SVM-Anova: SVM with univariate feature selection (original) (raw)
Note
Go to the endto download the full example code. or to run this example in your browser via JupyterLite or Binder
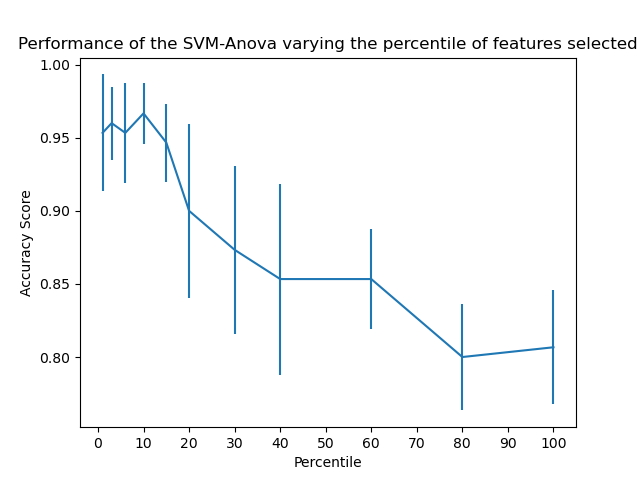
This example shows how to perform univariate feature selection before running a SVC (support vector classifier) to improve the classification scores. We use the iris dataset (4 features) and add 36 non-informative features. We can find that our model achieves best performance when we select around 10% of features.
Authors: The scikit-learn developers
SPDX-License-Identifier: BSD-3-Clause
Load some data to play with#
Create the pipeline#
Plot the cross-validation score as a function of percentile of features#
import matplotlib.pyplot as plt
from sklearn.model_selection import cross_val_score
score_means = list() score_stds = list() percentiles = (1, 3, 6, 10, 15, 20, 30, 40, 60, 80, 100)
for percentile in percentiles: clf.set_params(anova__percentile=percentile) this_scores = cross_val_score(clf, X, y) score_means.append(this_scores.mean()) score_stds.append(this_scores.std())
plt.errorbar(percentiles, score_means, np.array(score_stds)) plt.title("Performance of the SVM-Anova varying the percentile of features selected") plt.xticks(np.linspace(0, 100, 11, endpoint=True)) plt.xlabel("Percentile") plt.ylabel("Accuracy Score") plt.axis("tight") plt.show()
Total running time of the script: (0 minutes 0.313 seconds)
Related examples