miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN amplified neuroblastoma (original) (raw)
. Author manuscript; available in PMC: 2011 Apr 1.
Published in final edited form as: Nat Med. 2010 Sep 26;16(10):1134–1140. doi: 10.1038/nm.2227
Abstract
Inactivation of the p53 tumor-suppressor pathway occurs in many human cancers, however some cancers such as neuroblastoma and normal stem cells maintain wild-type p53. Here we describe a microRNA, miR-380-5p, that represses p53 expression via a conserved sequence in the p53 3′UTR. miR-380-5p is highly expressed in embryonic stem cells and neuroblastomas and high expression correlates with poor outcome in neuroblastomas with MYCN amplification. miR-380 overexpression cooperates with activated RAS to transform primary cells, form tumors in mice, and block oncogene induced senescence. In contrast, inhibition of endogenous miR-380-5p in embryonic stem or neuroblastoma cells results in induction of miR-380-5p targets including p53 and extensive apoptotic cell death. In vivo delivery of a miR-380-5p antagonist decreases tumor size in an orthotopic mouse model of neuroblastoma. We demonstrate a new mechanism of p53 regulation in cancer and stem cells and uncover a potential therapeutic target for neuroblastoma.
INTRODUCTION
The p53 tumor suppressor (TP53) is activated following a wide variety of genotoxic stresses, resulting in cell cycle arrest, apoptosis or senescence1,2. Inactivation of p53 is a common event in cancer3,4 and attempts to reactivate p53 function are being vigorously pursued as a cancer therapeutic5,6-8. However, in some cancers including more than 99% of primary neuroblastomas, TP53 remains wildtype9. After treatment with chemotherapy, alteration of p53 pathway components are frequently found to occur in neuroblastomas9. How wild-type p53 can be tolerated by neuroblastomas before treatment is unclear and suggests that other mechanisms exist to attenuate p53 function.
Changes in the level of p53 can modulate the p53-mediated transcription10,11 and the resulting biological output, such as apoptosis12-14. Even modest decreases in p53 abundance can lead to greatly reduced activation of a p53-reporter gene in vivo15. Interestingly, 50% of the tumors that form in Trp53+/− mice retain the wild-type p53 allele16, suggesting that a modest reduction in wild-type p53 protein promotes tumorigenesis.
The p53 mRNA has a complex 3′ untranslated region (3′ UTR) and highly conserved sequences within the 3′ UTR control p53 translation through poorly-understood mechanisms17. MicroRNAs are small non-coding RNAs that control gene expression by regulating mRNA translation and/or stability, typically by binding to regions of homology in the 3′ UTR of target mRNAs18. Several microRNAs with validated roles in the promotion or suppression of neoplasia have been identified19; 20.
Here we show that p53 is regulated in human cancer by miR-380-5p. We find that this microRNA is highly expressed in the majority of primary neuroblastomas and functions as a proto-oncogene in a mouse mammary transplant model. miR-380-5p is predicted to bind to a highly conserved region in the p53 3′UTR. Inhibition of miR-380-5p results in upregulation of p53 in embryonic stem (ES) and neuroblastoma cells and the induction of apoptosis, as well as diminished tumor growth in vivo. Our results identify a new therapeutic approach to reactivate p53 in neuroblastoma.
RESULTS
The p53 3′UTR contains two conserved potential binding sites for miR-380-5p
We identified a 104 bp region of high homology in the p53 3′UTR shared across human, mouse, rat and hamster species but not conserved in non-mammalian species (Fig. 1a). This corresponds to nucleotides (582-685) of the human TP53 3′UTR. Human and mouse p53 3′UTRs share 78% identity within this region. This is similar to the 84% identity found when comparing the coding portion of human TP53 exon 11 with the corresponding sequence from mouse, suggesting that this region of the 3′UTR may have functional importance. Using the miRanda algorithm21, we identified two predicted adjacent target sites for hsa-miR-380-5p within the conserved 3′UTR region (Fig. 1a), at a spacing previously reported to enhance cooperative repression22. Local RNA structure is proposed to regulate the efficiency of miRNA binding to target UTRs23,24. The sequence of both putative binding sites featured a preponderance of adjacent destabilizing structures (loops, single stranded regions and free ends) and only short stem structures, features preferred for miRNA:3′UTR interactions (Supplementary Table 1).
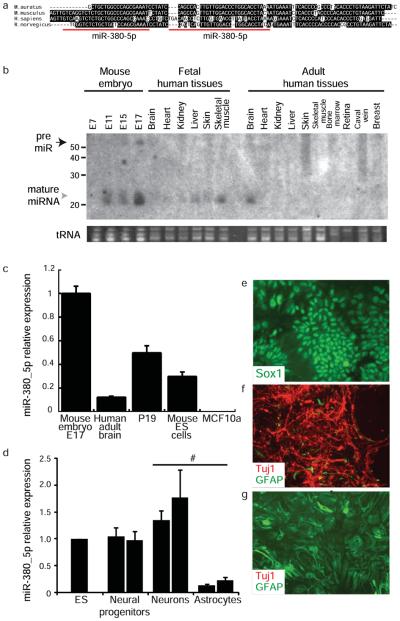
Fig. 1.
The p53 3′UTR contains binding sites for miR-380-5p, a developmentally restricted miRNA. (a) Alignment of human, mouse, rat and hamster p53 3′UTRs identifies a highly-conserved 104 bp region. The predicted miR-380-5p binding sites are indicated in red. (b) miR-380 northern blot of total RNA from mouse embryonic, human fetal and adult tissues. (c). miR-380-5p is highly expressed in normal brain, embryonic carcinoma (P19) and mouse ES cells by qRT-PCR. (d) qRT-PCR analysis showing miR-380-5p expression is maintained in ES cells differentiated to the neuronal lineage. (e). Neural progenitor cells express Sox1 (green). (f–g), differentiated cultures of neurons (f) and astrocytes (g) were costained for Tuj1 (red), an early neuronal marker, and GFAP (green), a marker of astrocytes. (c-d), error bars depict s.d., (d) independent biological replicates indicated by separate bars. #p<0.0002
Expression of miR-380 is developmentally restricted
miR-380-5p is encoded within a large miRNA cluster found in an imprinted region of human 14q3225. We detected abundant miR-380 expression in mouse embryonic and human fetal tissues, and the adult human brain, tissues in which p53 has important roles26 (Fig. 1b), but not in other adult tissues. miR-380-5p was also highly expressed in mouse ES cells and P19 embryonic carcinoma cells as determined by quantitative RT-PCR (Fig. 1c). Human breast MCF10A cells do not express detectable miR-380-5p and were used as a negative control line (Fig. 1c). miR-380-5p expression was maintained in mouse ES cells differentiated in culture to Sox1+ neural progenitors and Tuj1+ neurons but not in cultures containing predominantly Gfap+ astrocytes (Fig. 1d-g). Thus miR-380 is not simply a marker of undifferentiated cells but is also expressed through neuronal specification.
Endogenous miR-380-5p functions to suppress p53 and apoptosis in stem cells
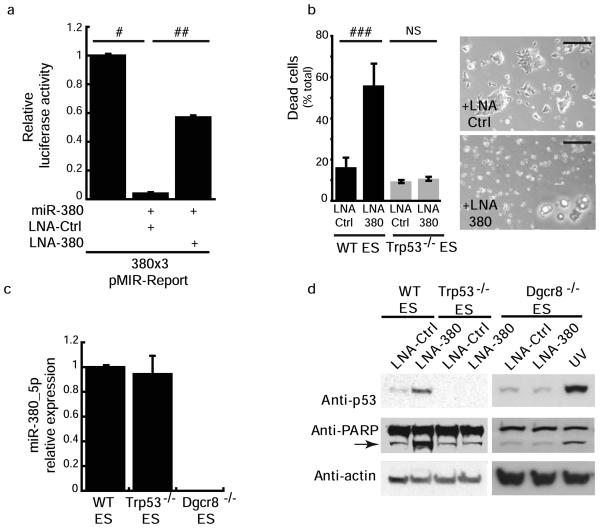
To examine the function of endogenous miR-380-5p, we utilized an LNA-modified antisense oligomer to inhibit miR-380-5p (LNA-380). Transfection of LNA-380, but not a control LNA, relieved repression of a luciferase reporter with three perfect miR-380-5p binding sites in the 3′UTR (Fig. 2a). Activity of a control reporter following transfection of miR-380-5p alone or with control LNA or LNA-380 was not changed (data not shown). Transfection of mouse ES cells with LNA-380 resulted in changes in ES cell morphology, a diminished colony size, increased number of non-adherent cells after 8 h and substantial cell death 24 h post-treatment (Fig. 2b). While _Trp53_−/− ES cells express similar levels of miR-380-5p to their wild-type counterparts, LNA-380 treatment of _Trp53_−/− ES cells did not induce cell death (Figs. 2b,c), demonstrating a requirement for p53 in cell death induced by LNA-380. Knockdown of miR-380-5p was accompanied by the up-regulation of p53 protein and the apoptotic marker, cleaved poly(ADP)-ribose polymerase (PARP) (Fig. 2d) in wild type ES cells but not _Trp53_−/− cells. This effect was observed over a range of LNA-380 concentrations and the phenotype was not observed in wild type ES cells treated with a variety of scrambled and other control LNAs (Supplementary Fig. 1 a-c). As an additional control, we tested whether mature microRNAs are required for induction of p53 by LNA-380. ES cells that are deficient in mature microRNA species (including miR-380-5p, Fig 2c), due to homologous deletion of DiGeorge syndrome critical region 8 (Dgcr8), remain responsive to genotoxic shock, and p53 is induced after UV irradiation (Fig. 2d). However treatment of _Dgcr8_−/− ES cells with LNA-380 did not induce p53 (Fig. 2d). Together these data show that inhibition of endogenous miR-380-5p in ES cells results in up-regulation of its targets including p53 and apoptotic cell death. At just 4 h after transfection of ES cells with LNA-380, cell morphology is indistinguishable from control LNA treated cells, however p53 protein is already increased (Supplementary Fig. 1d). This increase in p53 protein is not due to stabilization of the protein as the half-life of p53 is unchanged in LNA-380 treated cells compared to LNA-control treated cells (Supplementary Fig. 1e). In contrast, 4 h post UV irradiation, p53 protein was robustly stabilized (Supplementary Fig. 1e). Levels of the p53 regulators p19ARF, Mdm2 and Chek2 are unchanged by LNA-380 treatment (Supplementary Fig. 1f).
Fig. 2.
miR-380-5p is required for ES cell survival. (a) LNA-380 relieves miR-380-5p repression of the miR-380-5p Reporter. (b) ES Cell death is induced by knockdown of miR-380-5p in a p53 dependent manner. Right panel contains cell images 24 h post transfection with LNA-Ctrl or LNA-380. (c) qRT-PCR analysis of relative miR-380-5p expression in wild type (WT), _Trp53_−/− and _Dgcr8_−/− ES cells. (d) Western blots demonstrating p53 induction and PARP cleavage following knockdown of miR-380-5p by LNA-380 compared to LNA-ctrl transfected wild type, _Trp53_−/− and _Dgcr8_−/− ES cells, ultraviolet irradiated (UV). a, b, d results are from at least 3 independent experiments, c performed in triplicate, a–c, error bars depict s.d. #p < 0.00002, ##p < 0.00007, ###p < 0.0004, NS = not significant. Scale bars equal 200μm.
miR-380-5p levels decrease rapidly following UV-induced cellular stress
Treatment of ES cells with UV radiation leads to the rapid accumulation of p53. Interestingly, endogenous expression of miR-380-5p but not another miRNA from the same genomic cluster (miR-323) or an unrelated miRNA (miR-16) decreases in a manner inversely correlated with p53 protein expression (Supplementary Fig. 1g). While little is known about the transcriptional control and regulation of mature miR-380-5p stability, this suggests that ES cells have an inbuilt mechanism to minimize miR-380-5p levels in situations of cellular stress.
Ectopic expression of miR-380 is sufficient to suppress p53
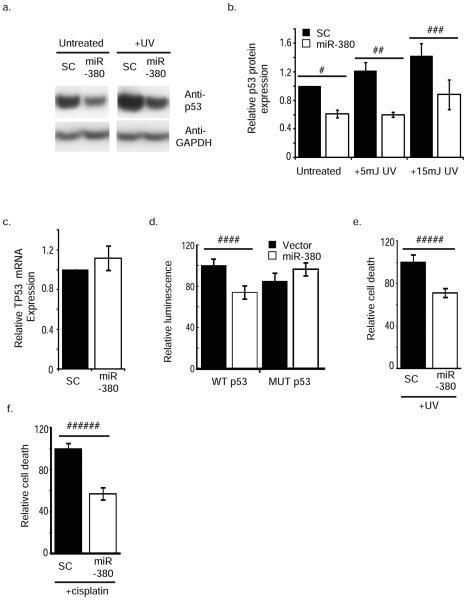
We transfected human MCF10A cells, which express wildtype p53 but not detectable miR-380-5p, with miR-380-5p or a non-targeting control miRNA (Supplementary Fig. 2a). Expression of miR-380-5p resulted in a significant ~ 40% decrease in basal p53 protein levels (Fig. 3a,b). UV irradiation led to a dose-dependent increase in p53 protein expression that was suppressed by the expression of miR-380-5p (Fig. 3a,b). There was no significant difference in p53 mRNA levels following miR-380-5p overexpression (Fig. 3c), suggesting a predominant role in the regulation of p53 translation rather than mRNA stability. For comparison, we transfected cells with miR-125b, which has recently been suggested to target p5327 but did not detect a significant effect of miR-125b on either p53 mRNA or protein levels (Supplementary Fig.2b,c). We obtained similar results using MCF7 and MCF10A cell lines that stably express miR-380 or a scrambled miRNA control (Supplementary Fig. 2 d-g). Together with our data from ES cells (Fig. 2, Supplementary Fig. 1), this suggests that miR-380-5p acts to directly regulate p53 translation rather than the stability of the mRNA or protein.
Fig. 3.
miR-380-5p targets p53 and decreases cell death following genotoxic stress. (a) Western blot demonstrating p53 expression in MCF10a cells transfected with either a scrambled (SC) miRNA or miR-380-5p. (b) p53 protein levels are significantly decreased by miR-380-5p transfection compared to SC, normalized to GAPDH. (c) qRT-PCR analysis of TP53 mRNA in MCF10a cells following expression of miR-380, normalized to GAPDH levels. (d) Decreased luciferase activity of p53 3′UTR reporters following expression of miR-380. (e–f) MCF10a cell populations stably expressing miR-380 or SC were treated with (e) UV or (f) cisplatin, and relative cell death was determined 24 h later. Error bars depict s.e.m.(b–c) or s.d. (d–f). a–b, d–f results are from at least three independent repeats, c, performed in triplicate. # p < 0.0009, ## p < 0.03, ### p < 0.002, #### p < 0.05, ##### p < 0.03, ###### p < 0.001.
We next asked if miR-380 expression directly regulates p53 expression via the conserved 104 bp element within its 3′UTR predicted to encode the two miR-380-5p binding sites (Fig. 1a). We generated luciferase reporter constructs that contain a single 104 bp wild-type p53 conserved element downstream of the luciferase open reading frame (WT-p53), or a reporter construct in which the nucleotides complimentary to the miR-380-5p seed sequences were deleted (MUT-p53) (Fig. 3d). These reporter constructs lack the remainder of the TP53 3′UTR that contains other previously identified translational regulatory elements28,29. When the WT-p53 reporter was co-transfected along with miR-380 into human embryonic kidney cells, luciferase activity was decreased ~ 30% compared to cells transfected with a control vector (Fig. 3d). Suppression of luciferase activity was not observed when the MUT-p53 reporter was co-transfected with miR-380. Thus miR-380-5p can directly attenuate translation via elements found in the p53 3′ UTR.
Expression of miR-380 attenuates cell death induced by genotoxic stress
We next asked if miR-380 over-expression can regulate apoptosis. Twenty-four hours following treatment with two different DNA damaging agents, UV light or cisplatin, significantly more cell death was observed in control cells than in those expressing miR-380 (Fig. 3e,f). No appreciable cell death was noted in the absence of UV or cisplatin treatment (data not shown).
miR-380 acts as an oncogene in vivo
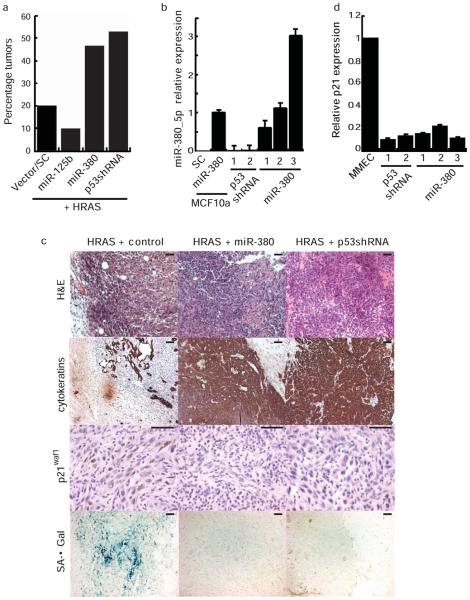
A stringent test of oncogene function in cancer is the ability to form tumors de novo in mouse models. Loss of p53 function cooperates with activated HRAS expression to induce a variety of tumors, including mammary cancers30; 31. We tested whether expression of miR-380 can similarly cooperate with activated HRAS to transform primary mouse mammary epithelial cells (MMECs). The unique biology of the mammary gland enables us to use a transplantation technique that rapidly generates transgenic mammary glands in vivo32 and can be used as an in vivo model of cellular transformation in a system with an intact p53 pathway. MMECs from naïve FVB/N mice were harvested and cultured for 72 h during which time cells were infected with retrovirus encoding activated HRAS and retrovirus encoding various small RNAs. Cells were subsequently transplanted into the cleared mammary fat pad of syngeneic mice (Supplementary Fig. 3a). Co-expression of activated HRAS with either miR-380 or a small hairpin RNA (shRNA p53) that specifically targets mouse Trp5333 resulted in down-regulation of a key transcriptional target of p53, p21waf1 this was in contrast to cells expressing activated HRAS and a scrambled control miRNA (Supplementary Fig. 3b). Mice receiving MMECs expressing activated HRAS plus a control retrovirus infrequently developed small tumors (Fig. 4a). In contrast, co-expression of activated HRAS with either miR-380 or shRNA p53 resulted in a substantially greater frequency of tumor formation (Fig. 4a). No significant differences in tumor latency or growth rates were observed between shRNA p53 and miR-380 groups (Supplementary Fig. 3c). Expression of miR-125b with activated HRAS resulted in even fewer tumors than the controls (Fig. 4a), consistent with a role for miR-125b in attenuating the proliferation of breast epithelium34. Elevated levels of miR-380-5p expression were observed in all tumors tested (Fig. 4b) and the average expression across these tumors was within the physiological range of expression observed in primary neuroblastomas (Supplementary Fig. 4a).
Fig. 4.
miR-380 prevents oncogene induced senescence and increases tumor incidence in a mouse mammary cancer model. (a) The incidence of palpable mammary tumors (1 tumor per mouse) arising from cells infected with the indicated miRNA retrovirus plus HRASV12 after 6 weeks is shown, vector/SC _n_=15, miR-125b _n_=10, miR-380 _n_=15, p53shRNA _n_=17. (b) qRT-PCR of mature miR-380-5p in tumors arising from cells infected with HRASV12 and miR-380 or p53-shRNA virus. MCF10a cells that stably express miR-380 or a scrambled miRNA are shown as controls. (c) Immunohistochemical staining of mammary tumors. (d) qRT-PCR for p21waf1 expression normalized to GAPDH in tumors arising from cells infected with HRASV12 and p53-shRNA or miR-380 retrovirus compared to the primary MMECs. b, d, error bars depict s.d., each column represents a separate tumor. Scale bars equal 100 μm.
miR-380 overcomes RAS-induced senescence in vivo
Tumors expressing activated HRAS plus control viral constructs were cystic in nature and mostly comprised of inflammatory cell infiltrates (Fig. 4c). In contrast, miR-380 or shRNA p53 expression together with activated HRAS resulted in solid rather than cystic tumors that were comprised predominantly of epithelial cells (Fig. 4c). Activated HRAS has been shown to induce senescence in a variety of tumor models in a p53-dependent manner 8,30. Tumor cells expressing activated HRAS plus control stain positive for senescence-associated beta-galactosidase (SA-_β_-gal). In contrast, tumors expressing HRAS plus miR-380 or shRNA p53 did not express senescence markers (Fig. 4c). p21waf1 was increased in wild-type mouse mammary epithelial cells and tumor cells expressing activated HRAS plus control. In contrast, p21waf1 expression was lower in tumor cells that co-express HRAS and miR-380 or shRNA p53 (Fig. 4c,d). These data are consistent with a role for miR-380 in promoting mammary tumorigenesis by suppressing the p53- and p21waf1-dependent oncogene-induced senescence program initiated by activated HRAS.
miR-380-5p is highly expressed in human and mouse neuroblastoma and predicts poor outcome for individuals with _MYCN_-amplified neuroblastoma
Virtually all neuroblastomas have wild-type p53 prior to treatment with chemotherapy, suggesting that the p53 pathway may be attenuated by another mechanism in these tumors35. We examined miR-380-5p and p53 expression in neuroblastoma cell lines and found that a majority express readily detectable levels of miR-380-5p and cell lines with wild-type p53 generally had low levels of p53 protein (Supplementary Fig 4b).
The MYCN gene is frequently amplified in human neuroblastomas and over-expression of MYCN in transgenic mice gives rise to neuroblastomas that recapitulate many features of the human disease36. Despite p53 being functional in these tumors, p53-driven apoptosis is minimal. Treatment of tumor-bearing mice with chemotherapy causes induction of p53, apoptosis and complete tumor remission37. The TH-MYCN transgenic mice are amongst the most widely used in vivo model of human neuroblastoma, having been used extensively as a preclinical model with a proven track record in therapeutic validation37-41. These tumors are diverse primary cancers that, unlike cell lines, have not been through experimental clonal selection and years of in vitro culture. Importantly, the mature miR-380-5p sequence and miR-380-5p target sequence in the p53 3′UTR are conserved between human and mouse. Using this transgenic neuroblastoma model, we found that miR-380-5p expression was on average 5-fold higher in primary tumors compared to benign neuroendocrine tissue taken from either wild type or _MYCN_-transgenic mice prior to the onset of disease (Fig. 5a).
Fig. 5.
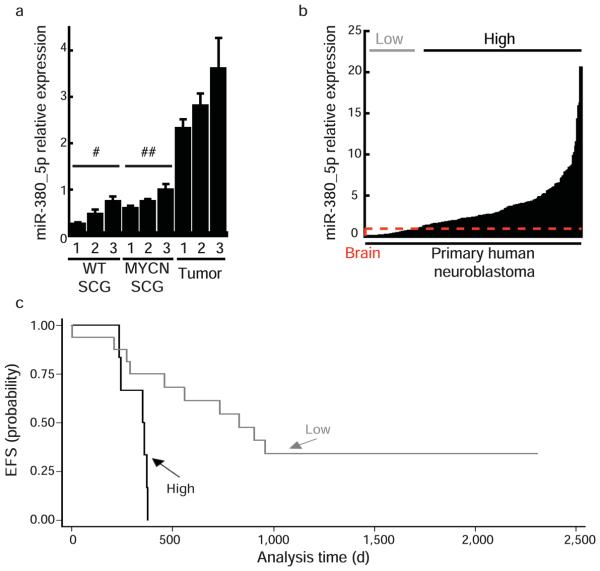
miR-380-5p is expressed in mouse and human neuroblastoma and is associated with poor outcome in subjects with MYCN amplification. (a) qRT-PCR for miR-380-5p expression in tumors and neuroendocrine ganglion (SCG) tissue from wild type (#) and transgenic (##)mice (b) miR-380-5p expression is detected by qRT-PCR in 203/205 primary human neuroblastoma samples taken before chemotherapy. miR-380-5p expression was normalized to RNU6B, normal human brain expression (indicated by red dashed line), ‘low’ and ‘high’ designate lowest quartile of miR-380-5p expression and the remainder, respectively. (c) Kaplan-Meier survival curves of event-free survival (EFS) in subgroups of subjects with neuroblastoma according to relative expression level of miR-380-5p, all with MYCN amplification (_n_=22). Subjects were dichotomized around the lower quartile of miR-380-5p expression. “High” _n_=6, mean miR-380-5p expression =3.19, s.e.m.= 0.83. “Low” _n_=16, mean miR-380-5p expression= 0.49, s.e.m.= 0.13. p=0.004. a, error bars depict s.d., #p<0.02, ##p<0.02.
We next examined miR-380-5p abundance in 205 primary human neuroblastoma samples collected from subjects prior to treatment with chemotherapy and compared to human brain, the highest expressing normal adult tissue (Fig. 1b). We found that miR-380-5p was readily detectable in 203 (99%) of 205 primary neuroblastomas (Fig. 5b), and substantially over-expressed, relative to brain, in 155 (76%) of 205 primary neuroblastoma samples (Fig. 5b). Expression of miR-380-5p did not correlate with individual age or tumour stage, however tumours with MYCN amplification had significantly lower expression of miR-380-5p (p<0.001). In these individuals, high miR-380-5p expression was associated with a significantly poorer outcome than those with low expression of miR-380-5p (Fig. 5c; P=0.004). In individuals without MYCN amplified tumors, miR-380-5p expression was not associated with subject clinical outcome (Supplementary Fig. 4c). Furthermore, by analyzing miR-380 expression in both primary and secondary tumours, we conclude that miR-380-5p expression is maintained in distant metastases (Supplementary Fig. 4d).
miR-380-5p attenuates p53 function in neuroblastoma
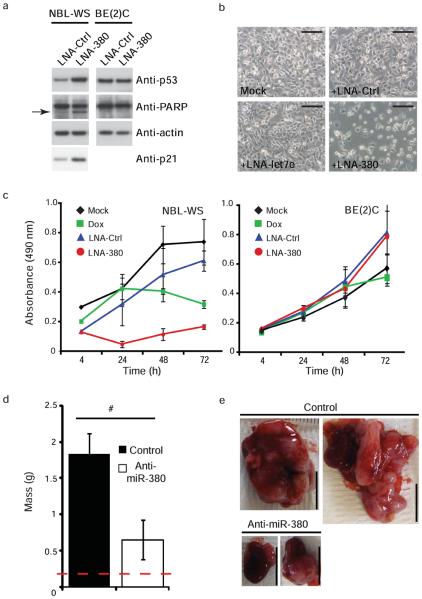
The association of miR-380-5p expression with poor outcome in human neuroblastoma samples suggested a functional role in tumorigenesis. Transfection of LNA-380 or two LNA-controls was performed in the NBL-WS human neuroblastoma cell line, which retains wildtype p53, and cell proliferation and p53 expression were examined. Knock-down of miR-380-5p resulted in a dramatic up-regulation of p53 and p21waf1 and PARP cleavage (Fig. 6a, Supplementary Fig. 4e). Diminished cellular viability following LNA-380 treatment was more pronounced than after treatment with the chemotherapeutic doxorubicin in these cells (Fig. 6b,c). Inhibition of miR-380-5p also resulted in p53 induction and impaired cellular proliferation in another p53-wildtype neuroblastoma cell line, SHEP (Supplementary Fig. 4f, g). In contrast, the BE(2)C line, taken from an individual at relapse and in which p53 has acquired an inactivating mutation35 did not demonstrate any changes in cell viability in response to LNA-380 transfection (Fig. 6a,c). We conclude that miR-380-5p expression plays a key role in suppressing p53 in human neuroblastoma.
Fig. 6.
Treatment with miR-380-5p antagonist induces p53-dependent cell death in neuroblastoma cells and decreased tumor growth in vivo. (a) Western blots demonstrating p53 and p21waf1 induction and PARP cleavage following knockdown of miR-380-5p by LNA-380 compared to LNA-Ctrl in NBL-WS cells (left panel) but not p53-mutant BE(2)C cells (right panel). Arrow indicates cleaved PARP. (b) Images of NBL-WS cells 24h after mock transfection (mock) or treatment with indicated LNAs, LNA-let7e included as an additional control. Scale bars equal 200 μm. (c) MTS assay demonstrating that knockdown of miR-380-5p by LNA-380 induces rapid loss of cell viability in NBL-WS cells (left graph) but not BE(2)C cells (right graph), doxorubicin treatment (dox).(d) Systemic treatment with miR-380 antagonist (anti-miR380) for 3 weeks results in decreased tumor size, mass depicted is the weight of the kidney (indicated by dashed red line) plus associated tumor (_n_=5 mice for each treatment). (e) Representative images of kidneys and associated neuroblastoma tumor mass from two different mice for each treatment group, scale bars depict 1cm. a-c, results are representative of at least three independent experiments, error bars depict s.e.m. #p<0.01.
Diminished tumor growth following miR-380-5p antagonist treatment in vivo
We next examined whether miR-380-5p antagonists could alter the growth of neuroblastoma in vivo in a relevant orthotopic model. For this purpose we chose a chimeric anti-miR that was modified at the 2′ position of the sugar with either a fluoro or a methoxyethyl group and a full phosphorothioate backbone. This design has been shown to produce potent inhibition of microRNAs in vivo 42, EGM unpublished observations. Primary tumors from transgenic TH-MYCN mice were orthotopically transplanted onto the kidney capsule of recipient Balb/c Nu/Nu mice. In this model, extensive neuroblastoma tumors form which envelop the kidney with a latency of approximately 3-5 weeks. Starting two days following transplant, mice received systemic treatment via intraperitoneal injection of chimeric anti-miRs designed to antagonize miR-380 (anti-miR380) or a control sequence twice weekly for 3 weeks (25 mg kg−1 per injection). Treatment with anti-miR380 led to dramatically decreased size and weight of neuroblastoma tumors (Fig. 6d, e; p=0.01). Anti-miR380 treated tumors also expressed diminished miR-380-5p and increased p21waf1 as compared to the control (Supplementary Fig. 5a,b). No toxicity was noted for mice treated with either anti-mir380 or control anti-miRs for three weeks. We conclude that systemic delivery of a miR-380 antagonist diminishes the growth of orthotopically transplanted primary neuroblastoma tumors.
DISCUSSION
We show that miR-380-5p is abundantly expressed in ES cells and provides a constitutive survival function by repressing expression of p53, which is likely just one of its many targets. ES cells exhibit an especially rapid rate of proliferation, lacking normal G1 and G2 phases, however they retain an intact p53/MDM2/ARF tumor suppressor response. Expression of miRNAs, such as miR-380-5p, may allow temporary and tunable repression of p53 in stem cells, thus permitting rapid cellular proliferation and self renewal, without the risks associated with irreversible loss of p53 function that is frequently found in cancer cells.
We have used neuroblastoma as a model disease in which to test the role of microRNA regulation of p53 in cancer. We show that nearly all neuroblastoma tumors express miR-380-5p and that high expression correlates with poor prognosis in individuals with MYCN amplified disease. In human neuroblastoma cell lines, inhibition of miR-380-5p increases p53 expression and induces apoptotic cell death (Supplementary Fig. 6). While p53 is the target we have studied most extensively, miR-380-5p is likely to target other mRNAs, some of which may also be important for control of proliferation or survival.
A rapidly growing body of evidence has identified miRNAs as potential targets for cancer therapy. For example, over-expression of miR-26 by adeno-associated virus in a MYC-driven liver cancer model can attenuate tumor formation43. Likewise, a transgenic switchable model of miR-21 expression induces miRNA-dependent tumor formation and can cause regression when miR-21 expression is inhibited44. The development of small miRNA antagonists has opened the possibility for the development of drug-like miRNA antagonists for cancer therapy. Inhibition of miR-10b by delivery of miRNA antagonists does not block tumor growth, but can attenuate breast cancer metastasis in animal models45. In the current study, we sought to confirm that miR-380 is also required by tumors in vivo. We find that treatment with anti-miR380 results in diminished tumor growth. To our knowledge, this is the first report of a systemically delivered, in vivo treatment inhibiting a miRNA, that has resulted in decreased primary tumor growth. We propose that miR-380-5p is a novel oncogene and a potential therapeutic target in p53 wild-type neuroblastoma which would be worth investigating in the clinic.
METHODS
Details of plasmids, cell culture conditions, immunoblotting and immunohistochemistry can be found in Supplementary Methods.
Bioinformatics
We determined local RNA structure as previously described23 using mFold46. Homo sapiens 3′UTR average –ΔG value of 16.2 ± 1.36 was calculated from the average of 30 3′ UTRs randomly selected from the Genbank database.
Orthotopic mammary transplant and tumorigenesis assays
We generated mice bearing mammary tumors by transplantation, essentially as described32 with the following modifications: Mouse mammary epithelial cells were harvested from female FVB/N mice and virally transduced. 500,000 cells were transplanted into the cleared inguinal fat pads of naive 3-4 week old female FVB/N mice. Mice were observed for tumor formation, tumor diameter was measured with calipers weekly and mice were sacrificed at 42 d following transplant, or sooner if they had reached the ethical endpoint of the experiment. Animal experimentation was approved by the Committee for Animal Research at the University of California, San Francisco. Tumor tissue was either embedded in OCT medium or fixed in 4% paraformaldehyde, paraffin embedded and processed for histology.
Transgenic Animal Experiments
TH-MYCN transgenic mice develop neuroblastomas with a median of ~100 d36. We microdissected control SCG tissue from freshly-sacrificed wild type 8–12 week old 129S6 or control TH-MYCN mice, all non-nerve tissue was removed and the tissue was snap frozen in liquid nitrogen. We prepared RNA from SCG or primary neuroblastoma tumors and performed qRT-PCR to determine miRNA expression.
Orthotopic Neuroblastoma Model and In vivo Therapeutics
We transplanted primary neuroblastoma tumors from TH-MYCN transgenic mice (2 mm3 per kidney capsule) into Balb/c Nu/Nu mice. Two days following transplant mice were treated with chemically-modified antisense oligonucleotides, designed to be complementary to miR-380-5p or a control anti-miR (Regulus Therapeutics, CA). We treated mice twice weekly (25 mg kg−1 per dose i.p.) for three weeks and sacrificed them a total of 4 weeks following transplant. The weight of the kidney plus the encompassing tumor or the contralateral kidney alone (not injected with tumor cells) was recorded. The median mass of the kidney alone was 0.17 g.
RNA expression analysis
We performed quantitative RT-PCR assays (Taqman, Applied Biosystems) as per the manufacturer's instructions using 25 ng (microRNA analysis) or 1 ug (p53, GAPDH, or p21) total RNA prepared using RNABee (Iso-Tex Diagnostic, Inc.), mirVana isolation kit (Ambion), Trizol (Invitrogen) or RNA prepared previously for the Children's Oncology Group protocol 904747. We analysed miR-380-5p expression in primary versus secondary disease by extracting RNA (Trizol) from snap frozen tissue provided by the Children's Hospital Westmead Tumour Biobank (NSW, Australia). Work with both these cohorts was approved by individual institutional review boards or the Children's Oncology Group Neuroblastoma Subcommittee. miR-380-5p expression was determined and expression normalized to miR-16, RNU6B, RNU19 or sno202 levels, which are readily detectable endogenous controls and do not change significantly in the samples tested. We either isolated or purchased (Ambion, Stratagene) total RNA samples containing the small RNA fraction, followed by Northern blot analysis for miR-380-5p or miR-380-3p as previously described48.
Luciferase assays
p53-3′UTR Reporters-The 104 bp conserved element from the human TP53 3′UTR or a mutant lacking the mir-380-5p seed sequences was cloned into a destabilized Firefly luciferase vector (Promega) to generate reporter constructs. We transfected 293T cells with the indicated miRNA expression vector, the reporter constructs and Renilla luciferase internal control vector and performed Dual-Glo reporter assays as indicated by the manufacturer (Promega). miR-380_5p Perfect Reporter-Three consecutive binding sites with perfect complementarity to miR-380-5p were cloned into the pMIR-Report Firefly luciferase vector (Ambion). We transfected NIH3T3 cells with 0.5 nM of the synthetic miRNA mimic (Ambion), the reporter constructs and Renilla luciferase internal control vector using Lipofectamine2000 (Invitrogen). We transfected 50 nM LNA oligos 24 h later and performed dual-Glo reporter assays after 48 h as indicated by the manufacturer.
Neuroblastoma clinical data set and statistical analysis
The neuroblastoma clinical data set has been described elsewhere47. Informed consent was obtained from all subjects. Researchers performing the miR-380-5p expression analysis were blind to all subject clinical characteristics and outcome of the subjects. We calculated Event-free survival time from the time of enrollment on protocol 9047 of the COG cohort to the time of the first occurrence of an event (relapse, progressive disease, secondary malignancy, or death), or to the date of last contact if no event occurred. We computed cumulative event-free survival by the Kaplan Meier method and compared between subgroups using the Log-rank test. The influence of selected factors on survival time was tested using the Cox proportional hazards model. Specific factors considered as being possibly associated with outcome included; MYCN amplification, age at diagnosis, neuroblastoma stage or miR-380-5p expression. We carried out statistical analyses using Stata, version 10.0, 2007 (StataCorp, College Station, Texas, USA).
Supplementary Material
1
ACKNOWLEDGMENTS
We gratefully thank J. M. Bishop, N. K. Hayward and R. L. Sutherland for their support of this project, the Children's Oncology Group and M. Grimmer for providing tumor samples, D. Lynch and J. Brugge for MCF10A cells expressing the ecotropic receptor, R. Jaenisch for _Trp53_−/− ES cells and S. Lowe for the p53 shRNA retrovirus.
This work was supported by grants from the US National Institutes of Health P50-CA58207, K08-CA104032, 1R01CA136717, 5R01DC005991 (to N.L.), R01CA102321, R01NS055750, P01CA081403 (to W.W.), K08NS48118 (to R.B.); the S.G. Komen Foundation; the UCSF Program for Breakthrough Biomedical Research (to A.G.); the G.W. Hooper Foundation; the Australian National Health and Medical Research Council (to T.P, M.H., M.N. and A. Swarbrick) and the Cancer Institute NSW (M.H. and M.N.). A. Swarbrick is a recipient of a Cancer Institute NSW early career development fellowship and R.L.J a US National Science Foundation fellowship. A.G. is a V-Foundation Scholar, A. Shaw is a Cancer Institute NSW Research Scholar and Y.P. is supported by an Australian Postgraduate Award from the Australian National Health and Medical Research Council.
Footnotes
AUTHOR CONTRIBUTIONS
SW, A Swarbrick and AG conceived and designed the experiments, discussed the results and wrote the manuscript. A Swarbrick, SW, A Shaw, YP, AN, AG, RJ, CS, CH, PL, AB, RB, NH, YC, LL performed experiments. LA and MN performed statistical analysis of human neuroblastoma dataset. AC provided patient samples and clinical data, EM provided anti-miRs for in vivo studies. MH, TP, WW, NL, CS supervised experiments or experimental design.
REFERENCES
- 1.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 4.Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 5.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7:979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 6.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 8.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tweddle DA, et al. The p53 pathway and its inactivation in neuroblastoma. Cancer Lett. 2003;197:93–98. doi: 10.1016/s0304-3835(03)00088-0. [DOI] [PubMed] [Google Scholar]
- 10.Yoon H, et al. Gene expression profiling of isogenic cells with different TP53 gene dosage reveals numerous genes that are affected by TP53 dosage and identifies CSPG2 as a direct target of p53. Proc Natl Acad Sci U S A. 2002;99:15632–15637. doi: 10.1073/pnas.242597299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao R, et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke AR, Gledhill S, Hooper ML, Bird CC, Wyllie AH. p53 dependence of early apoptotic and proliferative responses within the mouse intestinal epithelium following gamma-irradiation. Oncogene. 1994;9:1767–1773. [PubMed] [Google Scholar]
- 13.Clarke AR, et al. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 14.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 15.Gottlieb E, et al. Transgenic mouse model for studying the transcriptional activity of the p53 protein: age- and tissue-dependent changes in radiation-induced activation during embryogenesis. Embo J. 1997;16:1381–1390. doi: 10.1093/emboj/16.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkatachalam S, et al. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. Embo J. 1998;17:4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halaby MJ, Yang DQ. p53 translational control: a new facet of p53 regulation and its implication for tumorigenesis and cancer therapeutics. Gene. 2007;395:1–7. doi: 10.1016/j.gene.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 18.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 21.Enright AJ, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saetrom P, et al. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn DE, et al. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seitz H, et al. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tedeschi A, Di Giovanni S. The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO Rep. 2009;10:576–583. doi: 10.1038/embor.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le MT, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu L, Ma W, Benchimol S. A translation repressor element resides in the 3′ untranslated region of human p53 mRNA. Oncogene. 1999;18:6419–6424. doi: 10.1038/sj.onc.1203064. [DOI] [PubMed] [Google Scholar]
- 29.Mazan-Mamczarz K, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci U S A. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarkisian CJ, et al. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol. 2007;9:493. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- 31.Swarbrick A, Roy E, Allen T, Bishop JM. Id1 cooperates with oncogenic Ras to induce metastatic mammary carcinoma by subversion of the cellular senescence response. Proc Natl Acad Sci U S A. 2008;105:5402–5407. doi: 10.1073/pnas.0801505105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welm AL, Kim S, Welm BE, Bishop JM. MET and MYC cooperate in mammary tumorigenesis. Proc Natl Acad Sci U S A. 2005;102:4324–4329. doi: 10.1073/pnas.0500470102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hemann MT, et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 34.Scott GK, et al. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 35.Tweddle DA, Malcolm AJ, Bown N, Pearson AD, Lunec J. Evidence for the development of p53 mutations after cytotoxic therapy in a neuroblastoma cell line. Cancer Res. 2001;61:8–13. [PubMed] [Google Scholar]
- 36.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. Embo J. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chesler L, et al. Chemotherapy-induced apoptosis in a transgenic model of neuroblastoma proceeds through p53 induction. Neoplasia. 2008;10:1268–1274. doi: 10.1593/neo.08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu T, et al. Activation of tissue transglutaminase transcription by histone deacetylase inhibition as a therapeutic approach for Myc oncogenesis. Proc Natl Acad Sci U S A. 2007;104:18682–18687. doi: 10.1073/pnas.0705524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chesler L, et al. Inhibition of phosphatidylinositol 3-kinase destabilizes Mycn protein and blocks malignant progression in neuroblastoma. Cancer Res. 2006;66:8139–8146. doi: 10.1158/0008-5472.CAN-05-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hogarty MD, et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 2008;68:9735–9745. doi: 10.1158/0008-5472.CAN-07-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rounbehler RJ, et al. Targeting ornithine decarboxylase impairs development of MYCN-amplified neuroblastoma. Cancer Res. 2009;69:547–553. doi: 10.1158/0008-5472.CAN-08-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis S, et al. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res. 2009;37:70–77. doi: 10.1093/nar/gkn904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kota J, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010 doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 45.Ma L, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haber M, et al. Association of high-level MRP1 expression with poor clinical outcome in a large prospective study of primary neuroblastoma. J Clin Oncol. 2006;24:1546–1553. doi: 10.1200/JCO.2005.01.6196. [DOI] [PubMed] [Google Scholar]
- 48.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. Rna. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1