NR4A3 inhibits the tumor progression of hepatocellular carcinoma by inducing cell cycle G0/G1 phase arrest and upregulation of CDKN2AIP expression - PubMed (original) (raw)
. 2024 Oct 28;20(15):5850-5867.
doi: 10.7150/ijbs.95174. eCollection 2024.
Affiliations
- PMID: 39664575
- PMCID: PMC11628324
- DOI: 10.7150/ijbs.95174
NR4A3 inhibits the tumor progression of hepatocellular carcinoma by inducing cell cycle G0/G1 phase arrest and upregulation of CDKN2AIP expression
Xinge Zhao et al. Int J Biol Sci. 2024.
Abstract
Nuclear receptor subfamily 4 group A member 3 (NR4A3) is a member of the orphan nuclear receptor superfamily, and exhibits transcription factor activity by binding to sequence-specific DNA. Considering that the specific mechanism by which NR4A3 regulates gene transcription in HCC (hepatocellular carcinoma) has not yet been elucidated, our study aimed to explore the transcriptional role of NR4A3 in regulating the target gene CDKN2AIP (CDKN2A interacting protein), which will suppress the development of HCC. Our data show that NR4A3 is downregulated in human HCC tissues, and that low expression of NR4A3 is correlated with poor prognosis, indicating that NR4A3 could act as a tumor suppressor gene in HCC. NR4A3 overexpression suppresses cell proliferation, clone formation, cell cycle arrest at G0/G1 phase and tumor growth in vitro and in vivo and promote DNA damage. NR4A3 could directly regulate the expression of CDKN2AIP at the transcriptional level, suggesting that NR4A3 may play a role as a transcription factor in HCC and may serve as a potential biomarker for predicting prognosis for HCC patients.
Keywords: CDKN2AIP; NR4A3; cell cycle; hepatocellular carcinoma; tumor progression.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
Figure 1
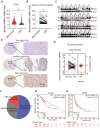
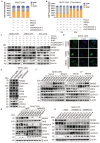
NR4A3 expression is down-regulated and associated with prognosis of HCC patients. (A) NR4A3 expression in HCC tissues and the corresponding normal tissues in The Cancer Genome Atlas (TCGA) datasets (log2 TPM, two-sided unpaired t-test). (B) NR4A3 mRNA expression in 56 paired HCC tissues and the adjacent matched noncancerous tissues were determined using qPCR. For qPCR, values were normalized with GAPDH. (C) The protein levels of NR4A3 in 28 paired HCC (T) and adjacent normal (N) samples were measured using western blot. β-actin was used as a loading control. (D) NR4A3 protein expression in 28 paired HCC tissues and the adjacent matched noncancerous tissues were quantified and analyzed. (E-F) The expression of NR4A3 in HCC tissues was cored, and representative IHC images showed the expression of NR4A3 in HCC tissues. (G-H) The correlation between NR4A3 expression and overall survival (G, n=364) and progression-free survival (H, n=370) of HCC patients in TCGA dataset was assessed by Kaplan-Meier survival analysis. The data were presented as mean ± SD. *p<0.05; **p<0.01.
Figure 2
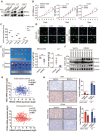
Overexpression of NR4A3 inhibits HCC cell proliferation and tumorigenicity. (A) Western blot showed that NR4A3 was efficiently up-regulated in MHCC-LM3, Li7, and Huh7 cells. (B) CCK8 assay revealed that the proliferation of HCC cells was inhibited by overexpression of NR4A3. (C) Colony formation assay showed that overexpression of NR4A3 inhibited the proliferation of HCC cells. The bar graph showed quantitative analysis data with three replicates. (D) Immunofluorescence showed overexpression of NR4A3 enhanced γ-H2AX foci. (E) In vivo growth assays showed the difference in tumor weight between NR4A3 overexpressed cells and the vector group in MHCC-LM3 (top panel) and Li7 (bottom panel) cell lines. (F) Western blot analysis showed that the expression of NR4A3 was significantly up-regulated in NR4A3-overexpressing tumor tissues collected from nude mice with tumor xenografts. β-actin was used as a loading control. (G) The correlation between NR4A3 and Ki67 or PCNA mRNA expression in HCC tissues was analyzed in The Cancer Genome Atlas (TCGA) dataset. (H) Immunohistochemical images of Ki67 and PCNA expressions in xenograft tumors derived from MHCC-LM3 and Li7 cells with NR4A3 overexpression. Original magnification: × 400. The positively stain area (in percentages) were analyzed. *, p<0.05; **, p<0.01.
Figure 3
Knockdown of NR4A3 promotes HCC cell proliferation and tumorigenicity. (A) Western blot analysis of NR4A3 protein in MHCC-97H, MHCC-97L and HCC-LY10 cells stably transfected with shRNA or shNC. (B) CCK8 assay revealed that the proliferation of HCC cells was promoted by down-regulation of NR4A3. (C) Colony formation assay showed that down-regulation of NR4A3 promoted the proliferation of HCC cells. The bar graph showed quantitative analysis data with three replicates. (D) Immunofluorescence showed knockout of NR4A3 decreased γ-H2AX foci. (E) In vivo growth assays showed the difference in tumor weight between sgNC and sgNR4A3 group in MHCC-97H (top panel) and between shNC and shNR4A3 groups in MHCC-97L (bottom panel) cell lines. (F) Western blot analysis showed that the expression of NR4A3 was obviously down-regulated in NR4A3-shRNA tumor tissues collected from nude mice with tumor xenografts. β-actin was used as a loading control. (G) Immunohistochemical images of Ki67 and PCNA expressions in xenograft tumors derived from MHCC-97H and MHCC-97L cells with NR4A3 knockout or knockdown. Original magnification: × 400. The positively stain area (in percentages) were analyzed. *, p<0.05; **, p<0.01.
Figure 4
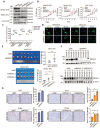
NR4A3 induces cell cycle arrest at the G0/G1 to S phase. The cell cycle distribution was showed in MHCC-LM3 (A), Li7 (B), and Huh7 (C) cells transfected with NR4A3 or the control vectors. (D) The cell cycle distribution of Li7 cells (vector/NR4A3) collected at 0, 12, and 24 h after synchronizing with 2 mM thymidine. The cell cycle distribution was showed in MHCC-97H (E), MHCC-97L (F) and HCC-LY10 (G) cells that were stably transfected with NR4A3-shRNAs or shNC vectors. (H) The cell cycle distribution of HCC-LY10 cells (Mock/shNC/shNR4A3-1/shNR4A3-2) collected at 0, 12, and 24 h after synchronizing with 2 mM thymidine. Western blot analysis of cell cycle associated proteins in the G1 phase (CDK6, CDK4, CyclinD1) and proliferation marker (PCNA) expression in NR4A3 overexpressing cells (I) and NR4A3 knockdown cells (K). (J) Western blot analysis showed that p53, p21 and γH2AX was efficiently up-regulated in NR4A3 up-regulated MHCC-LM3 and Li7 cells. (L) Western blot analysis showed that p53, p21 and γH2AX was efficiently decline in NR4A3 knockout MHCC-97H and MHCC-97L cells. β-actin was used as a loading control. *, p<0.05; **, p<0.01.
Figure 5
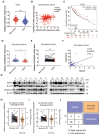
NR4A3 suppressed HCC progression by inducing CDKN2AIP expression. (A) The panel showed the overlap influenced signal pathway after over-expressing of NR4A3 in MHCC-LM3 cells. Western blot analysis of CDKN2AIP protein expressions examined in NR4A3 knockdown cells (B) and NR4A3 overexpressing cells (C). (D) Potential NR4A3-binding sites in the CDKN2AIP promoter identified with the database. (E) The panel is the sequence logo of NR4A3 potential binding site and mutant sites in the CDKN2AIP promoter. (F-G) Binding of NR4A3 to the CDKN2AIP promoter was performed by ChIP using the antibody against NR4A3 and negative control (IgG) in MHCC-97H and MHCC-97L cells. (H) Relative activities of the wide-type and mutant-type CDKN2AIP promoter after transfection of NR4A3 and vector into MHCC-LM3 and Huh7 cells analyzed using luciferase assay. Data are mean ± S.D. from experiments with three replicates. (I) Knockdown of CDKN2AIP reversed the inhibitory effect of NR4A3 on cell proliferation using CCK-8 assay. (J) Knockdown of CDKN2AIP reversed the inhibitory effect of NR4A3 on cell colony formation. (K) MHCC-LM3 cells stably overexpressing NR4A3 with knockdown of CDKN2AIP were injected into one flank of nude mice. Tumors were weighed. *, p<0.05; **, p<0.01.
Figure 6
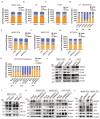
NR4A3 inhibits cell proliferation by up-regulating CDKN2AIP expression. (A) Knockdown of CDKN2AIP reversed the inhibitory effect of NR4A3 on cell cycle distribution was showed in MHCC-LM3 cells. (B) Knockdown of CDKN2AIP reversed the inhibitory effect of NR4A3 on cell cycle distribution collected at 0 and 24 h after synchronizing with 2 mM thymidine was showed in MHCC-LM3 cells. (C) Knockdown of CDKN2AIP reversed the inhibitory effect of NR4A3 on cell cycle-related proteins by western blot analysis. (D) Knockout of CDKN2AIP reversed γ-H2AX foci performed after overexpression of NR4A3 in MHCC-LM3 cells. (E) Knockout of CDKN2AIP reversed the protein expressions of cell cycle-related genes, PCNA and γH2AX examined in NR4A3-overexpressed MHCC-LM3 cells. Western blot analysis of cell cycle-related proteins in NR4A3-overexpressing tumor tissues (F) and in NR4A3-sgRNA (G)/shRNA tumor tissues (H) collected from nude mice with tumor xenografts. β-actin was used as a loading control. *, p<0.05; **, p<0.01.
Figure 7
Clinical correlation between CDKN2AIP and NR4A3. The mRNA expression level of CDKN2AIP in HCC and their adjacent noncancerous liver tissues were analyzed in TCGA database (A) and 56 HCC patients (D-E). The correlation between the mRNA levels of CDKN2AIP and NR4A3 analyzed in TCGA database (B) and 56 HCC patients (F). (C) The correlation between CDKN2AIP expression and overall survival (n=364) of HCC patients in TCGA dataset was assessed using Kaplan-Meier survival analysis. (G) The protein levels of NR4A3 in 42 paired HCC (T) and adjacent normal (N) samples (1-14) were measured using western blot. β-actin was used as a loading control. NR4A3 (H) and CDKN2AIP (I) protein expression in 42 paired HCC tissues and the adjacent matched noncancerous tissues were quantified and analyzed. (J) Correlations among NR4A3 and CDKN2AIP protein levels in 42 paired HCC tissues and adjacent normal samples were examined by western blot. Number represents the number of tissue cases. H, higher expression; L, lower expression. *, p<0.05; **, p<0.01.
Similar articles
- The novel role of LOC344887 in the enhancement of hepatocellular carcinoma progression via modulation of SHP1-regulated STAT3/HMGA2 signaling axis.
Lin YH, Chi HC, Wu MH, Liao CJ, Chen CY, Huang PS, Huang WC, Wang YW, Lin TK, Lai MW, Yeh CT, Lin KH. Lin YH, et al. Int J Biol Sci. 2024 Dec 2;20(15):6281-6296. doi: 10.7150/ijbs.99683. eCollection 2024. Int J Biol Sci. 2024. PMID: 39664573 Free PMC article. - PIWIL genes in hepatocellular carcinoma: a multi-omics approach uncovering dysregulated expression and ceRNA networks in mice.
Huang H, Lu R, Peng S, Huang S, Mo Y, Li G. Huang H, et al. BMC Genom Data. 2024 Nov 27;25(1):101. doi: 10.1186/s12863-024-01283-1. BMC Genom Data. 2024. PMID: 39604866 Free PMC article. - METTL3-Mediated m6A Modification of FMRP Drives Hepatocellular Carcinoma Progression and Indicates Poor Prognosis.
Fu S, Sun D, Wang Z, Zhu P, Ding W, Huang J, Guo X, Yang Y, Gu F. Fu S, et al. Cancer Biother Radiopharm. 2024 Dec;39(10):745-754. doi: 10.1089/cbr.2023.0186. Epub 2024 Sep 12. Cancer Biother Radiopharm. 2024. PMID: 39263746 - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Sequencing of systemic therapy in unresectable hepatocellular carcinoma: A systematic review and Bayesian network meta-analysis of randomized clinical trials.
Wang Q, Yu J, Sun X, Li J, Cao S, Han Y, Wang H, Yang Z, Li J, Hu C, Zhang Y, Jin L. Wang Q, et al. Crit Rev Oncol Hematol. 2024 Dec;204:104522. doi: 10.1016/j.critrevonc.2024.104522. Epub 2024 Sep 26. Crit Rev Oncol Hematol. 2024. PMID: 39332750
References
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA: a cancer journal for clinicians. 2021;71:7–33. - PubMed
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA: a cancer journal for clinicians. 2023;73:17–48. - PubMed
- Calderaro J, Ziol M, Paradis V, Zucman-Rossi J. Molecular and histological correlations in liver cancer. Journal of hepatology. 2019;71:616–30. - PubMed
- Islami F, Goding Sauer A, Miller KD, Siegel RL, Fedewa SA, Jacobs EJ. et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA: a cancer journal for clinicians. 2018;68:31–54. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous