 37530800 - APOBEC3: Friend or Foe in Human Papillomavirus Infection and Oncogenesis?
Warren CJ, Santiago ML, Pyeon D. Warren CJ, et al. Annu Rev Virol. 2022 Sep 29;9(1):375-395. doi: 10.1146/annurev-virology-092920-030354. Epub 2022 Jun 7. Annu Rev Virol. 2022. PMID: 35671565 Free PMC article. Review. - The efficacy of pegylated interferon alpha-2a and entecavir in HBeAg-positive children and adolescents with chronic hepatitis B.
He Y, Zhou Y, Wang H, Peng X, Chang Y, Hu P, Ren H, Xu H. He Y, et al. BMC Pediatr. 2022 Jul 20;22(1):426. doi: 10.1186/s12887-022-03482-0. BMC Pediatr. 2022. PMID: 35854256 Free PMC article.
References
- Allweiss L, Volz T, Giersch K, Kah J, Raffa G, Petersen J, Lohse AW, Beninati C, Pollicino T, Urban S et al (2018) Proliferation of primary human hepatocytes and prevention of hepatitis B virus reinfection efficiently deplete nuclear cccDNA in vivo. Gut 67: 542–552 - PubMed
- Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M, Levrero M (2012) IFN‐α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest 122: 529–537 - PMC - PubMed
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B 57: 289–300
- Bertoletti A, Ferrari C (2012) Innate and adaptive immune responses in chronic hepatitis B virus infections: towards restoration of immune control of viral infection. Gut 61: 1754–1764 - PubMed
- Bessho T, Sancar A (2000) Human DNA damage checkpoint protein hRAD9 Is a 3′ to 5′ exonuclease. J Biol Chem 275: 7451–7454 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases