Microcell-mediated chromosome transfer between non-identical human iPSCs - PubMed (original) (raw)
. 2024 Nov 5;35(4):102382.
doi: 10.1016/j.omtn.2024.102382. eCollection 2024 Dec 10.
Hitomaru Miyamoto 2, Kyotaro Yamazaki 2 3 4, Masaya Egawa 1, Hiroaki Kobayashi 2, Kanako Kazuki 5, Mitsuhiko Osaki 6, Teruhiko Suzuki 7, Shusei Hamamichi 5, Mitsuo Oshimura 5, Kazuma Tomizuka 1, Yasuhiro Kazuki 2 3 5
Affiliations
- PMID: 39634136
- PMCID: PMC11616053
- DOI: 10.1016/j.omtn.2024.102382
Microcell-mediated chromosome transfer between non-identical human iPSCs
Narumi Uno et al. Mol Ther Nucleic Acids. 2024.
Abstract
Microcell-mediated chromosome transfer (MMCT) is anticipated as a unique strategy to manipulate numbers of chromosomes, including the generation of hyperaneuploidy syndrome models with human induced pluripotent stem cells (hiPSCs). Mouse A9/Chinese hamster ovary (CHO) cell libraries of human monochromosomal hybrids as chromosome donor cells frequently exhibit chromosomal rearrangement in the components. The generation of a new A9/CHO library is time-consuming and laborious. Here, we developed an MMCT method using hiPSCs as chromosome donor and recipient cells, through micronucleation using paclitaxel and reversine. Membrane fusion during the MMCT was mediated through interactions between the ecotropic viral envelope transiently expressed in chromosome donor cells and mCAT-1 in chromosome recipient cells. This approach involved tagging Chr21 and ChrY by CRISPR-Cas9 and transferring human/mouse artificial chromosomes, Chr21, ChrX, and ChrY, wherein there are no previous reports demonstrating a full-length introduction. Thus, a strategy that combing CRISPR-Cas9-mediated chromosome tagging and MMCT from hiPSCs as chromosome donor cells to hiPSCs as recipient cells systematically produced isogenic disease model hiPSCs with hyperaneuploidy. This approach allows the study of rare diseases and promises to provide new insights into early developmental mechanisms by introducing a comprehensive set of influential chromosomes/regions into hiPSCs.
Keywords: Klinefelter’s syndrome; MT: Delivery Strategies; disease model; human artificial chromosome; induced pluripotent stem cell; microcell-mediated chromosome transfer; mouse artificial chromosome; paclitaxel; reversine; triple X syndrome; trisomy.
© 2024 The Author(s).
Conflict of interest statement
N.U., M. Oshimura, and Y.K. are inventors of patent applications based on the findings described in this paper. M. Oshimura is the CEO and shareholder of Trans Chromosomics Inc.
Figures
Graphical abstract
Figure 1
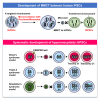
Optimization of treatment concentrations of paclitaxel and reversine for MMCT from hiPSC (A) Schematic illustration of micronucleation in hiPSCs with paclitaxel (PTX) and reversine (Rev) and application of Eco-MMCT using an ecotropic virus envelope and its receptor mCAT-1. (B) Giemsa-stained micronucler spreads of treated hiPSCs (201B7). Scale bars, 10 μm. (C) Number of micronuclei per cell under each treatment of hiPSCs. Line within the box marks the median. Box extends from the 25th percentile (Q1) to 75th percentile (Q3), representing the interquartile range (IQR). Whiskers extend from minimum to maximum values, n = 100 cells per group (∗∗p < 0.01, ANOVA). (D) Comparison of MMCT efficiency of MAC6 from hiPSCs (201B7-MAC6) to HT1080 or HFL1-HAC-iPSCs under various conditions. Blue bar indicates PEG-MMCT; red bar indicates Eco-MMCT (∗p < 0.05; Student’s t-test). Data are the mean ± SD (n = 3). The chromosome donor cells (CDCs), chromosome recipient cells (CRCs), and the transferred chromosomes are summarized in the frame at the top of the graph. (E) FISH analysis and karyotype of an HT1080 clone, transferred with MAC6. Fluorescence in situ hybridization (FISH) analysis of HT1080 transferred with MAC6. Blue indicates 4′,6-diamidino-2-phenylindole (DAPI) and red indicates mouse Cot-1 (MAC6).
Figure 2
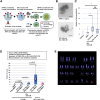
MMCT of endogenous ChrX from iPSCs to HT1080 (A) Schematic illustration of the relationship between filter pore size and microcell size and evaluation of the effect of filter pore size on microcell purification. Microcells containing ChrX might be larger than the 3-μm filter pore size and thus be lost during micronucleus purification. (B) Comparison of the minimum filter pore diameter used during microcell purification and the number of colonies obtained after MMCT. Data are the mean ± SD (n = 3). (C) Representative images of the drug resistance analysis of the hypoxanthine-guanine phospho ribosyl transferase (HPRT) gene on ChrX. The HPRT gene confers hypoxanthine-aminopterin-thymidine (HAT) resistance to cells and metabolizes 6-thioguanine (6TG) to produce cytotoxic metabolites, making it sensitive to 6TG. Thus, the HT1080 HPRT-KO cells transferred with ChrX recovered HAT resistance and 6TG sensitivity similar to wild-type HT1080 cells. Scale bars, 100 μm. (D and E) Multi-color FISH analysis of HT1080 HPRT-KO (XO) and a clone transferred with ChrX. Red frames indicate ChrX. (F) PCR with STS marker primers. STS markers showing polymorphisms on ChrX were selected for different band sizes between 201B7 and HT1080, and HT1080 clones acquired after ChrX transfer by MMCT were analyzed. The location of each STS marker is shown with the ideogram of ChrX. The results indicated that the clones +ChrX#3 and #4 might have an intact ChrX, but not +ChrX#2, which showed the loss of DXS1073. For Clone#1, DXS1227 might be defective, but it is not clear which of the STS marker polymorphisms, 201B7 or XX, was introduced.
Figure 3
Chromosome tagging by CRISPR-Cas9 for MMCT of targeted native Chr21 and Y (A) Schematic illustration of MMCT following chromosome tagging for Chr21. hiPSC 201B7 with a normal karyotype 46,XX was transferred with Chr21-tagged mCherryneo from hiPSCs 585A1 that were 46,XY. hiPSC 201B7 transferred with Chr21 showed trisomy 21 indicating Down syndrome. (B) Schematic diagram of chromosome tagging with mCherryneo on Chr21 by CRISPR-Cas9 is shown. The primer set detected 670 bp of the right homologous arm region. (C) PCR analysis of 21 G418-resistant clones to detect correct gene insertion. Negative control (N.C., 201B7), positive controls (P.C. 1 and 2, bulk samples from two replicate experiments, in which 201B7 were transfected with CRISPR-Cas9 and the plasmid vector containing mCherryneo). (D) Schematic illustration of MMCT following chromosome tagging for ChrY (upper diagram). hiPSC 201B7 with a normal karyotype 46,XX was transferred with ChrY-tagged GFPneo from HFL-1-iPSC that were 46,XY. hiPSC 201B7 transferred with a ChrY was XXY, indicating Klinefelter syndrome. (E) Schematic diagram of chromosome tagging with GFPneo on ChrY by CRISPR-Cas9. The primer set detected 935 bp of the left homologous arm region. (F) Representative images of a neo-resistant colony observed by microscopy (left, bright phase contrast; right, GFP fluorescence). Scale bars, 200 μm (white). (G) PCR analysis of the G418-resistant clones to detect correct gene insertion.
Figure 4
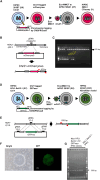
Transfer of native Chr21 using MMCT from hiPSCs to hiPSCs (A) MMCT efficiency of Basal-HAC from hiPSCs (201B7-Basal-HAC) or Chr21 from hiPSCs containing tagged-Chr21 to hiPSC 585A1. Data are the mean ± SD (n = 3). NS, not significant. (B) Representative images of a drug-resistant colony (left, bright phase contrast; right, mCherry fluorescence). Scale bars, 100 μm (white). (C) FISH of disomy Chr21 in 585A1 (left), trisomy Chr21 (center), and tetrasomy Chr21 in Chr21-transferred 585A1 (gray, DAPI; red, D21Z1; green, mCherryneo). (D–F) Quinacrine-Hoechst (QH) counter-stain karyotype images of 585A1, 585A1 Basal-HAC, and 585A1 trisomy chr21 clones. Red frames, Chr21.
Figure 5
Transfer of native ChrX and ChrY using MMCT from hiPSCs to hiPSCs (A) MMCT efficiency of ChrX from 201B7 (46,XX) to 201B7 HPRT-KO (46,XX) or ChrY from HFL-1-hY-GFPneo Y to 201B7 (46,XX). Data are the mean ± SD (n = 3). (B) PCR analysis of ChrY-specific genes. (C) FISH analysis of HFL1-SeV-2-1 iPSC providing ChrX and ChrY (gray, DAPI; red, dystrophin gene (DYS); green, GFPneo). Magenta arrows, ChrX; green arrows, ChrY. Enlarged images are images of ChrX and ChrY. (D) FISH analysis of HFL1-SeV-2-1 iPSC accepting ChrX and ChrY (gray, DAPI; red, DYS gene). (E and F) FISH analysis of clones with trisomy X (E) and XXY (F). Light blue, DAPI; red, DYS gene on ChrX; green, GFPneo on ChrY. (G) Disomy of exo-ChrY (gray, DAPI; red, DYS gene; green, GFPneo). (H) Fragmented ChrY showing the deletion of SRY by PCR analysis (gray, DAPI; red, DYS gene; green, GFPneo). Magenta arrows, ChrX; green arrows, ChrY.
Figure 6
Whole-genome CGH array analysis of the representative clones transferred with Chr21, ChrY, or ChrX This figure presents the CGH array analysis of representative clones that were transferred with Chr21 (585A1-21Exp.2#01), ChrX (201B7 HPRT-KO-X Exp1-1), or ChrY (201B7-YGFPneoExp3-1). The karyotype shown includes all human chromosomes from 1 to 22, X, and Y. Red lines indicate regions of amplification, while green lines indicate regions of deletion. (A) A summary ideogram of the whole genome in the analysis of 585A1-21Exp.2#01 compared with 585A1. (B) A summary ideogram of Chr21 from the CGH Analytics software for 585A1-21Exp.2#01. (C) A summary ideogram of the whole genome in the analysis of 201B7 HPRT-KO-X Exp1-1 compared with 201B7. (D) A summary ideogram of ChrX from 201B7 HPRT-KO-X Exp1-1. (E) A summary ideogram of the whole genome in the analysis of 201B7-YGFPneoExp3-1 compared with 201B7. (F) A summary ideogram of ChrY from 201B7-YGFPneoExp3-1. The graph shows the average log2 ratios, indicating the relative copy number changes of chromosomes. A value of 1 on the y axis represents a 2-fold increase in chromosome number and a value of −1 indicates a halving of the chromosome number (A–E).
Figure 7
Stability of transferred chromosomes and comparison of cell proliferation in long-term cultures (A) Percentage of cells with the three Chr21 copies in Chr21-transferred clones (585A1-21 Exp.2#01 and 585A1-21 Exp.3#04) at PDL 0 and PDL 30, with and without drug selection (n = 30 cells for each condition). (B) Percentage of cells with ChrY in ChrY-transferred clones (201B7-YGFPneo Exp.1-3 and 201B7-YGFPneo Exp.3-1) at PDL 0 and PDL 30, with and without drug selection. The red indicates the proportion of cells that do not retain the ChrY (n = 30 cells for each condition). (C) Percentage of cells with different karyotypes (48,XX,+X,+X; 47,XX,+X; 46,XX) in ChrX-transferred clones (201B7 HPRT-KO-X Exp.1-1 and 201B7 HPRT-KO-X Exp.2-1) at PDL 0 and PDL 30, with and without drug selection. Light blue indicates the proportion of cells retaining the transferred chromosome, gray indicates the proportion of cells with ChrX translocations, and indigo indicates the proportion of cells with four ChrX copies (n = 30 cells for each condition) (A–C). (D) Ratios of wild-type (WT) and mutant alleles (Mut #1 and Mut #2) of the HPRT gene in 201B7HPRT-KO-X Exp1-1, 201B7HPRT-KO-X Exp2-1, and 201B7HPRT-KO cell lines at PDL 30, with and without drug selection. WT is indicated in pink, Mut #1 in black, and Mut #2 in gray.
Figure 8
Schematic illustration of the advanced MMCT approach for human chromosomes using CRISPR-Cas9 chromosome tagging in hiPSCs compared with the A9/CHO cell library approach (A) The advanced MMCT is a straightforward approach for transferring a targeted chromosome into hiPSCs. First, a drug-selectable gene is inserted into the target chromosome using CRISPR-Cas9 through homologous recombination. For hiPSCs, an ecotropic virus envelope gene, which induces membrane fusion, is introduced into the CDCs, and the receptor gene (mCAT-1) is transiently introduced into the CRCs. PTX and Rev are used to induce micronucleation in the CDC. Microcells with the ecotropic virus envelope on their membrane can be obtained from this CDC, allowing chromosome transfer into the CRC. Then, the chromosome can be transferred to hiPSCs via MMCT. In this approach, hiPSCs with the introduced target chromosome can be established through the two-step chromosome and cell manipulation process. This advanced method reduces the overall process time to 2 months. (B) In the conventional MMCT approach using an A9/CHO cell library, the chromosome in human fibroblasts, which act as a chromosome resource, is tagged with a drug-selectable gene using the random integration method. The resulting drug-resistant clones are screened, and clones tagged on the target chromosome are selected. Next, whole-cell fusion is performed with A9/CHO cells to obtain A9/CHO hybrids containing the tagged target chromosome. To separate the target chromosome from the non-target human chromosomes derived from human fibroblasts, the target chromosome is introduced into the A9/CHO cells by MMCT. This results in the A9/CHO cell libraries of human monochromosomal hybrids. Finally, chromosomes from the A9/CHO cell libraries are introduced into hiPSCs using MMCT. In this approach, hiPSCs with the introduced target chromosome can be established through the four-step chromosome and cell manipulation process. The conventional method requires over 6 months to complete.
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Enabling Systemic Identification and Functionality Profiling for Cdc42 Homeostatic Modulators.
Malasala S, Azimian F, Chen YH, Twiss JL, Boykin C, Akhtar SN, Lu Q. Malasala S, et al. bioRxiv [Preprint]. 2024 Jan 8:2024.01.05.574351. doi: 10.1101/2024.01.05.574351. bioRxiv. 2024. PMID: 38260445 Free PMC article. Updated. Preprint. - Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.
Lee CC, Chen CW, Yen HK, Lin YP, Lai CY, Wang JL, Groot OQ, Janssen SJ, Schwab JH, Hsu FM, Lin WH. Lee CC, et al. Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924 - Platelet-rich therapies for musculoskeletal soft tissue injuries.
Moraes VY, Lenza M, Tamaoki MJ, Faloppa F, Belloti JC. Moraes VY, et al. Cochrane Database Syst Rev. 2014 Apr 29;2014(4):CD010071. doi: 10.1002/14651858.CD010071.pub3. Cochrane Database Syst Rev. 2014. PMID: 24782334 Free PMC article. Review. - Genedrive kit for detecting single nucleotide polymorphism m.1555A>G in neonates and their mothers: a systematic review and cost-effectiveness analysis.
Shabaninejad H, Kenny RP, Robinson T, Stoniute A, O'Keefe H, Still M, Thornton C, Pearson F, Beyer F, Meader N. Shabaninejad H, et al. Health Technol Assess. 2024 Oct;28(75):1-75. doi: 10.3310/TGAC4201. Health Technol Assess. 2024. PMID: 39487741 Free PMC article.
References
- Kugoh H., Mitsuya K., Meguro M., Shigenami K., Schulz T.C., Oshimura M. Mouse A9 cells containing single human chromosomes for analysis of genomic imprinting. DNA Res. 1999;6:165–172. - PubMed
- Inoue J., Mitsuya K., Maegawa S., Kugoh H., Kadota M., Okamura D., Shinohara T., Nishihara S., Takehara S., Yamauchi K., et al. Construction of 700 human/mouse A9 monochromosomal hybrids and analysis of imprinted genes on human chromosome 6. J. Hum. Genet. 2001;46:137–145. - PubMed
LinkOut - more resources
Full Text Sources