CRISPR interference interrogation of COPD GWAS genes reveals the functional significance of desmoplakin in iPSC-derived alveolar epithelial cells - PubMed (original) (raw)
. 2022 Jul 15;8(28):eabo6566.
doi: 10.1126/sciadv.abo6566. Epub 2022 Jul 13.
Rhiannon B Werder 1 2 3, Kristine M Abo 1 2, Jonathan Lindstrom-Vautrin 1, Carlos Villacorta-Martin 1, Jessie Huang 1 2, Anne Hinds 2, Nathan Boyer 4, Esther Bullitt 5, Marc Liesa 6 7, Edwin K Silverman 4, Darrell N Kotton 1 2, Michael H Cho 4, Xiaobo Zhou 4, Andrew A Wilson 1 2
Affiliations
- PMID: 35857525
- PMCID: PMC9278866
- DOI: 10.1126/sciadv.abo6566
CRISPR interference interrogation of COPD GWAS genes reveals the functional significance of desmoplakin in iPSC-derived alveolar epithelial cells
Rhiannon B Werder et al. Sci Adv. 2022.
Abstract
Genome-wide association studies (GWAS) have identified dozens of loci associated with chronic obstructive pulmonary disease (COPD) susceptibility; however, the function of associated genes in the cell type(s) affected in disease remains poorly understood, partly due to a lack of cell models that recapitulate human alveolar biology. Here, we apply CRISPR interference to interrogate the function of nine genes implicated in COPD by GWAS in induced pluripotent stem cell-derived type 2 alveolar epithelial cells (iAT2s). We find that multiple genes implicated by GWAS affect iAT2 function, including differentiation potential, maturation, and/or proliferation. Detailed characterization of the GWAS gene DSP demonstrates that it regulates iAT2 cell-cell junctions, proliferation, mitochondrial function, and response to cigarette smoke-induced injury. Our approach thus elucidates the biological function, as well as disease-relevant consequences of dysfunction, of genes implicated in COPD by GWAS in type 2 alveolar epithelial cells.
Figures
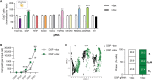
Fig. 1.. Overview of CRISPRi platform to interrogate COPD GWAS genes.
(A) Gene editing strategy to generate inducible CRISPRi iPSC/ESC lines with an NKX2-1–GFP and/or SFTPC-tdTomato reporter. (B) Schematic representation of dox-inducible dCas9-KRAB expression and lentiviral-delivered gRNA targeted to the transcriptional start site (TSS) of genes of interest (GOI) to mediate knockdown. (C) Effector genes at COPD genome-wide significant loci were identified using gene expression, methylation status, coding associations, deoxyribonuclease (DNase) hypersensitivity, chromatin interactions, and/or similarity in gene sets, as previously described (11). Genes were further filtered on the basis of expression during lung-directed differentiation of iPSCs, expression in differentiated iAT2s, and expression changes during human lung development. Expression levels of the final selected genes, FAM13A, DSP, TGFB2, MFAP2, RBMS3, SOX4, SOX9, HHIP, and ADGRG6, are shown in iPSCs, iPSC-lung progenitors, early (16 to 17.5 weeks of gestation) and late (20 to 21 weeks of gestation) human fetal lung (HFL), iAT2s grown as 3D alveolospheres or at air-liquid interface (ALI), primary pediatric (13-month-old male donor) and adult (32-year-old male donor) AT2s grown in CK + DCI with MRC-5 cells, and freshly isolated primary adult AT2s (26).
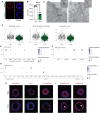
Fig. 2.. COPD GWAS candidate genes can be interrogated by CRISPRi.
(A) Schematic representation of proposed experiments to interrogate the function of genes of interest in iAT2s. iPSCs undergo directed differentiation to generate NKX2-1+ lung progenitors, which are then sorted and replated in distal medium (CK + DCI). iAT2s are harvested at either early or later time points to assess the effect of target genes on specific end points as indicated. (B) CRISPRi-iPSC–derived iAT2s were transduced with lentiviral-gRNA, and transduced cells were sorted 7 days later based on lenti-BFP expression. After recovery and expansion, iAT2s were treated with dox for 7 to 21 days to initiate CRISPRi-knockdown and harvested 7 days after passage. (C) Expression of each gene of interest following dox treatment as assessed by qRT-PCR, relative to control (−dox) iAT2s. (D) Expression of SFTPC following knockdown of each gene of interest as assessed by qRT-PCR relative to control (−dox) iAT2s. (E) Representative flow cytometry plots for iAT2s transduced with DSP gRNA treated without (−dox) or with (+dox) doxycycline showing NKX2-1–GFP and SFTPC-tdTomato expression. Cells were gated on nonfragmented single cells. (F) Representative live-cell imaging of iAT2s transduced with DSP gRNA treated without (−dox) or with (+dox) doxycycline (bright-field/SFTPC-tdTomato overlay; scale bar, 100 μm). (G) SFTPC expression following knockdown in RUES2 ST CRISPRi iAT2s. (H) Expression of SFTPA1 expression following knockdown of each gene of interest, assessed by qRT-PCR relative to control (−dox) iAT2s. NT, non-targeting gRNA. n = 3 experimental replicates of independent wells of a differentiation; error bars represent SD. Statistical significance was determined by unpaired, two-tailed Student’s t test; *P < 0.05 and **P < 0.005.
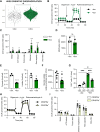
Fig. 3.. CRISPRi-knockdown of genes of interest modulates iAT2 proliferation.
(A) CRISPRi-iAT2s were transduced with lentiviral-gRNA as described in Fig. 2. Six days after passage, cells were treated with EdU for 24 hours. EdU incorporation was measured by flow cytometry, and the data were normalized to the control (−dox) iAT2s. (B) Graph showing yield per input iAT2 transduced with DSP gRNA and treated without (−dox) or with (+dox) doxycycline across five passages. (C) Single-cell RNA sequencing (scRNA-seq) showing cell cycle phase using UMAP of iAT2s transduced with DSP gRNA and treated without (−dox, black) or with (+dox, green) doxycycline. n = 3 experimental replicates of independent wells of a differentiation; error bars represent SD. Statistical significance was determined by unpaired, two-tailed Student’s t test; **P < 0.005, and ***P < 0.001.
Fig. 4.. DSP expression modulates iAT2 transcriptome.
(A) iAT2s were nucleofected with Cas9-gRNA RNP with gRNA targeting exon 1 of DSP. DSP expression was measured by qRT-PCR 48 hours after nucleofection. (B) Colony-forming efficiency was determined 2 weeks after nucleofection with RNP. n = 3 experimental replicates of independent wells of a differentiation. (C) iAT2s were transduced with lentiviral-gRNA targeting the TSS of DSP and sorted to obtain a pure population. iAT2s were then transduced with CRISPRa lentivirus (dCAS9-VP64). Cas9, (D) DSP, and (E) SFTPC expression were measured by qRT-PCR. (F) EdU incorporation was measured by flow cytometry, and the data were normalized to the control (-CRISPRa) iAT2s. n = 6 experimental replicates of independent wells of a differentiation. (G) UMAP of scRNA-seq, showing original identity (−dox = black, +dox = green). (H) Heatmap of scRNA-seq showing top 40 differentially expressed genes between –dox and +dox (green). (I) UMAP showing SFTPC and NAPSA expression. (J) Module score of AT2 maturation gene set (21). All error bars represent SD. Statistical significance was determined by unpaired, two-tailed Student’s t test; *P < 0.05, **P < 0.005, and ***P < 0.001.
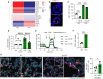
Fig. 5.. _DSP_-kd disrupts cell-cell junctions and cytoskeletal organization.
CRISPRi-iAT2s were transduced with DSP gRNA and treated without (−dox) or with (+dox) doxycycline. (A) Immunofluorescence staining of desmoplakin (DSP). DSP, red; nuclei, blue; scale bar, 10 μm. (B) Flow cytometry analysis of DSP expression, represented by mean fluorescence intensity (MFI). n = 3 experimental replicates of independent wells of a differentiation; error bars represent SD. Statistical significance was determined by unpaired, two-tailed Student’s t test; ***P < 0.001. (C) Transmission electron microscopy images showing tight junctions (TJ) and desmosomes (De); scale bar, 200 nm. (D) Violin plots showing module scores for KEGG (Kyoto Encyclopedia of Genes and Genomes) tight junction, cell adhesion molecules, and gap junction. (E) Dot plots showing expression of genes associated with desmosome, (F) adherens junction, and (G) TJ. Genes that were not expressed by iAT2s were excluded from dot plots. (H) Immunofluorescence staining of claudin-4, ZO-1, and E-cadherin. (I) Immunofluorescence staining of keratin-18 and α-tubulin. Arrows indicate disorganized filaments or tubules. Protein of interest, red; nuclei, blue; scale bar, 10 μm.
Fig. 6.. DSP expression influences mitochondrial respiration.
CRISPRi-iAT2s were transduced with DSP gRNA and treated without (−dox) or with (+dox) doxycycline (A to G). (A) Module score for KEGG oxidative phosphorylation of scRNA-seq. (B and C) Basal oxygen consumption rate (OCR) was measured, followed by injection of oligomycin, FCCP, and rotenone + antimycin A, as indicated. Data were normalized by cell count after the assay was complete. n = 6 technical replicates of a differentiation. (D) MitoTracker staining was quantified by flow cytometry, represented by MFI; n = 3 experimental replicates of independent wells of a differentiation. (E) Pyruvate or lactate was measured in the supernatant and normalized by cell count; n = 3 experimental replicates of independent wells of a differentiation. (F) Glucose uptake was determined using 2-DG and measuring luciferase; n = 3 experimental replicates of independent wells of two differentiations. (G) Cells were treated with 10 μM etomoxir or vehicle for 21 days. One day before harvest, cells were incubated with EdU. EdU incorporation was measured by flow cytometry. n = 3 experimental replicates of independent wells of a differentiation. (H and I) iAT2s were transduced with DSP gRNA ± CRISPRa lentivirus. Basal OCR was measured, followed by injection of oligomycin, FCCP, and rotenone + antimycin A, as indicated. Data were normalized by cell count after the assay was complete. n = 10 technical replicates of a differentiation. All error bars represent SD. Statistical significance was determined using unpaired, two-tailed Student’s t test or a one-way ANOVA with a Tukey multiple comparison test; *P < 0.05, **P < 0.005, and ***P < 0.001.
Fig. 7.. _DSP_-kd regulates proliferation through ERK-MAPK.
CRISPRi-iAT2s were transduced with DSP gRNA and treated without (−dox) or with (+dox) doxycycline. (A) PROGENγ (Pathway RespOnsive GENes) of scRNA-seq. (B) Immunofluorescence of phosphorylated (p)-ERK1/2 (red); scale bar, 10 μm. (C) Cells were treated with U0126 for 3 days and pulsed with EdU for 24 hours, and EdU was measured by flow cytometry. n = 3 experimental replicates of independent wells of a differentiation. (D and E) Cells were treated with U0126 for 7 days, and then basal OCR was measured, followed by injection of oligomycin, FCCP, and rotenone + antimycin A, as indicated. Data were normalized by cell count after the assay was complete. (F) KRAS expression was measured by qRT-PCR; n = 3 experimental replicates of independent wells of a differentiation. (G) Nkx2.1-Cre mice were crossed with Dspfl/fl mice. Mice were inoculated with 200 PFU of influenza (IAV) and injected with EdU 1 day before sacrifice at 10 days after infection. Pro-SPC (green), EdU (red), and nuclei (blue) were stained for immunofluorescence. Green arrows denote EdU- Pro-SPC+ cells; white arrows denote EdU+ Pro-SPC+ cells. EdU+ SPC+ cells were quantified in a blinded manner and normalized to total SPC+ cells. n = 4 mice per group.
Fig. 8.. DSP expression alters the cellular response to injury.
(A) Cells were plated in 2D and allowed to reach confluence before a scratch wound was made. Wound closure was calculated as a percentage of the initial wound over a 24-hour period. n = 3 experimental replicates of independent wells of a differentiation. (B) Cells were treated with TGF-β1 (1 ng/ml) for 48 hours before collecting RNA, and COL1A1 expression was measured by qRT-PCR. n = 3 experimental replicates of independent wells of a differentiation. (C) Cells were plated at ALI and then exposed to 5% cigarette smoke in a gas phase or humidified air. Cells were collected 2 hours after smoke exposure, and CLDN4, TJP1, and TJP3 were measured by qRT-PCR. n = 3 experimental replicates of independent ALIs of a differentiation. (D) Cells were exposed to 5% cigarette smoke in a gas phase or humidified air while plated at ALI and then replated in 3D Matrigel to reform alveolospheres over 2 weeks. Colony-forming efficiency was calculated relative to air-exposed cells. n = 3 experimental replicates of independent wells of two differentiations. All error bars represent SD. Statistical significance was determined using unpaired, two-tailed Student’s t test or a one-way ANOVA with a Tukey multiple comparison test; *P < 0.05, **P < 0.005, and ***P < 0.001.
Similar articles
- The COPD GWAS gene ADGRG6 instructs function and injury response in human iPSC-derived type II alveolar epithelial cells.
Werder RB, Berthiaume KA, Merritt C, Gallagher M, Villacorta-Martin C, Wang F, Bawa P, Malik V, Lyons SM, Basil MC, Morrisey EE, Kotton DN, Zhou X, Cho MH, Wilson AA. Werder RB, et al. Am J Hum Genet. 2023 Oct 5;110(10):1735-1749. doi: 10.1016/j.ajhg.2023.08.017. Epub 2023 Sep 20. Am J Hum Genet. 2023. PMID: 37734371 Free PMC article. - Genome-Wide Association Study: Functional Variant rs2076295 Regulates Desmoplakin Expression in Airway Epithelial Cells.
Hao Y, Bates S, Mou H, Yun JH, Pham B, Liu J, Qiu W, Guo F, Morrow JD, Hersh CP, Benway CJ, Gong L, Zhang Y, Rosas IO, Cho MH, Park JA, Castaldi PJ, Du F, Zhou X. Hao Y, et al. Am J Respir Crit Care Med. 2020 Nov 1;202(9):1225-1236. doi: 10.1164/rccm.201910-1958OC. Am J Respir Crit Care Med. 2020. PMID: 32551799 Free PMC article. - Identification of Functional Variants in the FAM13A Chronic Obstructive Pulmonary Disease Genome-Wide Association Study Locus by Massively Parallel Reporter Assays.
Castaldi PJ, Guo F, Qiao D, Du F, Naing ZZC, Li Y, Pham B, Mikkelsen TS, Cho MH, Silverman EK, Zhou X. Castaldi PJ, et al. Am J Respir Crit Care Med. 2019 Jan 1;199(1):52-61. doi: 10.1164/rccm.201802-0337OC. Am J Respir Crit Care Med. 2019. PMID: 30079747 Free PMC article. - Deciphering pathogenicity of variants of uncertain significance with CRISPR-edited iPSCs.
Guo H, Liu L, Nishiga M, Cong L, Wu JC. Guo H, et al. Trends Genet. 2021 Dec;37(12):1109-1123. doi: 10.1016/j.tig.2021.08.009. Epub 2021 Sep 8. Trends Genet. 2021. PMID: 34509299 Free PMC article. Review. - CRISPR/Cas9 Genome Editing: A Promising Tool for Therapeutic Applications of Induced Pluripotent Stem Cells.
Zhang Y, Sastre D, Wang F. Zhang Y, et al. Curr Stem Cell Res Ther. 2018;13(4):243-251. doi: 10.2174/1574888X13666180214124800. Curr Stem Cell Res Ther. 2018. PMID: 29446747 Review.
Cited by
- CRISPR/Cas9 gene editing: a novel strategy for fighting drug resistance in respiratory disorders.
Hussen BM, Najmadden ZB, Abdullah SR, Rasul MF, Mustafa SA, Ghafouri-Fard S, Taheri M. Hussen BM, et al. Cell Commun Signal. 2024 Jun 14;22(1):329. doi: 10.1186/s12964-024-01713-8. Cell Commun Signal. 2024. PMID: 38877530 Free PMC article. Review. - Nanomaterials-assisted gene editing and synthetic biology for optimizing the treatment of pulmonary diseases.
Lei L, Pan W, Shou X, Shao Y, Ye S, Zhang J, Kolliputi N, Shi L. Lei L, et al. J Nanobiotechnology. 2024 Jun 18;22(1):343. doi: 10.1186/s12951-024-02627-w. J Nanobiotechnology. 2024. PMID: 38890749 Free PMC article. Review. - Benchmarking methods integrating GWAS and single-cell transcriptomic data for mapping trait-cell type associations.
Li A, Lin T, Walker A, Tan X, Zhao R, Yao S, Sullivan PF, Hjerling-Leffler J, Wray NR, Zeng J. Li A, et al. medRxiv [Preprint]. 2025 Jun 5:2025.05.24.25328275. doi: 10.1101/2025.05.24.25328275. medRxiv. 2025. PMID: 40475144 Free PMC article. Preprint. - Advancing Treatment Strategies: A Comprehensive Review of Drug Delivery Innovations for Chronic Inflammatory Respiratory Diseases.
Wang J, Wang P, Shao Y, He D. Wang J, et al. Pharmaceutics. 2023 Aug 17;15(8):2151. doi: 10.3390/pharmaceutics15082151. Pharmaceutics. 2023. PMID: 37631365 Free PMC article. Review. - Lung repair and regeneration: Advanced models and insights into human disease.
Basil MC, Alysandratos KD, Kotton DN, Morrisey EE. Basil MC, et al. Cell Stem Cell. 2024 Apr 4;31(4):439-454. doi: 10.1016/j.stem.2024.02.009. Epub 2024 Mar 15. Cell Stem Cell. 2024. PMID: 38492572 Free PMC article. Review.
References
- World Health Organization, Chronic obstructive pulmonary disease (COPD) (WHO Fact Sheet, 2021); https://www.who.int/news-room/fact-sheets/detail/chronic-obstructive-pul....
- Regan E. A., Lynch D. A., Curran-Everett D., Curtis J. L., Austin J. H. M., Grenier P. A., Kauczor H.-U., Bailey W. C., DeMeo D. L., Casaburi R. H., Friedman P., Van Beek E. J. R., Hokanson J. E., Bowler R. P., Beaty T. H., Washko G. R., Han M. L. K., Kim V., Kim S. S., Yagihashi K., Washington L., McEvoy C. E., Tanner C., Mannino D. M., Make B. J., Silverman E. K., Crapop J. D.; Genetic Epidemiology of COPD (COPDGene) Investigators , Clinical and radiologic disease in smokers with normal spirometry. JAMA Intern. Med. 175, 1539–1549 (2015). - PMC - PubMed
- Lange P., Celli B., Agustí A., Jensen G. B., Divo M., Faner R., Guerra S., Marott J. L., Martinez F. D., Martinez-Camblor P., Meek P., Owen C. A., Petersen H., Pinto-Plata V., Schnohr P., Sood A., Soriano J. B., Tesfaigzi Y., Vestbo J., Lung-function trajectories leading to chronic obstructive pulmonary disease. N. Engl. J. Med. 373, 111–122 (2015). - PubMed
MeSH terms
Substances
Grants and funding
- R01 HL148667/HL/NHLBI NIH HHS/United States
- U01 TR001810/TR/NCATS NIH HHS/United States
- S10 OD028571/OD/NIH HHS/United States
- R01 HL147148/HL/NHLBI NIH HHS/United States
- R01 HL135142/HL/NHLBI NIH HHS/United States
- R01 DK117940/DK/NIDDK NIH HHS/United States
- R01 HL137927/HL/NHLBI NIH HHS/United States
- S10 OD021587/OD/NIH HHS/United States
- R01 HL127200/HL/NHLBI NIH HHS/United States
- F30 HL147426/HL/NHLBI NIH HHS/United States
- R01 DK101501/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous