KIF22 regulates mitosis and proliferation of chondrocyte cells - PubMed (original) (raw)
KIF22 regulates mitosis and proliferation of chondrocyte cells
Hiroka Kawaue et al. iScience. 2024.
Abstract
Point mutations in KIF22 have been linked to spondyloepimetaphyseal dysplasia with joint laxity, type 2 (SEMDJL2). Skeletal features of SEMDJL2 include short stature and joint laxity. Mechanisms underlying these limb abnormalities are unknown. Here in this manuscript, we have investigated the function of KIF22 in chondrocytes. Quantitative PCR and immunostaining revealed that Kif22 was highly expressed in proliferating-zone growth-plate chondrocytes. Kif22 knockdown resulted in defective mitotic spindle formation and reduced cell proliferation. Forced expression of SEMDJL-associated mutant Kif22 constructs likewise induced abnormal mitotic spindle morphology and reduced proliferation. Mice expressing a KIF22 truncation mutant had shorter growth plates and shorter tibial bones compared to wild-type mice. These results suggest that KIF22 regulates mitotic spindle formation in proliferating chondrocytes thereby linking the stunted longitudinal bone growth observed in SEMDJL2 to failures of chondrocyte division.
Keywords: Molecular biology; cell biology.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Graphical abstract
Figure 1
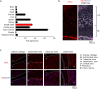
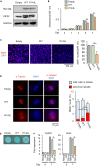
KIF22 is highly expressed in proliferating-zone chondrocytes of the growth plate (A) qPCR analysis of Kif22 mRNA in various organs and tissues of 5-week-old mice. Data are represented as mean ± SD; n = 3. Other statical data were shown in Table S1. (B) Immunofluorescent staining of KIF22 at the growth plate of tibia from 2-week-old mice. The proliferating zone (PZ) and hypertrophic zone (HZ) of the growth plate are indicated. Scale bar, 100 μm. (C) Immunofluorescent staining of KIF22 at the growth plate of tibia from 2-week-old mice. Images of articular cartilage, subarticular subchondral bone, trabecular bone below under the growth plate, and cortical bone.
Figure 2
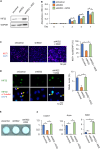
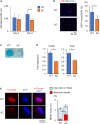
KIF22 is essential for cell proliferation in ATDC5 cells ATDC5 cells were infected with control shRNA adenovirus (shcontrol), Kif22 shRNA adenovirus (shKif22), and/or Myc-tagged-Kif22 overexpression adenovirus (Kif22). (A) Kif22 expression was examined by western blotting analysis. Uncropped image was shown in Figure S1. (B) ATDC5 cells were plated at 3,000 cells/cm2 in 96-well plates. CCK8 cell viability assay showing proliferation of ATDC5 cells. Data are represented as mean ± SD; n = 4. ∗; p < 0.05. Other statical data were shown in Table S1. (C) Ki67 immunofluorescence staining and quantification 48 h after infection. Scale bar, 50 μm. Data are represented as mean ± SD; n = 3. ∗; p < 0.05. Other statical data were shown in Table S1. (D) KIF22 and tubulin immunostaining 24 h after infection and cell synchronization. Nuclei were counterstained with DAPI. Mitotic nuclei (arrows) were quantified. Scale bar, 10 μm. Data are represented as mean ± SD; n = 4. ∗; p < 0.05. Other statical data were shown in Table S1. (E and F) Alcian blue staining (E) and qPCR analysis (F), 7 days after induction of differentiation. Data are represented as mean ± SD; n = 4. ∗; p < 0.05. Other statical data were shown in Table S1. More information is available at Figure S1.
Figure 3
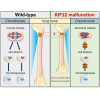
Kif22 mutant mice have a wider joint space and shorter growth plate (A) Schematic diagram of human KIF22, mouse KIF22 and the mouse KIF22 R144fs∗16 truncated C-terminal mutant (Kif22ΔC). Motor; motor domain, MTB; microtubule binding domain, DB; DNA binding domain. The red lines indicate the location of SEMDJL2 patient mutations and the corresponding amino acid in mouse. (B) Kif22 wild-type (WT) and Kif22 ΔC (ΔC) mRNA expression in primary chondrocytes from Kif22w/w and Kif22wt/ΔC mice were determined by qPCR using allele specific primers. Data are represented as mean ± SD; n = 3. ∗; p < 0.05. Other statical data were shown in Table S1. (C) Alcian blue and Alizarin red staining of skeletal preparations of 1-day-old Kif22 w/w and Kif22 wt/ΔC mice. Representative images of Kif22 w/w and Kif22 wt/ΔC was shown. Arrow indicates joint space between proximal tibia and distal femur. Scale increments are 1 mm. (D) Measurement of tibia length and (E) joint space from the skeletal preparations shown in (C). Each data are represented by a dot. The statistical data are also represented by box-and-whisker diagrams. ∗; p < 0.05. Other statical data were shown in Table S1. (F) von Kossa stained sections of proximal tibia from 8-week-old Kif22 w/w or Kif22 wt/ΔC mice. Representative images of Kif22 w/w and Kif22 wt/ΔC are shown. Yellow boxed region is shown magnified in lower panel. Low and high magnification scales are 200 μm and 50 μm, respectively. (G) Measurement of growth plate thickness in samples shown in (F). Each data are represented by a dot. The statistical data are also represented by box-and-whisker diagrams. ∗; p < 0.05. Other statical data were shown in Table S1.
Figure 4
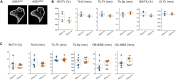
MicroCT and histomorphometric analysis of Kif22 mutant mice MicroCT analysis (A and B) and histomorphometric analysis (C) of tibia from 8-week-old mice was performed. (A) Representative microCT image of littermates. Parameters; Bone volume/Tissue volume (BV/TV), Trabecular number (Tb.N), Trabecular thickness (Tb.Th), Trabecular separation (Tb.Sp), Cortical bone area/Tissue area (BA/TA), Cortical thickness (Ct.th), Osteoblast number/Bone surface (OB.N/BS), Osteoclast number/Bone surface (OC.N/BS). Each data are represented by a dot. The statistical data are also represented by box-and-whisker diagrams. Other statical data were shown in Table S1.
Figure 5
Kif22 P143L mutation disturbs spindle fiber formation during mitosis ATDC5 stably expressing empty pcDNA3.1 plasmid (empty), myc-tagged wild-type Kif22 (WT) and Kif22 P143L (P143L) were generated. (A) Expression of endogenous and exogenous KIF22 proteins were determined by western blotting analysis. Uncropped image was shown in Figure S3. (B) CCK8 cell viability assay of ATDC5 cells. Data are represented as mean ± SD; n = 4. ∗; p < 0.05. Other statical data were shown in Table S1. (C) Ki67 immunofluorescence staining and quantification of WT and P143L at 24 h after seeding. Scale bar, 50 μm. Data are represented as mean ± SD; n = 4. ∗; p < 0.05. Other statical data were shown in Table S1. (D) Tubulin immunostaining of WT and P143L cells after cell synchronization. Nuclei were counterstained with DAPI. Total number of mitotic nuclei and abnormal spindles were quantified. Scale bar, 10 μm. (Data are represented as mean ± SD; n = 3. ∗; p < 0.05. Other statical data were shown in Table S1. (E) Alcian blue staining and (F) qPCR analysis of the indicated cells 7 days after induction of differentiation. Data are represented as mean ± SD; n = 3. ∗; p < 0.05. Other statical data were shown in Table S1. More information is available at Figure S2.
Figure 6
Spindle fiber formation and cell mitosis was disturbed in Kif22 wt/ΔC chondrocytes Chondrocytes were harvested from costal cartilage of P2 mice. (A) CCK-8 cell viability assay showing proliferation. Data are represented as mean ± SD; n = 4. ∗; p < 0.05. Other statical data were shown in Table S1. (B) Ki67 immunofluorescence staining and quantification of Kif22 w/w (WT) or Kif22 wt/ΔC (ΔC) primary chondrocytes 24 h after plating. Scale bar, 50 μm. Data are represented as mean ± SD; n = 3. ∗; p < 0.05. Other statical data were shown in Table S1. (C) Alcian blue staining and (D) qPCR analysis of WT or Kif22ΔC cells 7 days after plated with chondrogenic media. Data are represented as mean ± SD; n = 3. ∗; p < 0.05. Other statical data were shown in Table S1. (E) Tubulin immunostaining of WT or Kif22ΔC cells 24 h after plating and cell synchronization. Nuclei were counterstained with DAPI. Total number of mitotic nuclei and abnormal spindles were quantified. Scale bar, 10 μm. Data are represented as mean ± SD; n = 3. ∗; p < 0.05. Other statical data were shown in Table S1. More information is available at Figure S3.
Similar articles
- Identification of kinesin family member (KIF22) homozygous variants in spondyloepimetaphyseal dysplasia with joint laxity, lepdodactylic type and demonstration of proteoglycan biosynthesis impairment.
Dubail J, Rondeau S, Michot C, Baujat G, Capri Y, Thévenon J, Charpie M, Pejin Z, Phan G, Huber C, Cormier-Daire V. Dubail J, et al. J Bone Miner Res. 2024 Apr 19;39(3):287-297. doi: 10.1093/jbmr/zjad020. J Bone Miner Res. 2024. PMID: 38477767 - Whole-exome sequencing identifies mutations of KIF22 in spondyloepimetaphyseal dysplasia with joint laxity, leptodactylic type.
Min BJ, Kim N, Chung T, Kim OH, Nishimura G, Chung CY, Song HR, Kim HW, Lee HR, Kim J, Kang TH, Seo ME, Yang SD, Kim DH, Lee SB, Kim JI, Seo JS, Choi JY, Kang D, Kim D, Park WY, Cho TJ. Min BJ, et al. Am J Hum Genet. 2011 Dec 9;89(6):760-6. doi: 10.1016/j.ajhg.2011.10.015. Am J Hum Genet. 2011. PMID: 22152677 Free PMC article. - Spondyloepimetaphyseal dysplasia with joint laxity type 2: Aggregating the literature and reporting on the life of a 66-year-old man.
Beke A, da Costa Silveira K, Athey T, Kannu P. Beke A, et al. Am J Med Genet C Semin Med Genet. 2023 Jun;193(2):188-192. doi: 10.1002/ajmg.c.32053. Epub 2023 May 24. Am J Med Genet C Semin Med Genet. 2023. PMID: 37226647 - Recurrent dominant mutations affecting two adjacent residues in the motor domain of the monomeric kinesin KIF22 result in skeletal dysplasia and joint laxity.
Boyden ED, Campos-Xavier AB, Kalamajski S, Cameron TL, Suarez P, Tanackovic G, Andria G, Ballhausen D, Briggs MD, Hartley C, Cohn DH, Davidson HR, Hall C, Ikegawa S, Jouk PS, König R, Megarbané A, Nishimura G, Lachman RS, Mortier G, Rimoin DL, Rogers RC, Rossi M, Sawada H, Scott R, Unger S, Valadares ER, Bateman JF, Warman ML, Superti-Furga A, Bonafé L. Boyden ED, et al. Am J Hum Genet. 2011 Dec 9;89(6):767-72. doi: 10.1016/j.ajhg.2011.10.016. Am J Hum Genet. 2011. PMID: 22152678 Free PMC article. - Chondrocytes and longitudinal bone growth: the development of tibial dyschondroplasia.
Farquharson C, Jefferies D. Farquharson C, et al. Poult Sci. 2000 Jul;79(7):994-1004. doi: 10.1093/ps/79.7.994. Poult Sci. 2000. PMID: 10901201 Review.
References
- Min B.-J., Kim N., Chung T., Kim O.-H., Nishimura G., Chung C.Y., Song H.R., Kim H.W., Lee H.R., Kim J., et al. Whole-exome sequencing identifies mutations of KIF22 in spondyloepimetaphyseal dysplasia with joint laxity, leptodactylic type. Am. J. Hum. Genet. 2011;89:760–766. doi: 10.1016/j.ajhg.2011.10.015. - DOI - PMC - PubMed
- Boyden E.D., Campos-Xavier A.B., Kalamajski S., Cameron T.L., Suarez P., Tanackovic G., Andria G., Ballhausen D., Briggs M.D., Hartley C., et al. Recurrent dominant mutations affecting two adjacent residues in the motor domain of the monomeric kinesin KIF22 result in skeletal dysplasia and joint laxity. Am. J. Hum. Genet. 2011;89:767–772. doi: 10.1016/j.ajhg.2011.10.016. - DOI - PMC - PubMed
- Horev G., Ellegood J., Lerch J.P., Son Y.-E.E., Muthuswamy L., Vogel H., Krieger A.M., Buja A., Henkelman R.M., Wigler M., Mills A.A. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc. Natl. Acad. Sci. USA. 2011;108:17076–17081. doi: 10.1073/pnas.1114042108. - DOI - PMC - PubMed
- Portmann T., Yang M., Mao R., Panagiotakos G., Ellegood J., Dolen G., Bader P.L., Grueter B.A., Goold C., Fisher E., et al. Behavioral abnormalities and circuit defects in the basal ganglia of a mouse model of 16p11.2 deletion syndrome. Cell Rep. 2014;7:1077–1092. doi: 10.1016/j.celrep.2014.03.036. - DOI - PMC - PubMed
- Pucilowska J., Vithayathil J., Tavares E.J., Kelly C., Karlo J.C., Landreth G.E. The 16p11.2 deletion mouse model of autism exhibits altered cortical progenitor proliferation and brain cytoarchitecture linked to the ERK MAPK pathway. J. Neurosci. 2015;35:3190–3200. doi: 10.1523/JNEUROSCI.4864-13.2015. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials