Version 0.16.0 (March 22, 2015) — pandas 3.0.0rc0+31.g944c527c0a documentation (original) (raw)
This is a major release from 0.15.2 and includes a small number of API changes, several new features, enhancements, and performance improvements along with a large number of bug fixes. We recommend that all users upgrade to this version.
Highlights include:
DataFrame.assignmethod, see hereSeries.to_coo/from_coomethods to interact withscipy.sparse, see here- Backwards incompatible change to
Timedeltato conform the.secondsattribute withdatetime.timedelta, see here - Changes to the
.locslicing API to conform with the behavior of.ixsee here - Changes to the default for ordering in the
Categoricalconstructor, see here - Enhancement to the
.straccessor to make string operations easier, see here - The
pandas.tools.rplot,pandas.sandbox.qtpandasandpandas.rpymodules are deprecated. We refer users to external packages likeseaborn,pandas-qt andrpy2 for similar or equivalent functionality, see here
Check the API Changes and deprecations before updating.
What’s new in v0.16.0
New features#
DataFrame assign#
Inspired by dplyr’s mutate verb, DataFrame has a newassign() method. The function signature for assign is simply **kwargs. The keys are the column names for the new fields, and the values are either a value to be inserted (for example, a Series or NumPy array), or a function of one argument to be called on the DataFrame. The new values are inserted, and the entire DataFrame (with all original and new columns) is returned.
In [1]: iris = pd.read_csv('data/iris.data')
In [2]: iris.head() Out[2]: SepalLength SepalWidth PetalLength PetalWidth Name 0 5.1 3.5 1.4 0.2 Iris-setosa 1 4.9 3.0 1.4 0.2 Iris-setosa 2 4.7 3.2 1.3 0.2 Iris-setosa 3 4.6 3.1 1.5 0.2 Iris-setosa 4 5.0 3.6 1.4 0.2 Iris-setosa
[5 rows x 5 columns]
In [3]: iris.assign(sepal_ratio=iris['SepalWidth'] / iris['SepalLength']).head() Out[3]: SepalLength SepalWidth PetalLength PetalWidth Name sepal_ratio 0 5.1 3.5 1.4 0.2 Iris-setosa 0.686275 1 4.9 3.0 1.4 0.2 Iris-setosa 0.612245 2 4.7 3.2 1.3 0.2 Iris-setosa 0.680851 3 4.6 3.1 1.5 0.2 Iris-setosa 0.673913 4 5.0 3.6 1.4 0.2 Iris-setosa 0.720000
[5 rows x 6 columns]
Above was an example of inserting a precomputed value. We can also pass in a function to be evaluated.
In [4]: iris.assign(sepal_ratio=lambda x: (x['SepalWidth'] ...: / x['SepalLength'])).head() ...: Out[4]: SepalLength SepalWidth PetalLength PetalWidth Name sepal_ratio 0 5.1 3.5 1.4 0.2 Iris-setosa 0.686275 1 4.9 3.0 1.4 0.2 Iris-setosa 0.612245 2 4.7 3.2 1.3 0.2 Iris-setosa 0.680851 3 4.6 3.1 1.5 0.2 Iris-setosa 0.673913 4 5.0 3.6 1.4 0.2 Iris-setosa 0.720000
[5 rows x 6 columns]
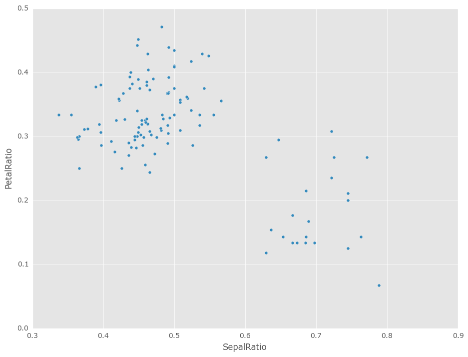
The power of assign comes when used in chains of operations. For example, we can limit the DataFrame to just those with a Sepal Length greater than 5, calculate the ratio, and plot
In [5]: iris = pd.read_csv('data/iris.data')
In [6]: (iris.query('SepalLength > 5') ...: .assign(SepalRatio=lambda x: x.SepalWidth / x.SepalLength, ...: PetalRatio=lambda x: x.PetalWidth / x.PetalLength) ...: .plot(kind='scatter', x='SepalRatio', y='PetalRatio')) ...: Out[6]: <Axes: xlabel='SepalRatio', ylabel='PetalRatio'>
See the documentation for more. (GH 9229)
Interaction with scipy.sparse#
Added SparseSeries.to_coo() and SparseSeries.from_coo() methods (GH 8048) for converting to and from scipy.sparse.coo_matrix instances (see here). For example, given a SparseSeries with MultiIndex we can convert to a scipy.sparse.coo_matrix by specifying the row and column labels as index levels:
s = pd.Series([3.0, np.nan, 1.0, 3.0, np.nan, np.nan]) s.index = pd.MultiIndex.from_tuples([(1, 2, 'a', 0), (1, 2, 'a', 1), (1, 1, 'b', 0), (1, 1, 'b', 1), (2, 1, 'b', 0), (2, 1, 'b', 1)], names=['A', 'B', 'C', 'D'])
s
SparseSeries
ss = s.to_sparse() ss
A, rows, columns = ss.to_coo(row_levels=['A', 'B'], column_levels=['C', 'D'], sort_labels=False)
A A.todense() rows columns
The from_coo method is a convenience method for creating a SparseSeriesfrom a scipy.sparse.coo_matrix:
from scipy import sparse A = sparse.coo_matrix(([3.0, 1.0, 2.0], ([1, 0, 0], [0, 2, 3])), shape=(3, 4)) A A.todense()
ss = pd.SparseSeries.from_coo(A) ss
String methods enhancements#
- Following new methods are accessible via
.straccessor to apply the function to each values. This is intended to make it more consistent with standard methods on strings. (GH 9282, GH 9352, GH 9386, GH 9387, GH 9439)
In [7]: s = pd.Series(['abcd', '3456', 'EFGH'])
In [8]: s.str.isalpha()
Out[8]:
0 True
1 False
2 True
Length: 3, dtype: bool
In [9]: s.str.find('ab')
Out[9]:
0 0
1 -1
2 -1
Length: 3, dtype: int64
- Series.str.pad() and Series.str.center() now accept
fillcharoption to specify filling character (GH 9352)
In [10]: s = pd.Series(['12', '300', '25'])
In [11]: s.str.pad(5, fillchar='_')
Out[11]:
0 ___12
1 __300
2 ___25
Length: 3, dtype: str - Added Series.str.slice_replace(), which previously raised
NotImplementedError(GH 8888)
In [12]: s = pd.Series(['ABCD', 'EFGH', 'IJK'])
In [13]: s.str.slice_replace(1, 3, 'X')
Out[13]:
0 AXD
1 EXH
2 IX
Length: 3, dtype: str
replaced with empty char
In [14]: s.str.slice_replace(0, 1)
Out[14]:
0 BCD
1 FGH
2 JK
Length: 3, dtype: str
Other enhancements#
- Reindex now supports
method='nearest'for frames or series with a monotonic increasing or decreasing index (GH 9258):
In [15]: df = pd.DataFrame({'x': range(5)})
In [16]: df.reindex([0.2, 1.8, 3.5], method='nearest')
Out[16]:
x
0.2 0
1.8 2
3.5 4
[3 rows x 1 columns]
This method is also exposed by the lower levelIndex.get_indexerandIndex.get_locmethods. - The
read_excel()function’s sheetname argument now accepts a list andNone, to get multiple or all sheets respectively. If more than one sheet is specified, a dictionary is returned. (GH 9450)
Returns the 1st and 4th sheet, as a dictionary of DataFrames.
pd.read_excel('path_to_file.xls', sheetname=['Sheet1', 3])
- Allow Stata files to be read incrementally with an iterator; support for long strings in Stata files. See the docs here (GH 9493:).
- Paths beginning with ~ will now be expanded to begin with the user’s home directory (GH 9066)
- Added time interval selection in
get_data_yahoo(GH 9071) - Added
Timestamp.to_datetime64()to complementTimedelta.to_timedelta64()(GH 9255) tseries.frequencies.to_offset()now acceptsTimedeltaas input (GH 9064)- Lag parameter was added to the autocorrelation method of
Series, defaults to lag-1 autocorrelation (GH 9192) Timedeltawill now acceptnanosecondskeyword in constructor (GH 9273)- SQL code now safely escapes table and column names (GH 8986)
- Added auto-complete for
Series.str.<tab>,Series.dt.<tab>andSeries.cat.<tab>(GH 9322) Index.get_indexernow supportsmethod='pad'andmethod='backfill'even for any target array, not just monotonic targets. These methods also work for monotonic decreasing as well as monotonic increasing indexes (GH 9258).Index.asofnow works on all index types (GH 9258).- A
verboseargument has been augmented inio.read_excel(), defaults to False. Set to True to print sheet names as they are parsed. (GH 9450) - Added
days_in_month(compatibility aliasdaysinmonth) property toTimestamp,DatetimeIndex,Period,PeriodIndex, andSeries.dt(GH 9572) - Added
decimaloption into_csvto provide formatting for non-‘.’ decimal separators (GH 781) - Added
normalizeoption forTimestampto normalized to midnight (GH 8794) - Added example for
DataFrameimport to R using HDF5 file andrhdf5library. See the documentation for more (GH 9636).
Backwards incompatible API changes#
Changes in timedelta#
In v0.15.0 a new scalar type Timedelta was introduced, that is a sub-class of datetime.timedelta. Mentioned here was a notice of an API change w.r.t. the .seconds accessor. The intent was to provide a user-friendly set of accessors that give the ‘natural’ value for that unit, e.g. if you had a Timedelta('1 day, 10:11:12'), then .seconds would return 12. However, this is at odds with the definition of datetime.timedelta, which defines .seconds as 10 * 3600 + 11 * 60 + 12 == 36672.
So in v0.16.0, we are restoring the API to match that of datetime.timedelta. Further, the component values are still available through the .components accessor. This affects the .seconds and .microseconds accessors, and removes the .hours, .minutes, .milliseconds accessors. These changes affect TimedeltaIndex and the Series .dt accessor as well. (GH 9185, GH 9139)
Previous behavior
In [2]: t = pd.Timedelta('1 day, 10:11:12.100123')
In [3]: t.days Out[3]: 1
In [4]: t.seconds Out[4]: 12
In [5]: t.microseconds Out[5]: 123
New behavior
In [17]: t = pd.Timedelta('1 day, 10:11:12.100123')
In [18]: t.days Out[18]: 1
In [19]: t.seconds Out[19]: 36672
In [20]: t.microseconds Out[20]: 100123
Using .components allows the full component access
In [21]: t.components Out[21]: Components(days=1, hours=10, minutes=11, seconds=12, milliseconds=100, microseconds=123, nanoseconds=0)
In [22]: t.components.seconds Out[22]: 12
Indexing changes#
The behavior of a small sub-set of edge cases for using .loc have changed (GH 8613). Furthermore we have improved the content of the error messages that are raised:
- Slicing with
.locwhere the start and/or stop bound is not found in the index is now allowed; this previously would raise aKeyError. This makes the behavior the same as.ixin this case. This change is only for slicing, not when indexing with a single label.
In [23]: df = pd.DataFrame(np.random.randn(5, 4),
....: columns=list('ABCD'),
....: index=pd.date_range('20130101', periods=5))
....:
In [24]: df
Out[24]:
A B C D
2013-01-01 0.469112 -0.282863 -1.509059 -1.135632
2013-01-02 1.212112 -0.173215 0.119209 -1.044236
2013-01-03 -0.861849 -2.104569 -0.494929 1.071804
2013-01-04 0.721555 -0.706771 -1.039575 0.271860
2013-01-05 -0.424972 0.567020 0.276232 -1.087401
[5 rows x 4 columns]
In [25]: s = pd.Series(range(5), [-2, -1, 1, 2, 3])
In [26]: s
Out[26]:
-2 0
-1 1
1 2
2 3
3 4
Length: 5, dtype: int64
Previous behavior
In [4]: df.loc['2013-01-02':'2013-01-10']
KeyError: 'stop bound [2013-01-10] is not in the [index]'
In [6]: s.loc[-10:3]
KeyError: 'start bound [-10] is not the [index]'
New behavior
In [27]: df.loc['2013-01-02':'2013-01-10']
Out[27]:
A B C D
2013-01-02 1.212112 -0.173215 0.119209 -1.044236
2013-01-03 -0.861849 -2.104569 -0.494929 1.071804
2013-01-04 0.721555 -0.706771 -1.039575 0.271860
2013-01-05 -0.424972 0.567020 0.276232 -1.087401
[4 rows x 4 columns]
In [28]: s.loc[-10:3]
Out[28]:
-2 0
-1 1
1 2
2 3
3 4
Length: 5, dtype: int64
- Allow slicing with float-like values on an integer index for
.ix. Previously this was only enabled for.loc:
Previous behavior
In [8]: s.ix[-1.0:2]
TypeError: the slice start value [-1.0] is not a proper indexer for this index type (Int64Index)
New behavior
In [2]: s.ix[-1.0:2]
Out[2]:
-1 1
1 2
2 3
dtype: int64
- Provide a useful exception for indexing with an invalid type for that index when using
.loc. For example trying to use.locon an index of typeDatetimeIndexorPeriodIndexorTimedeltaIndex, with an integer (or a float).
Previous behavior
In [4]: df.loc[2:3]
KeyError: 'start bound [2] is not the [index]'
New behavior
In [4]: df.loc[2:3]
TypeError: Cannot do slice indexing on <class 'pandas.tseries.index.DatetimeIndex'> with <type 'int'> keys
Categorical changes#
In prior versions, Categoricals that had an unspecified ordering (meaning no ordered keyword was passed) were defaulted as ordered Categoricals. Going forward, the ordered keyword in the Categorical constructor will default to False. Ordering must now be explicit.
Furthermore, previously you could change the ordered attribute of a Categorical by just setting the attribute, e.g. cat.ordered=True; This is now deprecated and you should use cat.as_ordered() or cat.as_unordered(). These will by default return a new object and not modify the existing object. (GH 9347, GH 9190)
Previous behavior
In [3]: s = pd.Series([0, 1, 2], dtype='category')
In [4]: s Out[4]: 0 0 1 1 2 2 dtype: category Categories (3, int64): [0 < 1 < 2]
In [5]: s.cat.ordered Out[5]: True
In [6]: s.cat.ordered = False
In [7]: s Out[7]: 0 0 1 1 2 2 dtype: category Categories (3, int64): [0, 1, 2]
New behavior
In [29]: s = pd.Series([0, 1, 2], dtype='category')
In [30]: s Out[30]: 0 0 1 1 2 2 Length: 3, dtype: category Categories (3, int64): [0, 1, 2]
In [31]: s.cat.ordered Out[31]: False
In [32]: s = s.cat.as_ordered()
In [33]: s Out[33]: 0 0 1 1 2 2 Length: 3, dtype: category Categories (3, int64): [0 < 1 < 2]
In [34]: s.cat.ordered Out[34]: True
you can set in the constructor of the Categorical
In [35]: s = pd.Series(pd.Categorical([0, 1, 2], ordered=True))
In [36]: s Out[36]: 0 0 1 1 2 2 Length: 3, dtype: category Categories (3, int64): [0 < 1 < 2]
In [37]: s.cat.ordered Out[37]: True
For ease of creation of series of categorical data, we have added the ability to pass keywords when calling .astype(). These are passed directly to the constructor.
In [54]: s = pd.Series(["a", "b", "c", "a"]).astype('category', ordered=True)
In [55]: s Out[55]: 0 a 1 b 2 c 3 a dtype: category Categories (3, object): [a < b < c]
In [56]: s = (pd.Series(["a", "b", "c", "a"]) ....: .astype('category', categories=list('abcdef'), ordered=False))
In [57]: s Out[57]: 0 a 1 b 2 c 3 a dtype: category Categories (6, object): [a, b, c, d, e, f]
Other API changes#
Index.duplicatednow returnsnp.array(dtype=bool)rather thanIndex(dtype=object)containingboolvalues. (GH 8875)DataFrame.to_jsonnow returns accurate type serialisation for each column for frames of mixed dtype (GH 9037)
Previously data was coerced to a common dtype before serialisation, which for example resulted in integers being serialised to floats:
In [2]: pd.DataFrame({'i': [1,2], 'f': [3.0, 4.2]}).to_json()
Out[2]: '{"f":{"0":3.0,"1":4.2},"i":{"0":1.0,"1":2.0}}'
Now each column is serialised using its correct dtype:
In [2]: pd.DataFrame({'i': [1,2], 'f': [3.0, 4.2]}).to_json()
Out[2]: '{"f":{"0":3.0,"1":4.2},"i":{"0":1,"1":2}}'DatetimeIndex,PeriodIndexandTimedeltaIndex.summarynow output the same format. (GH 9116)TimedeltaIndex.freqstrnow output the same string format asDatetimeIndex. (GH 9116)- Bar and horizontal bar plots no longer add a dashed line along the info axis. The prior style can be achieved with matplotlib’s
axhlineoraxvlinemethods (GH 9088). Seriesaccessors.dt,.catand.strnow raiseAttributeErrorinstead ofTypeErrorif the series does not contain the appropriate type of data (GH 9617). This follows Python’s built-in exception hierarchy more closely and ensures that tests likehasattr(s, 'cat')are consistent on both Python 2 and 3.Seriesnow supports bitwise operation for integral types (GH 9016). Previously even if the input dtypes were integral, the output dtype was coerced tobool.
Previous behavior
In [2]: pd.Series([0, 1, 2, 3], list('abcd')) | pd.Series([4, 4, 4, 4], list('abcd'))
Out[2]:
a True
b True
c True
d True
dtype: bool
New behavior. If the input dtypes are integral, the output dtype is also integral and the output values are the result of the bitwise operation.
In [2]: pd.Series([0, 1, 2, 3], list('abcd')) | pd.Series([4, 4, 4, 4], list('abcd'))
Out[2]:
a 4
b 5
c 6
d 7
dtype: int64- During division involving a
SeriesorDataFrame,0/0and0//0now givenp.naninstead ofnp.inf. (GH 9144, GH 8445)
Previous behavior
In [2]: p = pd.Series([0, 1])
In [3]: p / 0
Out[3]:
0 inf
1 inf
dtype: float64
In [4]: p // 0
Out[4]:
0 inf
1 inf
dtype: float64
New behavior
In [38]: p = pd.Series([0, 1])
In [39]: p / 0
Out[39]:
0 NaN
1 inf
Length: 2, dtype: float64
In [40]: p // 0
Out[40]:
0 NaN
1 inf
Length: 2, dtype: float64 Series.values_countsandSeries.describefor categorical data will now putNaNentries at the end. (GH 9443)Series.describefor categorical data will now give counts and frequencies of 0, notNaN, for unused categories (GH 9443)- Due to a bug fix, looking up a partial string label with
DatetimeIndex.asofnow includes values that match the string, even if they are after the start of the partial string label (GH 9258).
Old behavior:
In [4]: pd.to_datetime(['2000-01-31', '2000-02-28']).asof('2000-02')
Out[4]: Timestamp('2000-01-31 00:00:00')
Fixed behavior:
In [41]: pd.to_datetime(['2000-01-31', '2000-02-28']).asof('2000-02')
Out[41]: Timestamp('2000-02-28 00:00:00')
To reproduce the old behavior, simply add more precision to the label (e.g., use2000-02-01instead of2000-02).
Deprecations#
- The
rplottrellis plotting interface is deprecated and will be removed in a future version. We refer to external packages likeseaborn for similar but more refined functionality (GH 3445). The documentation includes some examples how to convert your existing code fromrplotto seaborn here. - The
pandas.sandbox.qtpandasinterface is deprecated and will be removed in a future version. We refer users to the external package pandas-qt. (GH 9615) - The
pandas.rpyinterface is deprecated and will be removed in a future version. Similar functionality can be accessed through the rpy2 project (GH 9602) - Adding
DatetimeIndex/PeriodIndexto anotherDatetimeIndex/PeriodIndexis being deprecated as a set-operation. This will be changed to aTypeErrorin a future version..union()should be used for the union set operation. (GH 9094) - Subtracting
DatetimeIndex/PeriodIndexfrom anotherDatetimeIndex/PeriodIndexis being deprecated as a set-operation. This will be changed to an actual numeric subtraction yielding aTimeDeltaIndexin a future version..difference()should be used for the differencing set operation. (GH 9094)
Removal of prior version deprecations/changes#
DataFrame.pivot_tableandcrosstab’srowsandcolskeyword arguments were removed in favor ofindexandcolumns(GH 6581)DataFrame.to_excelandDataFrame.to_csvcolskeyword argument was removed in favor ofcolumns(GH 6581)- Removed
convert_dummiesin favor ofget_dummies(GH 6581) - Removed
value_rangein favor ofdescribe(GH 6581)
Performance improvements#
- Fixed a performance regression for
.locindexing with an array or list-like (GH 9126:). DataFrame.to_json30x performance improvement for mixed dtype frames. (GH 9037)- Performance improvements in
MultiIndex.duplicatedby working with labels instead of values (GH 9125) - Improved the speed of
nuniqueby callinguniqueinstead ofvalue_counts(GH 9129, GH 7771) - Performance improvement of up to 10x in
DataFrame.countandDataFrame.dropnaby taking advantage of homogeneous/heterogeneous dtypes appropriately (GH 9136) - Performance improvement of up to 20x in
DataFrame.countwhen using aMultiIndexand thelevelkeyword argument (GH 9163) - Performance and memory usage improvements in
mergewhen key space exceedsint64bounds (GH 9151) - Performance improvements in multi-key
groupby(GH 9429) - Performance improvements in
MultiIndex.sortlevel(GH 9445) - Performance and memory usage improvements in
DataFrame.duplicated(GH 9398) - Cythonized
Period(GH 9440) - Decreased memory usage on
to_hdf(GH 9648)
Bug fixes#
- Changed
.to_htmlto remove leading/trailing spaces in table body (GH 4987) - Fixed issue using
read_csvon s3 with Python 3 (GH 9452) - Fixed compatibility issue in
DatetimeIndexaffecting architectures wherenumpy.int_defaults tonumpy.int32(GH 8943) - Bug in Panel indexing with an object-like (GH 9140)
- Bug in the returned
Series.dt.componentsindex was reset to the default index (GH 9247) - Bug in
Categorical.__getitem__/__setitem__with listlike input getting incorrect results from indexer coercion (GH 9469) - Bug in partial setting with a DatetimeIndex (GH 9478)
- Bug in groupby for integer and datetime64 columns when applying an aggregator that caused the value to be changed when the number was sufficiently large (GH 9311, GH 6620)
- Fixed bug in
to_sqlwhen mapping aTimestampobject column (datetime column with timezone info) to the appropriate sqlalchemy type (GH 9085). - Fixed bug in
to_sqldtypeargument not accepting an instantiated SQLAlchemy type (GH 9083). - Bug in
.locpartial setting with anp.datetime64(GH 9516) - Incorrect dtypes inferred on datetimelike looking
Series& on.xsslices (GH 9477) - Items in
Categorical.unique()(ands.unique()ifsis of dtypecategory) now appear in the order in which they are originally found, not in sorted order (GH 9331). This is now consistent with the behavior for other dtypes in pandas. - Fixed bug on big endian platforms which produced incorrect results in
StataReader(GH 8688). - Bug in
MultiIndex.has_duplicateswhen having many levels causes an indexer overflow (GH 9075, GH 5873) - Bug in
pivotandunstackwherenanvalues would break index alignment (GH 4862, GH 7401, GH 7403, GH 7405, GH 7466, GH 9497) - Bug in left
joinon MultiIndex withsort=Trueor null values (GH 9210). - Bug in
MultiIndexwhere inserting new keys would fail (GH 9250). - Bug in
groupbywhen key space exceedsint64bounds (GH 9096). - Bug in
unstackwithTimedeltaIndexorDatetimeIndexand nulls (GH 9491). - Bug in
rankwhere comparing floats with tolerance will cause inconsistent behaviour (GH 8365). - Fixed character encoding bug in
read_stataandStataReaderwhen loading data from a URL (GH 9231). - Bug in adding
offsets.Nanoto other offsets raisesTypeError(GH 9284) - Bug in
DatetimeIndexiteration, related to (GH 8890), fixed in (GH 9100) - Bugs in
resamplearound DST transitions. This required fixing offset classes so they behave correctly on DST transitions. (GH 5172, GH 8744, GH 8653, GH 9173, GH 9468). - Bug in binary operator method (eg
.mul()) alignment with integer levels (GH 9463). - Bug in boxplot, scatter and hexbin plot may show an unnecessary warning (GH 8877)
- Bug in subplot with
layoutkw may show unnecessary warning (GH 9464) - Bug in using grouper functions that need passed through arguments (e.g. axis), when using wrapped function (e.g.
fillna), (GH 9221) DataFramenow properly supports simultaneouscopyanddtypearguments in constructor (GH 9099)- Bug in
read_csvwhen using skiprows on a file with CR line endings with the c engine. (GH 9079) isnullnow detectsNaTinPeriodIndex(GH 9129)- Bug in groupby
.nth()with a multiple column groupby (GH 8979) - Bug in
DataFrame.whereandSeries.wherecoerce numerics to string incorrectly (GH 9280) - Bug in
DataFrame.whereandSeries.whereraiseValueErrorwhen string list-like is passed. (GH 9280) - Accessing
Series.strmethods on with non-string values now raisesTypeErrorinstead of producing incorrect results (GH 9184) - Bug in
DatetimeIndex.__contains__when index has duplicates and is not monotonic increasing (GH 9512) - Fixed division by zero error for
Series.kurt()when all values are equal (GH 9197) - Fixed issue in the
xlsxwriterengine where it added a default ‘General’ format to cells if no other format was applied. This prevented other row or column formatting being applied. (GH 9167) - Fixes issue with
index_col=Falsewhenusecolsis also specified inread_csv. (GH 9082) - Bug where
wide_to_longwould modify the input stub names list (GH 9204) - Bug in
to_sqlnot storing float64 values using double precision. (GH 9009) SparseSeriesandSparsePanelnow accept zero argument constructors (same as their non-sparse counterparts) (GH 9272).- Regression in merging
Categoricalandobjectdtypes (GH 9426) - Bug in
read_csvwith buffer overflows with certain malformed input files (GH 9205) - Bug in groupby MultiIndex with missing pair (GH 9049, GH 9344)
- Fixed bug in
Series.groupbywhere grouping onMultiIndexlevels would ignore the sort argument (GH 9444) - Fix bug in
DataFrame.Groupbywheresort=Falseis ignored in the case of Categorical columns. (GH 8868) - Fixed bug with reading CSV files from Amazon S3 on python 3 raising a TypeError (GH 9452)
- Bug in the Google BigQuery reader where the ‘jobComplete’ key may be present but False in the query results (GH 8728)
- Bug in
Series.values_countswith excludingNaNfor categorical typeSerieswithdropna=True(GH 9443) - Fixed missing numeric_only option for
DataFrame.std/var/sem(GH 9201) - Support constructing
PanelorPanel4Dwith scalar data (GH 8285) Seriestext representation disconnected frommax_rows/max_columns(GH 7508).Seriesnumber formatting inconsistent when truncated (GH 8532).
Previous behavior
In [2]: pd.options.display.max_rows = 10
In [3]: s = pd.Series([1,1,1,1,1,1,1,1,1,1,0.9999,1,1]*10)
In [4]: s
Out[4]:
0 1
1 1
2 1
...
127 0.9999
128 1.0000
129 1.0000
Length: 130, dtype: float64
New behavior
0 1.0000
1 1.0000
2 1.0000
3 1.0000
4 1.0000
...
125 1.0000
126 1.0000
127 0.9999
128 1.0000
129 1.0000
dtype: float64
- A Spurious
SettingWithCopyWarning was generated when setting a new item in a frame in some cases (GH 8730)
The following would previously report aSettingWithCopyWarning.
In [42]: df1 = pd.DataFrame({'x': pd.Series(['a', 'b', 'c']),
....: 'y': pd.Series(['d', 'e', 'f'])})
....:
In [43]: df2 = df1[['x']]
In [44]: df2['y'] = ['g', 'h', 'i']
Contributors#
A total of 60 people contributed patches to this release. People with a “+” by their names contributed a patch for the first time.
- Aaron Toth +
- Alan Du +
- Alessandro Amici +
- Artemy Kolchinsky
- Ashwini Chaudhary +
- Ben Schiller
- Bill Letson
- Brandon Bradley +
- Chau Hoang +
- Chris Reynolds
- Chris Whelan +
- Christer van der Meeren +
- David Cottrell +
- David Stephens
- Ehsan Azarnasab +
- Garrett-R +
- Guillaume Gay
- Jake Torcasso +
- Jason Sexauer
- Jeff Reback
- John McNamara
- Joris Van den Bossche
- Joschka zur Jacobsmühlen +
- Juarez Bochi +
- Junya Hayashi +
- K.-Michael Aye
- Kerby Shedden +
- Kevin Sheppard
- Kieran O’Mahony
- Kodi Arfer +
- Matti Airas +
- Min RK +
- Mortada Mehyar
- Robert +
- Scott E Lasley
- Scott Lasley +
- Sergio Pascual +
- Skipper Seabold
- Stephan Hoyer
- Thomas Grainger
- Tom Augspurger
- TomAugspurger
- Vladimir Filimonov +
- Vyomkesh Tripathi +
- Will Holmgren
- Yulong Yang +
- behzad nouri
- bertrandhaut +
- bjonen
- cel4 +
- clham
- hsperr +
- ischwabacher
- jnmclarty
- josham +
- jreback
- omtinez +
- roch +
- sinhrks
- unutbu