Wide Data via Lasso and Parallel Computing - MATLAB & Simulink (original) (raw)
This example shows how to use lasso along with cross validation to identify important predictors.
Load the sample data.
Lasso and elastic net are especially well suited for wide data, that is, data with more predictors than observations with lasso and elastic net. There are redundant predictors in this type of data. You can use lasso along with cross validation to identify important predictors.
Compute the default lasso fit.
[b fitinfo] = lasso(NIR,octane);
Plot the number of predictors in the fitted lasso regularization as a function of Lambda, using a logarithmic x -axis.
lassoPlot(b,fitinfo,'PlotType','Lambda','XScale','log');
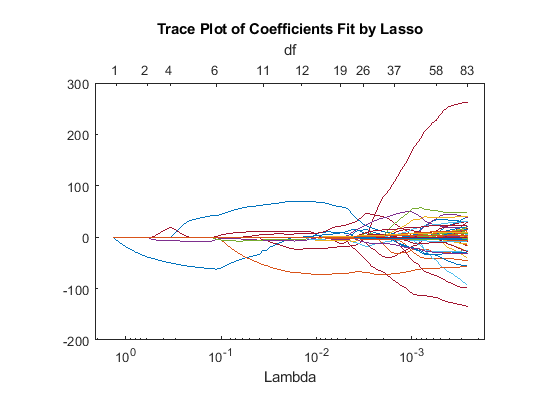
It is difficult to tell which value of Lambda is appropriate. To determine a good value, try fitting with cross validation.
tic [b fitinfo] = lasso(NIR,octane,'CV',10); toc
Elapsed time is 1.309120 seconds.
Plot the result.
lassoPlot(b,fitinfo,'PlotType','Lambda','XScale','log');

Display the suggested value of Lambda.
Display the Lambda with minimal MSE.
Examine the quality of the fit for the suggested value of Lambda.
lambdaindex = fitinfo.Index1SE; mse = fitinfo.MSE(lambdaindex) df = fitinfo.DF(lambdaindex)
The fit uses just 11 of the 401 predictors and achieves a small cross-validated MSE.
Examine the plot of cross-validated MSE.
lassoPlot(b,fitinfo,'PlotType','CV'); % Use a log scale for MSE to see small MSE values better set(gca,'YScale','log');

As Lambda increases (toward the left), MSE increases rapidly. The coefficients are reduced too much and they do not adequately fit the responses. As Lambda decreases, the models are larger (have more nonzero coefficients). The increasing MSE suggests that the models are overfitted.
The default set of Lambda values does not include values small enough to include all predictors. In this case, there does not appear to be a reason to look at smaller values. However, if you want smaller values than the default, use the LambdaRatio parameter, or supply a sequence of Lambda values using the Lambda parameter. For details, see the lasso reference page.
Cross validation can be slow. If you have a Parallel Computing Toolbox™ license, speed the computation of cross-validated lasso estimate using parallel computing. Start a parallel pool.
Starting parallel pool (parpool) using the 'Processes' profile ... 13-Nov-2024 15:35:53: Job Queued. Waiting for parallel pool job with ID 1 to start ... Connected to parallel pool with 4 workers.
mypool =
ProcessPool with properties:
Connected: true
NumWorkers: 4
Busy: false
Cluster: Processes (Local Cluster)
AttachedFiles: {}
AutoAddClientPath: true
FileStore: [1x1 parallel.FileStore]
ValueStore: [1x1 parallel.ValueStore]
IdleTimeout: 30 minutes (30 minutes remaining)
SpmdEnabled: trueSet the parallel computing option and compute the lasso estimate.
opts = statset('UseParallel',true); tic; [b fitinfo] = lasso(NIR,octane,'CV',10,'Options',opts); toc
Elapsed time is 1.829342 seconds.
Computing in parallel using two workers is faster on this problem.
Stop parallel pool.
Parallel pool using the 'Processes' profile is shutting down.
See Also
lasso | lassoglm | fitrlinear | lassoPlot | ridge