Computational tools — pandas 0.18.1 documentation (original) (raw)
Statistical Functions¶
Percent Change¶
Series, DataFrame, and Panel all have a method pct_change to compute the percent change over a given number of periods (using fill_method to fill NA/null values before computing the percent change).
In [1]: ser = pd.Series(np.random.randn(8))
In [2]: ser.pct_change() Out[2]: 0 NaN 1 -1.602976 2 4.334938 3 -0.247456 4 -2.067345 5 -1.142903 6 -1.688214 7 -9.759729 dtype: float64
In [3]: df = pd.DataFrame(np.random.randn(10, 4))
In [4]: df.pct_change(periods=3) Out[4]: 0 1 2 3 0 NaN NaN NaN NaN 1 NaN NaN NaN NaN 2 NaN NaN NaN NaN 3 -0.218320 -1.054001 1.987147 -0.510183 4 -0.439121 -1.816454 0.649715 -4.822809 5 -0.127833 -3.042065 -5.866604 -1.776977 6 -2.596833 -1.959538 -2.111697 -3.798900 7 -0.117826 -2.169058 0.036094 -0.067696 8 2.492606 -1.357320 -1.205802 -1.558697 9 -1.012977 2.324558 -1.003744 -0.371806
Covariance¶
The Series object has a method cov to compute covariance between series (excluding NA/null values).
In [5]: s1 = pd.Series(np.random.randn(1000))
In [6]: s2 = pd.Series(np.random.randn(1000))
In [7]: s1.cov(s2) Out[7]: 0.00068010881743110698
Analogously, DataFrame has a method cov to compute pairwise covariances among the series in the DataFrame, also excluding NA/null values.
Note
Assuming the missing data are missing at random this results in an estimate for the covariance matrix which is unbiased. However, for many applications this estimate may not be acceptable because the estimated covariance matrix is not guaranteed to be positive semi-definite. This could lead to estimated correlations having absolute values which are greater than one, and/or a non-invertible covariance matrix. See Estimation of covariance matricesfor more details.
In [8]: frame = pd.DataFrame(np.random.randn(1000, 5), columns=['a', 'b', 'c', 'd', 'e'])
In [9]: frame.cov() Out[9]: a b c d e a 1.000882 -0.003177 -0.002698 -0.006889 0.031912 b -0.003177 1.024721 0.000191 0.009212 0.000857 c -0.002698 0.000191 0.950735 -0.031743 -0.005087 d -0.006889 0.009212 -0.031743 1.002983 -0.047952 e 0.031912 0.000857 -0.005087 -0.047952 1.042487
DataFrame.cov also supports an optional min_periods keyword that specifies the required minimum number of observations for each column pair in order to have a valid result.
In [10]: frame = pd.DataFrame(np.random.randn(20, 3), columns=['a', 'b', 'c'])
In [11]: frame.ix[:5, 'a'] = np.nan
In [12]: frame.ix[5:10, 'b'] = np.nan
In [13]: frame.cov() Out[13]: a b c a 1.210090 -0.430629 0.018002 b -0.430629 1.240960 0.347188 c 0.018002 0.347188 1.301149
In [14]: frame.cov(min_periods=12) Out[14]: a b c a 1.210090 NaN 0.018002 b NaN 1.240960 0.347188 c 0.018002 0.347188 1.301149
Correlation¶
Several methods for computing correlations are provided:
| Method name | Description |
|---|---|
| pearson (default) | Standard correlation coefficient |
| kendall | Kendall Tau correlation coefficient |
| spearman | Spearman rank correlation coefficient |
All of these are currently computed using pairwise complete observations.
Note
Please see the caveats associated with this method of calculating correlation matrices in thecovariance section.
In [15]: frame = pd.DataFrame(np.random.randn(1000, 5), columns=['a', 'b', 'c', 'd', 'e'])
In [16]: frame.ix[::2] = np.nan
Series with Series
In [17]: frame['a'].corr(frame['b']) Out[17]: 0.013479040400098789
In [18]: frame['a'].corr(frame['b'], method='spearman') Out[18]: -0.0072898851595406388
Pairwise correlation of DataFrame columns
In [19]: frame.corr() Out[19]: a b c d e a 1.000000 0.013479 -0.049269 -0.042239 -0.028525 b 0.013479 1.000000 -0.020433 -0.011139 0.005654 c -0.049269 -0.020433 1.000000 0.018587 -0.054269 d -0.042239 -0.011139 0.018587 1.000000 -0.017060 e -0.028525 0.005654 -0.054269 -0.017060 1.000000
Note that non-numeric columns will be automatically excluded from the correlation calculation.
Like cov, corr also supports the optional min_periods keyword:
In [20]: frame = pd.DataFrame(np.random.randn(20, 3), columns=['a', 'b', 'c'])
In [21]: frame.ix[:5, 'a'] = np.nan
In [22]: frame.ix[5:10, 'b'] = np.nan
In [23]: frame.corr() Out[23]: a b c a 1.000000 -0.076520 0.160092 b -0.076520 1.000000 0.135967 c 0.160092 0.135967 1.000000
In [24]: frame.corr(min_periods=12) Out[24]: a b c a 1.000000 NaN 0.160092 b NaN 1.000000 0.135967 c 0.160092 0.135967 1.000000
A related method corrwith is implemented on DataFrame to compute the correlation between like-labeled Series contained in different DataFrame objects.
In [25]: index = ['a', 'b', 'c', 'd', 'e']
In [26]: columns = ['one', 'two', 'three', 'four']
In [27]: df1 = pd.DataFrame(np.random.randn(5, 4), index=index, columns=columns)
In [28]: df2 = pd.DataFrame(np.random.randn(4, 4), index=index[:4], columns=columns)
In [29]: df1.corrwith(df2) Out[29]: one -0.125501 two -0.493244 three 0.344056 four 0.004183 dtype: float64
In [30]: df2.corrwith(df1, axis=1) Out[30]: a -0.675817 b 0.458296 c 0.190809 d -0.186275 e NaN dtype: float64
Data ranking¶
The rank method produces a data ranking with ties being assigned the mean of the ranks (by default) for the group:
In [31]: s = pd.Series(np.random.np.random.randn(5), index=list('abcde'))
In [32]: s['d'] = s['b'] # so there's a tie
In [33]: s.rank() Out[33]: a 5.0 b 2.5 c 1.0 d 2.5 e 4.0 dtype: float64
rank is also a DataFrame method and can rank either the rows (axis=0) or the columns (axis=1). NaN values are excluded from the ranking.
In [34]: df = pd.DataFrame(np.random.np.random.randn(10, 6))
In [35]: df[4] = df[2][:5] # some ties
In [36]: df Out[36]: 0 1 2 3 4 5 0 -0.904948 -1.163537 -1.457187 0.135463 -1.457187 0.294650 1 -0.976288 -0.244652 -0.748406 -0.999601 -0.748406 -0.800809 2 0.401965 1.460840 1.256057 1.308127 1.256057 0.876004 3 0.205954 0.369552 -0.669304 0.038378 -0.669304 1.140296 4 -0.477586 -0.730705 -1.129149 -0.601463 -1.129149 -0.211196 5 -1.092970 -0.689246 0.908114 0.204848 NaN 0.463347 6 0.376892 0.959292 0.095572 -0.593740 NaN -0.069180 7 -1.002601 1.957794 -0.120708 0.094214 NaN -1.467422 8 -0.547231 0.664402 -0.519424 -0.073254 NaN -1.263544 9 -0.250277 -0.237428 -1.056443 0.419477 NaN 1.375064
In [37]: df.rank(1) Out[37]: 0 1 2 3 4 5 0 4.0 3.0 1.5 5.0 1.5 6.0 1 2.0 6.0 4.5 1.0 4.5 3.0 2 1.0 6.0 3.5 5.0 3.5 2.0 3 4.0 5.0 1.5 3.0 1.5 6.0 4 5.0 3.0 1.5 4.0 1.5 6.0 5 1.0 2.0 5.0 3.0 NaN 4.0 6 4.0 5.0 3.0 1.0 NaN 2.0 7 2.0 5.0 3.0 4.0 NaN 1.0 8 2.0 5.0 3.0 4.0 NaN 1.0 9 2.0 3.0 1.0 4.0 NaN 5.0
rank optionally takes a parameter ascending which by default is true; when false, data is reverse-ranked, with larger values assigned a smaller rank.
rank supports different tie-breaking methods, specified with the methodparameter:
- average : average rank of tied group
- min : lowest rank in the group
- max : highest rank in the group
- first : ranks assigned in the order they appear in the array
Window Functions¶
Warning
Prior to version 0.18.0, pd.rolling_*, pd.expanding_*, and pd.ewm* were module level functions and are now deprecated. These are replaced by using the Rolling, Expanding and EWM. objects and a corresponding method call.
The deprecation warning will show the new syntax, see an example hereYou can view the previous documentationhere
For working with data, a number of windows functions are provided for computing common window or rolling statistics. Among these are count, sum, mean, median, correlation, variance, covariance, standard deviation, skewness, and kurtosis.
Note
The API for window statistics is quite similar to the way one works with GroupBy objects, see the documentation here
We work with rolling, expanding and exponentially weighted data through the corresponding objects, Rolling, Expanding and EWM.
In [38]: s = pd.Series(np.random.randn(1000), index=pd.date_range('1/1/2000', periods=1000))
In [39]: s = s.cumsum()
In [40]: s
Out[40]:
2000-01-01 -0.268824
2000-01-02 -1.771855
2000-01-03 -0.818003
2000-01-04 -0.659244
2000-01-05 -1.942133
2000-01-06 -1.869391
2000-01-07 0.563674
...
2002-09-20 -68.233054
2002-09-21 -66.765687
2002-09-22 -67.457323
2002-09-23 -69.253182
2002-09-24 -70.296818
2002-09-25 -70.844674
2002-09-26 -72.475016
Freq: D, dtype: float64
These are created from methods on Series and DataFrame.
In [41]: r = s.rolling(window=60)
In [42]: r Out[42]: Rolling [window=60,center=False,axis=0]
These object provide tab-completion of the avaible methods and properties.
In [14]: r. r.agg r.apply r.count r.exclusions r.max r.median r.name r.skew r.sum r.aggregate r.corr r.cov r.kurt r.mean r.min r.quantile r.std r.var
Generally these methods all have the same interface. They all accept the following arguments:
- window: size of moving window
- min_periods: threshold of non-null data points to require (otherwise result is NA)
- center: boolean, whether to set the labels at the center (default is False)
Warning
The freq and how arguments were in the API prior to 0.18.0 changes. These are deprecated in the new API. You can simply resample the input prior to creating a window function.
For example, instead of s.rolling(window=5,freq='D').max() to get the max value on a rolling 5 Day window, one could use s.resample('D').max().rolling(window=5).max(), which first resamples the data to daily data, then provides a rolling 5 day window.
We can then call methods on these rolling objects. These return like-indexed objects:
In [43]: r.mean()
Out[43]:
2000-01-01 NaN
2000-01-02 NaN
2000-01-03 NaN
2000-01-04 NaN
2000-01-05 NaN
2000-01-06 NaN
2000-01-07 NaN
...
2002-09-20 -62.694135
2002-09-21 -62.812190
2002-09-22 -62.914971
2002-09-23 -63.061867
2002-09-24 -63.213876
2002-09-25 -63.375074
2002-09-26 -63.539734
Freq: D, dtype: float64
In [44]: s.plot(style='k--') Out[44]: <matplotlib.axes._subplots.AxesSubplot at 0x11ce37110>
In [45]: r.mean().plot(style='k') Out[45]: <matplotlib.axes._subplots.AxesSubplot at 0x11ce37110>
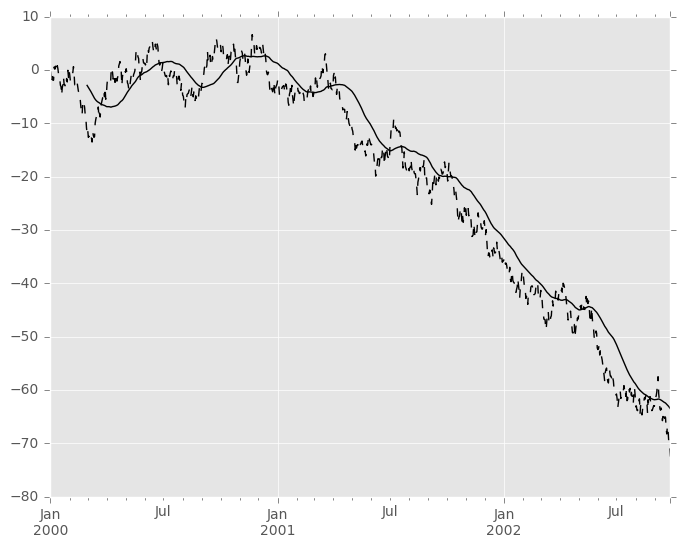
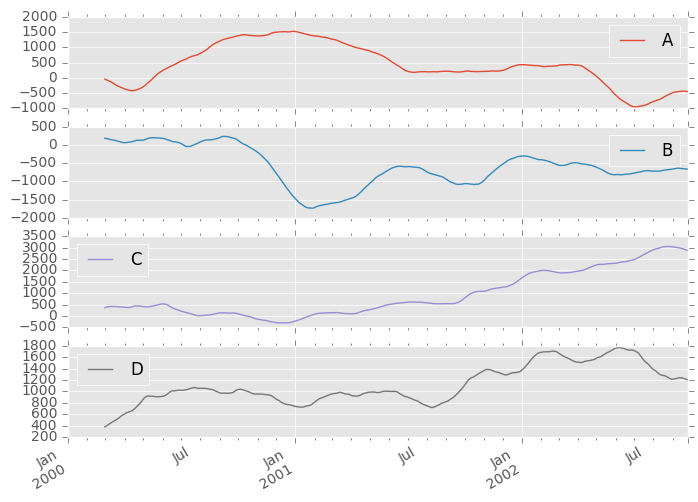
They can also be applied to DataFrame objects. This is really just syntactic sugar for applying the moving window operator to all of the DataFrame’s columns:
In [46]: df = pd.DataFrame(np.random.randn(1000, 4), ....: index=pd.date_range('1/1/2000', periods=1000), ....: columns=['A', 'B', 'C', 'D']) ....:
In [47]: df = df.cumsum()
In [48]: df.rolling(window=60).sum().plot(subplots=True) Out[48]: array([<matplotlib.axes._subplots.AxesSubplot object at 0x11cdc2890>, <matplotlib.axes._subplots.AxesSubplot object at 0x120dfef50>, <matplotlib.axes._subplots.AxesSubplot object at 0x120cd4110>, <matplotlib.axes._subplots.AxesSubplot object at 0x1127f6e90>], dtype=object)
Method Summary¶
We provide a number of the common statistical functions:
| Method | Description |
|---|---|
| count() | Number of non-null observations |
| sum() | Sum of values |
| mean() | Mean of values |
| median() | Arithmetic median of values |
| min() | Minimum |
| max() | Maximum |
| std() | Bessel-corrected sample standard deviation |
| var() | Unbiased variance |
| skew() | Sample skewness (3rd moment) |
| kurt() | Sample kurtosis (4th moment) |
| quantile() | Sample quantile (value at %) |
| apply() | Generic apply |
| cov() | Unbiased covariance (binary) |
| corr() | Correlation (binary) |
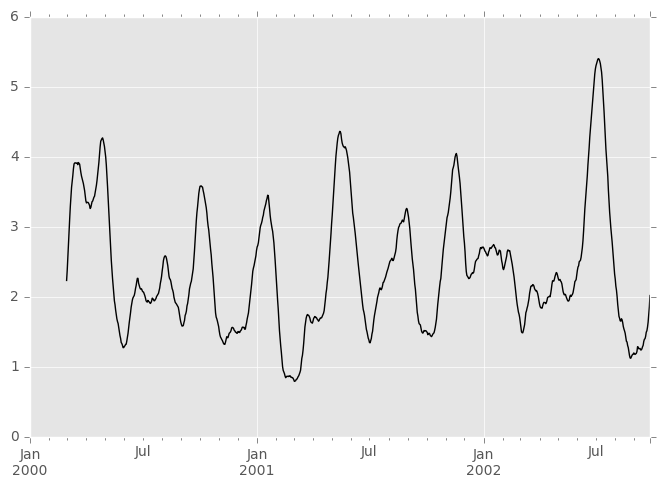
The apply() function takes an extra func argument and performs generic rolling computations. The func argument should be a single function that produces a single value from an ndarray input. Suppose we wanted to compute the mean absolute deviation on a rolling basis:
In [49]: mad = lambda x: np.fabs(x - x.mean()).mean()
In [50]: s.rolling(window=60).apply(mad).plot(style='k') Out[50]: <matplotlib.axes._subplots.AxesSubplot at 0x112f25950>
Rolling Windows¶
Passing win_type to .rolling generates a generic rolling window computation, that is weighted according the win_type. The following methods are available:
| Method | Description |
|---|---|
| sum() | Sum of values |
| mean() | Mean of values |
The weights used in the window are specified by the win_type keyword. The list of recognized types are:
- boxcar
- triang
- blackman
- hamming
- bartlett
- parzen
- bohman
- blackmanharris
- nuttall
- barthann
- kaiser (needs beta)
- gaussian (needs std)
- general_gaussian (needs power, width)
- slepian (needs width).
In [51]: ser = pd.Series(np.random.randn(10), index=pd.date_range('1/1/2000', periods=10))
In [52]: ser.rolling(window=5, win_type='triang').mean() Out[52]: 2000-01-01 NaN 2000-01-02 NaN 2000-01-03 NaN 2000-01-04 NaN 2000-01-05 -1.037870 2000-01-06 -0.767705 2000-01-07 -0.383197 2000-01-08 -0.395513 2000-01-09 -0.558440 2000-01-10 -0.672416 Freq: D, dtype: float64
Note that the boxcar window is equivalent to mean().
In [53]: ser.rolling(window=5, win_type='boxcar').mean() Out[53]: 2000-01-01 NaN 2000-01-02 NaN 2000-01-03 NaN 2000-01-04 NaN 2000-01-05 -0.841164 2000-01-06 -0.779948 2000-01-07 -0.565487 2000-01-08 -0.502815 2000-01-09 -0.553755 2000-01-10 -0.472211 Freq: D, dtype: float64
In [54]: ser.rolling(window=5).mean() Out[54]: 2000-01-01 NaN 2000-01-02 NaN 2000-01-03 NaN 2000-01-04 NaN 2000-01-05 -0.841164 2000-01-06 -0.779948 2000-01-07 -0.565487 2000-01-08 -0.502815 2000-01-09 -0.553755 2000-01-10 -0.472211 Freq: D, dtype: float64
For some windowing functions, additional parameters must be specified:
In [55]: ser.rolling(window=5, win_type='gaussian').mean(std=0.1) Out[55]: 2000-01-01 NaN 2000-01-02 NaN 2000-01-03 NaN 2000-01-04 NaN 2000-01-05 -1.309989 2000-01-06 -1.153000 2000-01-07 0.606382 2000-01-08 -0.681101 2000-01-09 -0.289724 2000-01-10 -0.996632 Freq: D, dtype: float64
Note
For .sum() with a win_type, there is no normalization done to the weights for the window. Passing custom weights of [1, 1, 1] will yield a different result than passing weights of [2, 2, 2], for example. When passing awin_type instead of explicitly specifying the weights, the weights are already normalized so that the largest weight is 1.
In contrast, the nature of the .mean() calculation is such that the weights are normalized with respect to each other. Weights of [1, 1, 1] and [2, 2, 2] yield the same result.
Centering Windows¶
By default the labels are set to the right edge of the window, but acenter keyword is available so the labels can be set at the center.
In [56]: ser.rolling(window=5).mean() Out[56]: 2000-01-01 NaN 2000-01-02 NaN 2000-01-03 NaN 2000-01-04 NaN 2000-01-05 -0.841164 2000-01-06 -0.779948 2000-01-07 -0.565487 2000-01-08 -0.502815 2000-01-09 -0.553755 2000-01-10 -0.472211 Freq: D, dtype: float64
In [57]: ser.rolling(window=5, center=True).mean() Out[57]: 2000-01-01 NaN 2000-01-02 NaN 2000-01-03 -0.841164 2000-01-04 -0.779948 2000-01-05 -0.565487 2000-01-06 -0.502815 2000-01-07 -0.553755 2000-01-08 -0.472211 2000-01-09 NaN 2000-01-10 NaN Freq: D, dtype: float64
Binary Window Functions¶
cov() and corr() can compute moving window statistics about two Series or any combination of DataFrame/Series orDataFrame/DataFrame. Here is the behavior in each case:
- two Series: compute the statistic for the pairing.
- DataFrame/Series: compute the statistics for each column of the DataFrame with the passed Series, thus returning a DataFrame.
- DataFrame/DataFrame: by default compute the statistic for matching column names, returning a DataFrame. If the keyword argument pairwise=True is passed then computes the statistic for each pair of columns, returning aPanel whose items are the dates in question (see the next section).
For example:
In [58]: df2 = df[:20]
In [59]: df2.rolling(window=5).corr(df2['B']) Out[59]: A B C D 2000-01-01 NaN NaN NaN NaN 2000-01-02 NaN NaN NaN NaN 2000-01-03 NaN NaN NaN NaN 2000-01-04 NaN NaN NaN NaN 2000-01-05 -0.262853 1.0 0.334449 0.193380 2000-01-06 -0.083745 1.0 -0.521587 -0.556126 2000-01-07 -0.292940 1.0 -0.658532 -0.458128 ... ... ... ... ... 2000-01-14 0.519499 1.0 -0.687277 0.192822 2000-01-15 0.048982 1.0 0.167669 -0.061463 2000-01-16 0.217190 1.0 0.167564 -0.326034 2000-01-17 0.641180 1.0 -0.164780 -0.111487 2000-01-18 0.130422 1.0 0.322833 0.632383 2000-01-19 0.317278 1.0 0.384528 0.813656 2000-01-20 0.293598 1.0 0.159538 0.742381
[20 rows x 4 columns]
Computing rolling pairwise covariances and correlations¶
In financial data analysis and other fields it’s common to compute covariance and correlation matrices for a collection of time series. Often one is also interested in moving-window covariance and correlation matrices. This can be done by passing the pairwise keyword argument, which in the case ofDataFrame inputs will yield a Panel whose items are the dates in question. In the case of a single DataFrame argument the pairwise argument can even be omitted:
Note
Missing values are ignored and each entry is computed using the pairwise complete observations. Please see the covariance section for caveats associated with this method of calculating covariance and correlation matrices.
In [60]: covs = df[['B','C','D']].rolling(window=50).cov(df[['A','B','C']], pairwise=True)
In [61]: covs[df.index[-50]] Out[61]: A B C B 2.667506 1.671711 1.938634 C 8.513843 1.938634 10.556436 D -7.714737 -1.434529 -7.082653
In [62]: correls = df.rolling(window=50).corr()
In [63]: correls[df.index[-50]] Out[63]: A B C D A 1.000000 0.604221 0.767429 -0.776170 B 0.604221 1.000000 0.461484 -0.381148 C 0.767429 0.461484 1.000000 -0.748863 D -0.776170 -0.381148 -0.748863 1.000000
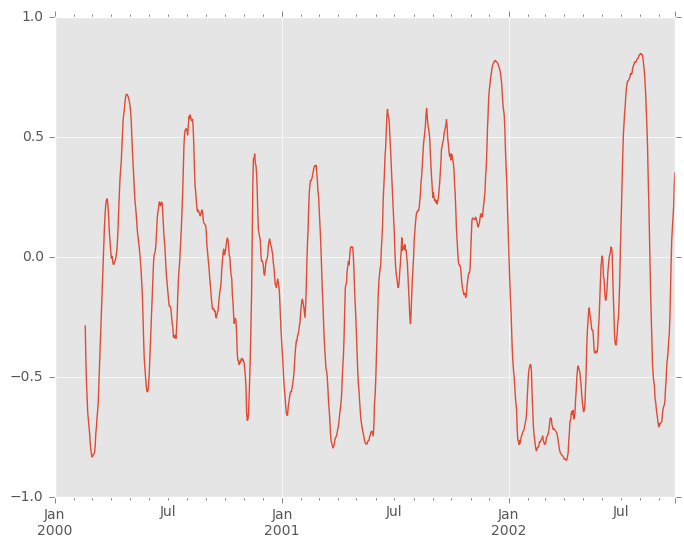
You can efficiently retrieve the time series of correlations between two columns using .loc indexing:
In [64]: correls.loc[:, 'A', 'C'].plot() Out[64]: <matplotlib.axes._subplots.AxesSubplot at 0x11ea521d0>
Aggregation¶
Once the Rolling, Expanding or EWM objects have been created, several methods are available to perform multiple computations on the data. This is very similar to a .groupby(...).agg seen here.
In [65]: dfa = pd.DataFrame(np.random.randn(1000, 3), ....: index=pd.date_range('1/1/2000', periods=1000), ....: columns=['A', 'B', 'C']) ....:
In [66]: r = dfa.rolling(window=60,min_periods=1)
In [67]: r Out[67]: Rolling [window=60,min_periods=1,center=False,axis=0]
We can aggregate by passing a function to the entire DataFrame, or select a Series (or multiple Series) via standard getitem.
In [68]: r.aggregate(np.sum) Out[68]: A B C 2000-01-01 0.314226 -0.001675 0.071823 2000-01-02 1.206791 0.678918 -0.267817 2000-01-03 1.421701 0.600508 -0.445482 2000-01-04 1.912539 -0.759594 1.146974 2000-01-05 2.919639 -0.061759 -0.743617 2000-01-06 2.665637 1.298392 -0.803529 2000-01-07 2.513985 1.923089 -1.928308 ... ... ... ... 2002-09-20 1.447669 -12.360302 2.734381 2002-09-21 1.871783 -13.896542 3.086102 2002-09-22 2.540658 -12.594402 3.162542 2002-09-23 2.974674 -12.727703 3.861005 2002-09-24 1.391366 -13.584590 3.790683 2002-09-25 2.027313 -15.083214 3.377896 2002-09-26 1.290363 -13.569459 3.809884
[1000 rows x 3 columns]
In [69]: r['A'].aggregate(np.sum)
Out[69]:
2000-01-01 0.314226
2000-01-02 1.206791
2000-01-03 1.421701
2000-01-04 1.912539
2000-01-05 2.919639
2000-01-06 2.665637
2000-01-07 2.513985
...
2002-09-20 1.447669
2002-09-21 1.871783
2002-09-22 2.540658
2002-09-23 2.974674
2002-09-24 1.391366
2002-09-25 2.027313
2002-09-26 1.290363
Freq: D, Name: A, dtype: float64
In [70]: r[['A','B']].aggregate(np.sum) Out[70]: A B 2000-01-01 0.314226 -0.001675 2000-01-02 1.206791 0.678918 2000-01-03 1.421701 0.600508 2000-01-04 1.912539 -0.759594 2000-01-05 2.919639 -0.061759 2000-01-06 2.665637 1.298392 2000-01-07 2.513985 1.923089 ... ... ... 2002-09-20 1.447669 -12.360302 2002-09-21 1.871783 -13.896542 2002-09-22 2.540658 -12.594402 2002-09-23 2.974674 -12.727703 2002-09-24 1.391366 -13.584590 2002-09-25 2.027313 -15.083214 2002-09-26 1.290363 -13.569459
[1000 rows x 2 columns]
As you can see, the result of the aggregation will have the selected columns, or all columns if none are selected.
Applying multiple functions at once¶
With windowed Series you can also pass a list or dict of functions to do aggregation with, outputting a DataFrame:
In [71]: r['A'].agg([np.sum, np.mean, np.std]) Out[71]: sum mean std 2000-01-01 0.314226 0.314226 NaN 2000-01-02 1.206791 0.603396 0.408948 2000-01-03 1.421701 0.473900 0.365959 2000-01-04 1.912539 0.478135 0.298925 2000-01-05 2.919639 0.583928 0.350682 2000-01-06 2.665637 0.444273 0.464115 2000-01-07 2.513985 0.359141 0.479828 ... ... ... ... 2002-09-20 1.447669 0.024128 1.034827 2002-09-21 1.871783 0.031196 1.031417 2002-09-22 2.540658 0.042344 1.026341 2002-09-23 2.974674 0.049578 1.030021 2002-09-24 1.391366 0.023189 1.024793 2002-09-25 2.027313 0.033789 1.022099 2002-09-26 1.290363 0.021506 1.024751
[1000 rows x 3 columns]
If a dict is passed, the keys will be used to name the columns. Otherwise the function’s name (stored in the function object) will be used.
In [72]: r['A'].agg({'result1' : np.sum, ....: 'result2' : np.mean}) ....: Out[72]: result2 result1 2000-01-01 0.314226 0.314226 2000-01-02 0.603396 1.206791 2000-01-03 0.473900 1.421701 2000-01-04 0.478135 1.912539 2000-01-05 0.583928 2.919639 2000-01-06 0.444273 2.665637 2000-01-07 0.359141 2.513985 ... ... ... 2002-09-20 0.024128 1.447669 2002-09-21 0.031196 1.871783 2002-09-22 0.042344 2.540658 2002-09-23 0.049578 2.974674 2002-09-24 0.023189 1.391366 2002-09-25 0.033789 2.027313 2002-09-26 0.021506 1.290363
[1000 rows x 2 columns]
On a widowed DataFrame, you can pass a list of functions to apply to each column, which produces an aggregated result with a hierarchical index:
In [73]: r.agg([np.sum, np.mean])
Out[73]:
A B C
sum mean sum mean sum mean
2000-01-01 0.314226 0.314226 -0.001675 -0.001675 0.071823 0.071823
2000-01-02 1.206791 0.603396 0.678918 0.339459 -0.267817 -0.133908
2000-01-03 1.421701 0.473900 0.600508 0.200169 -0.445482 -0.148494
2000-01-04 1.912539 0.478135 -0.759594 -0.189899 1.146974 0.286744
2000-01-05 2.919639 0.583928 -0.061759 -0.012352 -0.743617 -0.148723
2000-01-06 2.665637 0.444273 1.298392 0.216399 -0.803529 -0.133921
2000-01-07 2.513985 0.359141 1.923089 0.274727 -1.928308 -0.275473
... ... ... ... ... ... ...
2002-09-20 1.447669 0.024128 -12.360302 -0.206005 2.734381 0.045573
2002-09-21 1.871783 0.031196 -13.896542 -0.231609 3.086102 0.051435
2002-09-22 2.540658 0.042344 -12.594402 -0.209907 3.162542 0.052709
2002-09-23 2.974674 0.049578 -12.727703 -0.212128 3.861005 0.064350
2002-09-24 1.391366 0.023189 -13.584590 -0.226410 3.790683 0.063178
2002-09-25 2.027313 0.033789 -15.083214 -0.251387 3.377896 0.056298
2002-09-26 1.290363 0.021506 -13.569459 -0.226158 3.809884 0.063498
[1000 rows x 6 columns]
Passing a dict of functions has different behavior by default, see the next section.
Applying different functions to DataFrame columns¶
By passing a dict to aggregate you can apply a different aggregation to the columns of a DataFrame:
In [74]: r.agg({'A' : np.sum, ....: 'B' : lambda x: np.std(x, ddof=1)}) ....: Out[74]: A B 2000-01-01 0.314226 NaN 2000-01-02 1.206791 0.482437 2000-01-03 1.421701 0.417825 2000-01-04 1.912539 0.851468 2000-01-05 2.919639 0.837474 2000-01-06 2.665637 0.935441 2000-01-07 2.513985 0.867770 ... ... ... 2002-09-20 1.447669 1.084259 2002-09-21 1.871783 1.088368 2002-09-22 2.540658 1.084707 2002-09-23 2.974674 1.084936 2002-09-24 1.391366 1.079268 2002-09-25 2.027313 1.091334 2002-09-26 1.290363 1.060255
[1000 rows x 2 columns]
The function names can also be strings. In order for a string to be valid it must be implemented on the windowed object
In [75]: r.agg({'A' : 'sum', 'B' : 'std'}) Out[75]: A B 2000-01-01 0.314226 NaN 2000-01-02 1.206791 0.482437 2000-01-03 1.421701 0.417825 2000-01-04 1.912539 0.851468 2000-01-05 2.919639 0.837474 2000-01-06 2.665637 0.935441 2000-01-07 2.513985 0.867770 ... ... ... 2002-09-20 1.447669 1.084259 2002-09-21 1.871783 1.088368 2002-09-22 2.540658 1.084707 2002-09-23 2.974674 1.084936 2002-09-24 1.391366 1.079268 2002-09-25 2.027313 1.091334 2002-09-26 1.290363 1.060255
[1000 rows x 2 columns]
Furthermore you can pass a nested dict to indicate different aggregations on different columns.
In [76]: r.agg({'A' : ['sum','std'], 'B' : ['mean','std'] })
Out[76]:
A B
sum std mean std
2000-01-01 0.314226 NaN -0.001675 NaN
2000-01-02 1.206791 0.408948 0.339459 0.482437
2000-01-03 1.421701 0.365959 0.200169 0.417825
2000-01-04 1.912539 0.298925 -0.189899 0.851468
2000-01-05 2.919639 0.350682 -0.012352 0.837474
2000-01-06 2.665637 0.464115 0.216399 0.935441
2000-01-07 2.513985 0.479828 0.274727 0.867770
... ... ... ... ...
2002-09-20 1.447669 1.034827 -0.206005 1.084259
2002-09-21 1.871783 1.031417 -0.231609 1.088368
2002-09-22 2.540658 1.026341 -0.209907 1.084707
2002-09-23 2.974674 1.030021 -0.212128 1.084936
2002-09-24 1.391366 1.024793 -0.226410 1.079268
2002-09-25 2.027313 1.022099 -0.251387 1.091334
2002-09-26 1.290363 1.024751 -0.226158 1.060255
[1000 rows x 4 columns]
Expanding Windows¶
A common alternative to rolling statistics is to use an expanding window, which yields the value of the statistic with all the data available up to that point in time.
These follow a similar interface to .rolling, with the .expanding method returning an Expanding object.
As these calculations are a special case of rolling statistics, they are implemented in pandas such that the following two calls are equivalent:
In [77]: df.rolling(window=len(df), min_periods=1).mean()[:5] Out[77]: A B C D 2000-01-01 -1.388345 3.317290 0.344542 -0.036968 2000-01-02 -1.123132 3.622300 1.675867 0.595300 2000-01-03 -0.628502 3.626503 2.455240 1.060158 2000-01-04 -0.768740 3.888917 2.451354 1.281874 2000-01-05 -0.824034 4.108035 2.556112 1.140723
In [78]: df.expanding(min_periods=1).mean()[:5] Out[78]: A B C D 2000-01-01 -1.388345 3.317290 0.344542 -0.036968 2000-01-02 -1.123132 3.622300 1.675867 0.595300 2000-01-03 -0.628502 3.626503 2.455240 1.060158 2000-01-04 -0.768740 3.888917 2.451354 1.281874 2000-01-05 -0.824034 4.108035 2.556112 1.140723
These have a similar set of methods to .rolling methods.
Method Summary¶
| Function | Description |
|---|---|
| count() | Number of non-null observations |
| sum() | Sum of values |
| mean() | Mean of values |
| median() | Arithmetic median of values |
| min() | Minimum |
| max() | Maximum |
| std() | Unbiased standard deviation |
| var() | Unbiased variance |
| skew() | Unbiased skewness (3rd moment) |
| kurt() | Unbiased kurtosis (4th moment) |
| quantile() | Sample quantile (value at %) |
| apply() | Generic apply |
| cov() | Unbiased covariance (binary) |
| corr() | Correlation (binary) |
Aside from not having a window parameter, these functions have the same interfaces as their .rolling counterparts. Like above, the parameters they all accept are:
- min_periods: threshold of non-null data points to require. Defaults to minimum needed to compute statistic. No NaNs will be output oncemin_periods non-null data points have been seen.
- center: boolean, whether to set the labels at the center (default is False)
Note
The output of the .rolling and .expanding methods do not return aNaN if there are at least min_periods non-null values in the current window. This differs from cumsum, cumprod, cummax, andcummin, which return NaN in the output wherever a NaN is encountered in the input.
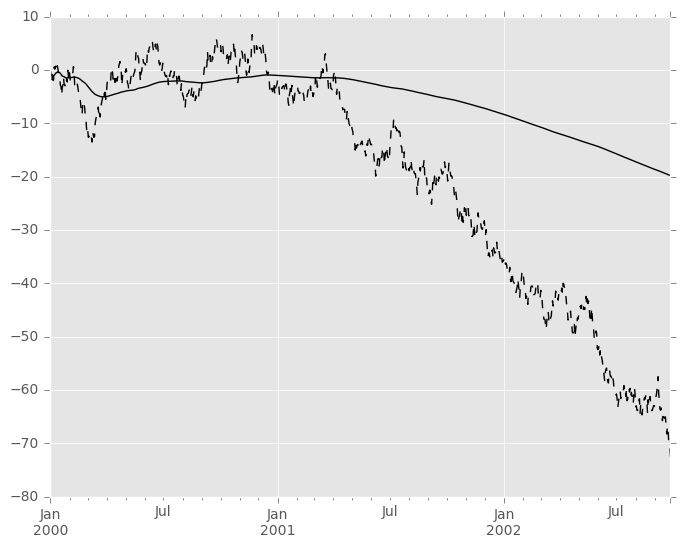
An expanding window statistic will be more stable (and less responsive) than its rolling window counterpart as the increasing window size decreases the relative impact of an individual data point. As an example, here is themean() output for the previous time series dataset:
In [79]: s.plot(style='k--') Out[79]: <matplotlib.axes._subplots.AxesSubplot at 0x118390f10>
In [80]: s.expanding().mean().plot(style='k') Out[80]: <matplotlib.axes._subplots.AxesSubplot at 0x118390f10>
Exponentially Weighted Windows¶
A related set of functions are exponentially weighted versions of several of the above statistics. A similar interface to .rolling and .expanding is accessed thru the .ewm method to receive an EWM object. A number of expanding EW (exponentially weighted) methods are provided:
| Function | Description |
|---|---|
| mean() | EW moving average |
| var() | EW moving variance |
| std() | EW moving standard deviation |
| corr() | EW moving correlation |
| cov() | EW moving covariance |
In general, a weighted moving average is calculated as
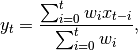
where 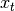 is the input and
is the input and 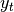 is the result.
is the result.
The EW functions support two variants of exponential weights. The default, adjust=True, uses the weights 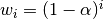 which gives
which gives
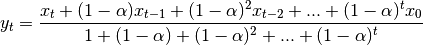
When adjust=False is specified, moving averages are calculated as
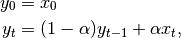
which is equivalent to using weights
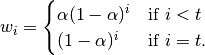
Note
These equations are sometimes written in terms of 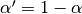 , e.g.
, e.g.
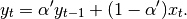
The difference between the above two variants arises because we are dealing with series which have finite history. Consider a series of infinite history:
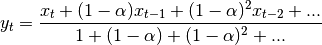
Noting that the denominator is a geometric series with initial term equal to 1 and a ratio of 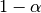 we have
we have
![y_t &= \frac{x_t + (1 - \alpha)x_{t-1} + (1 - \alpha)^2 x_{t-2} + ...}
{\frac{1}{1 - (1 - \alpha)}}\\
&= [x_t + (1 - \alpha)x_{t-1} + (1 - \alpha)^2 x_{t-2} + ...] \alpha \\
&= \alpha x_t + [(1-\alpha)x_{t-1} + (1 - \alpha)^2 x_{t-2} + ...]\alpha \\
&= \alpha x_t + (1 - \alpha)[x_{t-1} + (1 - \alpha) x_{t-2} + ...]\alpha\\
&= \alpha x_t + (1 - \alpha) y_{t-1}](https://pandas.pydata.org/pandas-docs/version/0.18.1/_images/math/2be73bffdf3cefb77c1cafd822ff2dafccd03a99.png)
which shows the equivalence of the above two variants for infinite series. When adjust=True we have 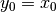 and from the last representation above we have
and from the last representation above we have 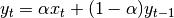 , therefore there is an assumption that
, therefore there is an assumption that 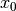 is not an ordinary value but rather an exponentially weighted moment of the infinite series up to that point.
is not an ordinary value but rather an exponentially weighted moment of the infinite series up to that point.
One must have 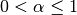 , and while since version 0.18.0 it has been possible to pass
, and while since version 0.18.0 it has been possible to pass 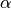 directly, it’s often easier to think about either the span, center of mass (com) or half-lifeof an EW moment:
directly, it’s often easier to think about either the span, center of mass (com) or half-lifeof an EW moment:
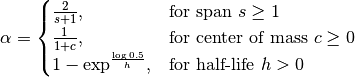
One must specify precisely one of span, center of mass, half-lifeand alpha to the EW functions:
- Span corresponds to what is commonly called an “N-day EW moving average”.
- Center of mass has a more physical interpretation and can be thought of in terms of span:
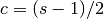 .
. - Half-life is the period of time for the exponential weight to reduce to one half.
- Alpha specifies the smoothing factor directly.
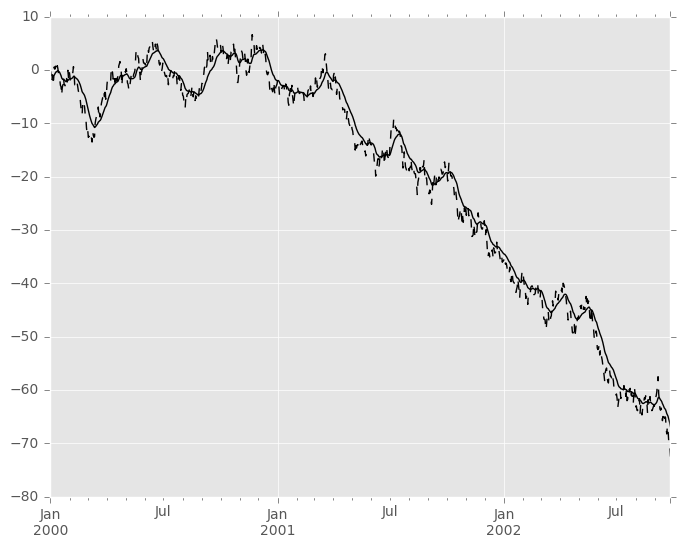
Here is an example for a univariate time series:
In [81]: s.plot(style='k--') Out[81]: <matplotlib.axes._subplots.AxesSubplot at 0x1132e2dd0>
In [82]: s.ewm(span=20).mean().plot(style='k') Out[82]: <matplotlib.axes._subplots.AxesSubplot at 0x1132e2dd0>
EWM has a min_periods argument, which has the same meaning it does for all the .expanding and .rolling methods: no output values will be set until at least min_periods non-null values are encountered in the (expanding) window. (This is a change from versions prior to 0.15.0, in which the min_periodsargument affected only the min_periods consecutive entries starting at the first non-null value.)
EWM also has an ignore_na argument, which deterines how intermediate null values affect the calculation of the weights. When ignore_na=False (the default), weights are calculated based on absolute positions, so that intermediate null values affect the result. When ignore_na=True (which reproduces the behavior in versions prior to 0.15.0), weights are calculated by ignoring intermediate null values. For example, assuming adjust=True, if ignore_na=False, the weighted average of 3, NaN, 5 would be calculated as
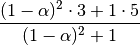
Whereas if ignore_na=True, the weighted average would be calculated as
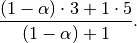
The var(), std(), and cov() functions have a bias argument, specifying whether the result should contain biased or unbiased statistics. For example, if bias=True, ewmvar(x) is calculated asewmvar(x) = ewma(x**2) - ewma(x)**2; whereas if bias=False (the default), the biased variance statistics are scaled by debiasing factors
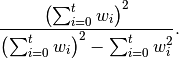
(For 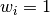 , this reduces to the usual
, this reduces to the usual 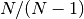 factor, with
factor, with 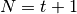 .) See Weighted Sample Variancefor further details.
.) See Weighted Sample Variancefor further details.