Structural basis for activation of DNMT1 - PubMed (original) (raw)
. 2022 Nov 21;13(1):7130.
doi: 10.1038/s41467-022-34779-4.
Hiroki Onoda # 1 2, Kosuke Yamaguchi 3, Satomi Kori 1, Shun Matsuzawa 1, Yoshie Chiba 4, Shota Tanimoto 4, Sae Yoshimi 1, Hiroki Sato 1, Atsushi Yamagata 5, Mikako Shirouzu 5, Naruhiko Adachi 6, Jafar Sharif 7, Haruhiko Koseki 7 8, Atsuya Nishiyama 4, Makoto Nakanishi 4, Pierre-Antoine Defossez 3, Kyohei Arita 9
Affiliations
- PMID: 36414620
- PMCID: PMC9681727
- DOI: 10.1038/s41467-022-34779-4
Structural basis for activation of DNMT1
Amika Kikuchi et al. Nat Commun. 2022.
Abstract
DNMT1 is an essential enzyme that maintains genomic DNA methylation, and its function is regulated by mechanisms that are not yet fully understood. Here, we report the cryo-EM structure of human DNMT1 bound to its two natural activators: hemimethylated DNA and ubiquitinated histone H3. We find that a hitherto unstudied linker, between the RFTS and CXXC domains, plays a key role for activation. It contains a conserved α-helix which engages a crucial "Toggle" pocket, displacing a previously described inhibitory linker, and allowing the DNA Recognition Helix to spring into the active conformation. This is accompanied by large-scale reorganization of the inhibitory RFTS and CXXC domains, allowing the enzyme to gain full activity. Our results therefore provide a mechanistic basis for the activation of DNMT1, with consequences for basic research and drug design.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
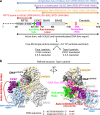
Fig. 1. Cryo-EM single particle analysis of DNMT1:H3Ub2:DNAmCG/fCG.
a Domain architecture of DNMT1, with amino acid numbers indicated. b Cryo-EM map of the DNMT1:H3Ub2:DNAmCG/fCG ternary complex superposed on the cartoon model. Disordered Auto-Inhibitory Linker and N-terminal portion of BAH1 domain are shown as dotted lines.
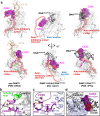
Fig. 2. Spatial rearrangement of the CXXC domain upon activation.
a Comparison of the overall structure of apo-DNMT1 (left, PDB:
4WXX
), DNMT1:H3Ub2:DNAmCG/fCG ternary complex (center, this study), and DNMT1:DNACG/CG binary complex (right, PDB:
3PTA
). Upper and lower panels show the side and front views of DNMT1, respectively. RFTS domain, Activating Helix, CXXC domain, Auto-Inhibitory Linker, and DNA Recognition Helix were colored light orange, light green, purple, red, and pink, respectively. CXXC domain and Auto-Inhibitory Linker are also exhibited as the transparent surface model. The disordered regions of BAH1 and Auto-Inhibitory Linker in DNMT1:H3Ub2:DNAmCG/fCG ternary complex are shown as dotted line. The red surface in the CXXC domain indicates R681-K683, which binds the major groove of unmethylated DNACG/CG. b Detail of the interaction between the CXXC domain and catalytic domain showing gray cartoon and stick models in the ternary complex. c Detail of the interaction between the CXXC domain and BAH1 domain showing light-purple cartoon and stick models in the ternary complex. d Model structure of CXXC domain bound to DNACG/CG in the ternary complex. CXXC:DNACG/CG in PDB:
3PTA
is superimposed on the CXXC domain in the ternary complex. The red surface in the CXXC domain indicates the binding surface for binding to the major groove of unmethylated DNACG/CG.
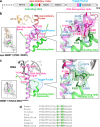
Fig. 3. Activation of DNMT1 by a pair of phenylalanine residues in the Activating Helix.
a Domain structure of DNMT1. b Structure around Activating Helix (green), DNA Recognition Helix (pink), and Toggle Pocket (pale cyan) of apo-DNMT1 (PDB:
4WXX
). The Toggle Pocket is highlighted by a light blue circle. The right panel is a magnified figure of the Toggle Pocket. Residues in Activating Helix and DNA Recognition Helix that are involved in binding to the Toggle Pocket are shown as green and pink stick models, respectively. Residues for the formation of the Toggle Pocket are shown as a stick with a transparent sphere model. c Structure around Activating Helix (green), DNA Recognition Helix (pink), and Toggle Pocket (pale cyan) of the ternary complex. The color scheme is the same as Fig. 3b. d Multiple sequence alignment of DNMT1 around the Activating Helix. Phenylalanines 628, 631, and 632 are highlighted.
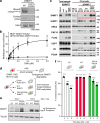
Fig. 4. Functional assays demonstrate the importance of the Activating Helix.
a Pull-down assay using immobilized hemimethylated DNA. Three independent experiments were performed. b in vitro DNA methylation activity of DNMT1 WT (black square: mean value, white square: three independent experiment values) and F631A/F632A mutant (blacks circle: mean value, white circle: three independent experiment values) for hemimethylated DNA. The vertical axis indicates the turnover frequency of the methylation reaction of DNMT1 (15 nM) after 1 h. Michaelis–Menten curve is shown as lines. Data were presented as mean values ± SD for n = 3. c Interphase extracts depleted with either control or xDNMT1 antibodies were incubated with control buffer, purified recombinant WT xDNMT1, and F506A/F507A. The chromatin fractions were isolated at the indicated times and their bound proteins, as well as inputs, were analyzed by immunoblotting. The gel image is representative of n = 3 independent experiments. d Experimental scheme in human HCT116 colon cancer cells: the endogenous DNMT1 protein is tagged with an auxin-inducible degron (AID) tag, causing the protein to be degraded after the addition of the auxin analog IAA. In this background, rescue vectors are added that encode either WT or FF/AA versions of DNMT1. The empty vector is used as a negative control. e Western blotting shows that endogenous DNMT1 is degraded upon IAA addition, whereas exogenous WT and FF/AA DNMT1 are not degraded. The repetitive of the experiment is n = 1. f Measurement of global DNA levels by luminometric methylation assay (LUMA). Cells expressing WT DNMT1 maintain global DNA methylation when endogenous DNMT1 is degraded, whereas the FF/AA mutant does not support DNA methylation maintenance. Data represent the mean ± SD of n = 3 independent experiments. Source data of Figs. 4a–c, 4e, f are provided as a Source Data file.
Fig. 5. Schematic representation of the DNMT1 activation mechanism.
Left and right figures show the auto-inhibitory and activation states of DNMT1, respectively.
Similar articles
- Enhanced processivity of Dnmt1 by monoubiquitinated histone H3.
Mishima Y, Brueckner L, Takahashi S, Kawakami T, Otani J, Shinohara A, Takeshita K, Garvilles RG, Watanabe M, Sakai N, Takeshima H, Nachtegael C, Nishiyama A, Nakanishi M, Arita K, Nakashima K, Hojo H, Suetake I. Mishima Y, et al. Genes Cells. 2020 Jan;25(1):22-32. doi: 10.1111/gtc.12732. Epub 2019 Dec 3. Genes Cells. 2020. PMID: 31680384 - Mechanism studies of the activation of DNA methyltransferase DNMT1 triggered by histone H3 ubiquitination, revealed by multi-scale molecular dynamics simulations.
Sun J, Liu F, Yuan L, Pang NN, Zhu B, Yang N. Sun J, et al. Sci China Life Sci. 2023 Feb;66(2):313-323. doi: 10.1007/s11427-021-2179-8. Epub 2022 Oct 20. Sci China Life Sci. 2023. PMID: 36271982 - Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance.
Ishiyama S, Nishiyama A, Saeki Y, Moritsugu K, Morimoto D, Yamaguchi L, Arai N, Matsumura R, Kawakami T, Mishima Y, Hojo H, Shimamura S, Ishikawa F, Tajima S, Tanaka K, Ariyoshi M, Shirakawa M, Ikeguchi M, Kidera A, Suetake I, Arita K, Nakanishi M. Ishiyama S, et al. Mol Cell. 2017 Oct 19;68(2):350-360.e7. doi: 10.1016/j.molcel.2017.09.037. Mol Cell. 2017. PMID: 29053958 - Molecular coupling of DNA methylation and histone methylation.
Hashimoto H, Vertino PM, Cheng X. Hashimoto H, et al. Epigenomics. 2010 Oct;2(5):657-69. doi: 10.2217/epi.10.44. Epigenomics. 2010. PMID: 21339843 Free PMC article. Review. - Regulation of maintenance DNA methylation via histone ubiquitylation.
Nishiyama A, Yamaguchi L, Nakanishi M. Nishiyama A, et al. J Biochem. 2016 Jan;159(1):9-15. doi: 10.1093/jb/mvv113. Epub 2015 Nov 20. J Biochem. 2016. PMID: 26590302 Free PMC article. Review.
Cited by
- Structural insight into the DNMT1 reaction cycle by cryo-electron microscopy.
De I, Weidenhausen J, Concha N, Müller CW. De I, et al. PLoS One. 2024 Sep 3;19(9):e0307850. doi: 10.1371/journal.pone.0307850. eCollection 2024. PLoS One. 2024. PMID: 39226277 Free PMC article. - CDCA7 is an evolutionarily conserved hemimethylated DNA sensor in eukaryotes.
Wassing IE, Nishiyama A, Shikimachi R, Jia Q, Kikuchi A, Hiruta M, Sugimura K, Hong X, Chiba Y, Peng J, Jenness C, Nakanishi M, Zhao L, Arita K, Funabiki H. Wassing IE, et al. Sci Adv. 2024 Aug 23;10(34):eadp5753. doi: 10.1126/sciadv.adp5753. Epub 2024 Aug 23. Sci Adv. 2024. PMID: 39178260 Free PMC article. - Translation efficiency covariation across cell types is a conserved organizing principle of mammalian transcriptomes.
Liu Y, Hoskins I, Geng M, Zhao Q, Chacko J, Qi K, Persyn L, Wang J, Zheng D, Zhong Y, Rao S, Park D, Cenik ES, Agarwal V, Ozadam H, Cenik C. Liu Y, et al. bioRxiv [Preprint]. 2024 Aug 11:2024.08.11.607360. doi: 10.1101/2024.08.11.607360. bioRxiv. 2024. PMID: 39149359 Free PMC article. Preprint. - Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Shao Z, Lu J, Khudaverdyan N, Song J. Shao Z, et al. Nat Commun. 2024 Aug 9;15(1):6815. doi: 10.1038/s41467-024-51246-4. Nat Commun. 2024. PMID: 39122718 Free PMC article. - Structure of human DPPA3 bound to the UHRF1 PHD finger reveals its functional and structural differences from mouse DPPA3.
Shiraishi N, Konuma T, Chiba Y, Hokazono S, Nakamura N, Islam MH, Nakanishi M, Nishiyama A, Arita K. Shiraishi N, et al. Commun Biol. 2024 Jun 19;7(1):746. doi: 10.1038/s42003-024-06434-9. Commun Biol. 2024. PMID: 38898124 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials