scipy.stats.normal_inverse_gamma — SciPy v1.16.0 Manual (original) (raw)
scipy.stats.normal_inverse_gamma = <scipy.stats._multivariate.normal_inverse_gamma_gen object>[source]#
Normal-inverse-gamma distribution.
The normal-inverse-gamma distribution is the conjugate prior of a normal distribution with unknown mean and variance.
Parameters:
mu, lmbda, a, barray_like
Shape parameters of the distribution. See notes.
seed{None, int, np.random.RandomState, np.random.Generator}, optional
Used for drawing random variates. If seed is None, the RandomState singleton is used. If seed is an int, a new RandomState instance is used, seeded with seed. If seed is already a RandomState or Generator instance, then that object is used. Default is None.
Methods
Notes
The probability density function of normal_inverse_gamma is:
\[f(x, \sigma^2; \mu, \lambda, \alpha, \beta) = \frac{\sqrt{\lambda}}{\sqrt{2 \pi \sigma^2}} \frac{\beta^\alpha}{\Gamma(\alpha)} \left( \frac{1}{\sigma^2} \right)^{\alpha + 1} \exp \left(- \frac{2 \beta + \lambda (x - \mu)^2} {2 \sigma^2} \right)\]
where all parameters are real and finite, and \(\sigma^2 > 0\),\(\lambda > 0\), \(\alpha > 0\), and \(\beta > 0\).
Methods normal_inverse_gamma.pdf and normal_inverse_gamma.logpdfaccept x and s2 for arguments \(x\) and \(\sigma^2\). All methods accept mu, lmbda, a, and b for shape parameters\(\mu\), \(\lambda\), \(\alpha\), and \(\beta\), respectively.
Added in version 1.15.
References
Examples
Suppose we wish to investigate the relationship between the normal-inverse-gamma distribution and the inverse gamma distribution.
import numpy as np from scipy import stats import matplotlib.pyplot as plt rng = np.random.default_rng() mu, lmbda, a, b = 0, 1, 20, 20 norm_inv_gamma = stats.normal_inverse_gamma(mu, lmbda, a, b) inv_gamma = stats.invgamma(a, scale=b)
One approach is to compare the distribution of the s2 elements of random variates against the PDF of an inverse gamma distribution.
_, s2 = norm_inv_gamma.rvs(size=10000, random_state=rng) bins = np.linspace(s2.min(), s2.max(), 50) plt.hist(s2, bins=bins, density=True, label='Frequency density') s2 = np.linspace(s2.min(), s2.max(), 300) plt.plot(s2, inv_gamma.pdf(s2), label='PDF') plt.xlabel(r'$\sigma^2$') plt.ylabel('Frequency density / PMF') plt.show()
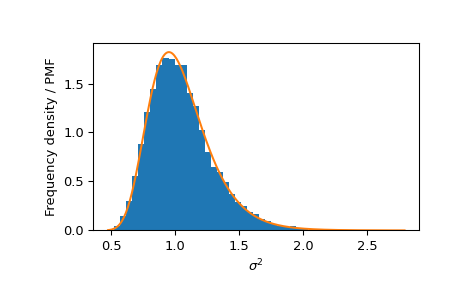
Similarly, we can compare the marginal distribution of s2 against an inverse gamma distribution.
from scipy.integrate import quad_vec from scipy import integrate s2 = np.linspace(0.5, 3, 6) res = quad_vec(lambda x: norm_inv_gamma.pdf(x, s2), -np.inf, np.inf)[0] np.allclose(res, inv_gamma.pdf(s2)) True
The sample mean is comparable to the mean of the distribution.
x, s2 = norm_inv_gamma.rvs(size=10000, random_state=rng) x.mean(), s2.mean() (np.float64(-0.005254750127304425), np.float64(1.050438111436508)) norm_inv_gamma.mean() (np.float64(0.0), np.float64(1.0526315789473684))
Similarly, for the variance:
x.var(ddof=1), s2.var(ddof=1) (np.float64(1.0546150578185023), np.float64(0.061829865266330754)) norm_inv_gamma.var() (np.float64(1.0526315789473684), np.float64(0.061557402277623886))