numpy.cov — NumPy v1.13 Manual (original) (raw)
numpy. cov(m, y=None, rowvar=True, bias=False, ddof=None, fweights=None, aweights=None)[source]¶
Estimate a covariance matrix, given data and weights.
Covariance indicates the level to which two variables vary together. If we examine N-dimensional samples, ![X = [x_1, x_2, ... x_N]^T](https://docs.scipy.org/doc/numpy-1.13.0/_images/math/8d14fb48024fc0aaeed58a8b7e012bf142402c41.png) , then the covariance matrix element
, then the covariance matrix element 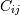 is the covariance of
is the covariance of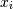 and
and 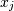 . The element
. The element 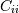 is the variance of
is the variance of 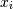 .
.
See the notes for an outline of the algorithm.
| Parameters: | m : array_like A 1-D or 2-D array containing multiple variables and observations. Each row of m represents a variable, and each column a single observation of all those variables. Also see rowvar below. y : array_like, optional An additional set of variables and observations. y has the same form as that of m. rowvar : bool, optional If rowvar is True (default), then each row represents a variable, with observations in the columns. Otherwise, the relationship is transposed: each column represents a variable, while the rows contain observations. bias : bool, optional Default normalization (False) is by (N - 1), where N is the number of observations given (unbiased estimate). If bias is True, then normalization is by N. These values can be overridden by using the keyword ddof in numpy versions >= 1.5. ddof : int, optional If not None the default value implied by bias is overridden. Note that ddof=1 will return the unbiased estimate, even if both_fweights_ and aweights are specified, and ddof=0 will return the simple average. See the notes for the details. The default value is None. New in version 1.5. fweights : array_like, int, optional 1-D array of integer freguency weights; the number of times each observation vector should be repeated. New in version 1.10. aweights : array_like, optional 1-D array of observation vector weights. These relative weights are typically large for observations considered “important” and smaller for observations considered less “important”. If ddof=0 the array of weights can be used to assign probabilities to observation vectors. New in version 1.10. |
|---|---|
| Returns: | out : ndarray The covariance matrix of the variables. |
See also
Normalized covariance matrix
Notes
Assume that the observations are in the columns of the observation array m and let f = fweights and a = aweights for brevity. The steps to compute the weighted covariance are as follows:
w = f * a v1 = np.sum(w) v2 = np.sum(w * a) m -= np.sum(m * w, axis=1, keepdims=True) / v1 cov = np.dot(m * w, m.T) * v1 / (v1**2 - ddof * v2)
Note that when a == 1, the normalization factorv1 / (v1**2 - ddof * v2) goes over to 1 / (np.sum(f) - ddof)as it should.
Examples
Consider two variables, 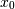 and
and 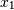 , which correlate perfectly, but in opposite directions:
, which correlate perfectly, but in opposite directions:
x = np.array([[0, 2], [1, 1], [2, 0]]).T x array([[0, 1, 2], [2, 1, 0]])
Note how 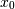 increases while
increases while 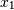 decreases. The covariance matrix shows this clearly:
decreases. The covariance matrix shows this clearly:
np.cov(x) array([[ 1., -1.], [-1., 1.]])
Note that element 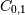 , which shows the correlation between
, which shows the correlation between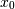 and
and 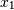 , is negative.
, is negative.
Further, note how x and y are combined:
x = [-2.1, -1, 4.3] y = [3, 1.1, 0.12] X = np.vstack((x,y)) print(np.cov(X)) [[ 11.71 -4.286 ] [ -4.286 2.14413333]] print(np.cov(x, y)) [[ 11.71 -4.286 ] [ -4.286 2.14413333]] print(np.cov(x)) 11.71