A Phosphorylated Cytoplasmic Autoantigen, GW182, Associates with a Unique Population of Human mRNAs within Novel Cytoplasmic Speckles (original) (raw)
Abstract
A novel human cellular structure has been identified that contains a unique autoimmune antigen and multiple messenger RNAs. This complex was discovered using an autoimmune serum from a patient with motor and sensory neuropathy and contains a protein of 182 kDa. The gene and cDNA encoding the protein indicated an open reading frame with glycine-tryptophan (GW) repeats and a single RNA recognition motif. Both the patient's serum and a rabbit serum raised against the recombinant GW protein costained discrete cytoplasmic speckles designated as GW bodies (GWBs) that do not overlap with the Golgi complex, endosomes, lysosomes, or peroxisomes. The mRNAs associated with GW182 represent a clustered set of transcripts that are presumed to reside within the GW complexes. We propose that the GW ribonucleoprotein complex is involved in the posttranscriptional regulation of gene expression by sequestering a specific subset of gene transcripts involved in cell growth and homeostasis.
INTRODUCTION
Historically human autoantibodies have been used to discover, identify, and understand the function of novel cellular constituents and macromolecules (Tan, 1991; von Muhlen and Tan, 1995). For example, small nuclear ribonucleoproteins (snRNPs) and the spliceosome were initially elucidated through the use of human autoantibodies (Lerner and Steitz, 1981; Tan, 1991). In addition, a number of unique nucleolar (Reimer et al., 1987), Golgi complex (Chan and Fritzler, 1998), and endosome (Waite et al., 1998; Selak et al., 1999) proteins have been identified using human autoantibodies.
Several known autoantigens such as SS-A/Ro, SS-B/La, Sm, Hu, and Nova bind distinct RNAs and, in concert with other proteins, form macromolecular complexes that are key components of gene expression at multiple levels including transcription, translation, transport, and stability (Keene, 1999; Brennan and Steitz, 2001; Keene, 2001; Musunuru and Darnell, 2001). Proteins associated with mRNA are of interest because it is thought that messenger ribonucleoproteins may be involved in stabilizing, degrading, or in regulating translation of the mRNAs upon exit from the nucleus (Brennan and Steitz, 2001; Keene, 2001). One of the approaches to identification of proteins involved in mRNA processing, referred to as ribonomics, involves biochemically or immunologically recovering protein/RNA complexes, followed by microarray analysis to determine their structural and functional relationships (Tenenbaum et al., 2000). Using this approach, some of the mRNA subsets and profiles have been deciphered for proteins such as eIF-4E, PABP, and ELAV/Hu (Tenenbaum et al., 2000) as well as Fragile X Mental Retardation RNA-binding protein (FMRP;Brown et al., 2001). It has been proposed that the expression of structurally related mRNAs is coordinated by multitargeted RNA binding proteins in order to regulate complex processes (Keene, 2001). It is not known whether expression of certain clustered mRNAs might be coordinated and if multiple mRNAs reside together within a physical particle (Keene, 2001).
In this study, we used sera from a patient with a motor and sensory polyneuropathy to identify a novel 182-kDa protein that we designate as GW182. This protein localizes to a novel cytoplasmic domain that appears distinct from endosomes, lysosomes, peroxisomes, or the Golgi complex. We discuss the possible role of these autoimmune mRNP complexes in posttranscriptional gene expression.
MATERIALS AND METHODS
Patient Serum and Antibodies
All human sera used in this study were obtained from serum banks at the Advanced Diagnostics Laboratory, University of Calgary and the W.M. Keck Autoimmune Disease Center, The Scripps Research Institute. The index human serum was obtained from a 49-year-old Caucasian woman with a mixed motor and sensory polyneuropathy. The selection of this serum was based on its unique reactivity to an apparently novel cytoplasmic domain.
cDNA Cloning and Analysis
An HeLa uni-ZAP XR cDNA expression library (catalogue no. 937216; Stratagene, La Jolla, CA) was screened with the index human serum using techniques described in detail elsewhere (Fritzler et al., 1993, 1995). A single positive clone identified as 5.1 was subcloned into pBS SK+ plasmid, and the complete nucleotide sequence was determined in both strands using BigDye terminator sequencing and a semiautomated sequencer (Model 377; Applied Biosystems Inc., Foster City, CA). To search for similar known sequences, the nucleotide and deduced amino acid sequences of 5.1 were analyzed using BLAST search (Altschul et al., 1990) and ExPASy Proteomics tools on the World Wide Web Internet server (http://www.expasy.org/tools). An EST clone KIAA1460 (GenBank accession no. AB040893) with 99% identity to clone 5.1 was obtained from the cDNA bank section at the Kazusa DNA Research Institute (Chiba, Japan).
Confirmation of the 5′ Sequence of GW182 and Construction of Full-Length GW182
PCR amplification was conducted on HeLa marathon ready cDNA (CLONTECH, Palo Alto, CA) and isolated HeLa genomic DNA using primer 1: 5′-TTTCTCGAGGATCCGCCATGGATGCTGATTCTGCCTCCAGTTCT-3′, and primer 2: 5′-TTATCTATACATGCCCCTGAGTT-3′ (also see Figure 2). The annealing temperature for PCR amplification was 55°C. The PCR products obtained were purified and sequenced using the aforementioned primers. The full-length cDNA for GW182 was constructed by subcloning the 5′ sequence derived from the above PCR into the KIAA1460 cDNA that encodes the 3′ sequence.
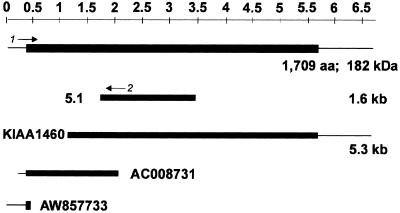
Figure 2.
A representation of the 5.1 cDNA derived from expression cloning, the EST clone KIAA1460 (GenBank accession no.AB040893), the overlapping genomic fragment AC008731, and EST AW857733whose combined sequences represent the predicted full-length clone of the GW182 protein. The upstream region was verified using primers 1 and 2, in the predicted 5′-UTR and in the clone 5.1 using HeLa mRNA. Solid boxes represent open reading frame. The full-length protein is 1709 aa and 182 kDa in size.
Recombinant Protein Production and Generation of Rabbit Antibody
The cDNA insert from clone 5.1 was subcloned into the expression vector pET28 (Novagen, Madison, WI) and transformed to_E_s_cherichia coli_ JM109(DE3) for recombinant protein production. The N-terminal 6× histidine fusion recombinant protein from 1-liter culture was purified using Ni2+ affinity chromatography (QIAGEN, Valencia, CA) by following the manufacturer's instructions. Two New Zealand White female rabbits were immunized by subcutaneous injections of 0.5 mg of the purified protein in an equal volume of Freund's complete adjuvant (Difco Laboratories, Detroit, MI; Fritzler et al., 1995). After 4 weeks, the animals were boosted with 0.5 mg of the same recombinant protein in Freund's incomplete adjuvant (Difco). Preimmune rabbit sera before immunization and sera obtained after the booster injections were analyzed by immunoprecipitation and immunofluorescence. The postimmune rabbit serum was referred to a rabbit–anti-GW182p, where p is the antibody raised to the partial length protein of GW182.
Cell Labeling and Immunoprecipitation
HeLa cell (ATCC, Rockville, MD) extracts were prepared after incubation of cells overnight in the presence of either [32P]orthophosphate (ICN Biomedicals, Costa Mesa, CA) or [35S]methionine (Trans35S label; ICN) as previously described (Chan_et al.,_ 1986; Bestagno et al., 1987; Chan and Tan, 1987). The lysis buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1.0% Nonidet P-40) was supplemented with Complete protease inhibitor cocktail (Boehringer Mannheim, Indianapolis, IN). Immunoprecipitation (IP) reactions were prepared by combining 100 μl 10% protein A-Sepharose beads (catalogue no. P-3391; Sigma, St. Louis, MO), 10 μl human serum, 500 μl NET2 buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS, 0.02% sodium azide), and 50–100 μl of labeled cell extract. After 1 h of incubation at 4–8°C, the Sepharose beads were washed five times in NET2, and the proteins were eluted in 10 μl of sample buffer (Laemmli, 1970). The proteins were analyzed by 10 or 12.5% SDS-PAGE (Laemmli, 1970) as described. RNA-protein complexes were immunoprecipitated, and the purified RNA was analyzed by autoradiography after removal of proteins with phenol/chloroform and precipitation with isoamyl alcohol. The proteins were analyzed on a 10% gel SDS-PAGE, and RNA was analyzed in 7 M urea gels containing 10% ployacrylamide.
Immunoprecipitation of mRNP Complexes
HeLa S3 cells ( catalogue no. CCL-2.2; ATCC) were grown in suspension to a confluence of approximately 1 × 106/ml. After centrifugation, the cells were washed with cold PBS and resuspended in approximately two pellet volumes of polysome lysis buffer containing 100 mM KCl, 5 mM MgCl2, 10 mM Hepes, pH 7.0, and 0.5% Nonidet P-40. One millimolar DTT, 100 units/ml RNase OUT (GIBCO/BRL, Rockville, MD), 0.2% vanadyl ribonuclease complexes (GIBCO/BRL), 0.2 mM PMSF, 1 mg/ml pepstatin A, 5 mg/ml bestatin, and 20 mg/ml leupeptin were added at the time of use. The resuspended cells were incubated on ice for 5 min and then frozen and stored at −100°C. At the time of use the cell lysate was thawed and centrifuged at 16,000 × _g_in a microfuge for 10 min at 4°C. The supernatant from the cell lysate typically contained approximately 30 mg/ml total protein.
IP reactions were performed essentially as described by Tenenbaum_et al._ (2000). Protein A-Sepharose beads were swollen 1:5 vol/vol in NT2 buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.5% Nonidet P-40 supplemented with 5% BSA. A 300-μl aliquot of the 1:5 vol/vol preswollen protein A bead slurry was used per IP reaction and incubated overnight at 4°C with excess immunoprecipitating anti-GW182 antibodies (typically 10–20 μl depending on the titer of the reagent). The antibody-coated beads were washed with ice-cold NT2 buffer and resuspended in 900 μl of NT2 buffer supplemented with 100 units/ml RNase OUT, 0.2% vanadyl ribonuclease complex, 1 mM DTT, and 20 mM EDTA. The beads were briefly mixed, and then 100 μl of the cell lysate containing mRNPs was added and immediately centrifuged. A 100-μl aliquot was removed and used to represent total cellular mRNA. The IP reactions were tumbled at room temperature for 2 h and then washed six times with ice-cold NT2 buffer. Washed beads were resuspended in 100 μl NT2 buffer supplemented with 0.1% SDS and 30 μg proteinase-K and incubated for 30 min in a 55°C water bath. The immunoprecipitated mRNA was isolated by phenol-chloroform-iso-amyl alcohol extraction and ethanol precipitation.
Probing of cDNA Arrays
cDNA array analysis was performed by using Atlas Human 1.2 Arrays (CLONTECH) that contain 1200 cDNA segments spotted on a nylon membrane. Probing of cDNA arrays was performed as described in the_CLONTECH_ Atlas cDNA Expression Arrays User Manual (PT3140-1). Briefly, total RNA or GW-associated RNA was isolated and used to produce [α-32P]dATP reverse-transcribed radiolabeled probes using a pooled set of primers complementary to the genes represented on the cDNA array. The radiolabeled probes were purified by passage over CHROMA SPIN-200 columns (CLONTECH). Radiolabeled probes were hybridized to the cDNA array membranes according to the manufacturer's directions and then washed and visualized using a phosphorimaging screen (Molecular Dynamics, Sunnyvale, CA).
Analysis of cDNA Arrays
Phosphorimages were scanned by using the Molecular Dynamics STORM 860 System at 100-μm resolution and stored as .gel files. Images were analyzed by using ATLASIMAGE 2.1 software (CLONTECH). A default external background setting was used in conjunction with a background-based signal threshold to determine gene signal significance. The signal for a gene was considered significantly above background if the adjusted intensity (total signal minus background) was more than twofold the background signal. Comparisons of multiple cDNA array images were performed by using an average of all of the gene signals on a specific array (global normalization) to normalize the signal intensity between arrays (Tenenbaum et al., 2000). cDNA array images and overlays were prepared by using Adobe Photoshop 5.0.2 (Adobe Systems, San Jose, CA).
In Vitro Transcription/Translation and Immunoprecipitation
Clone 5.1 and the full-length GW182 construct were used as templates for in vitro transcription and translation (TnT; Promega, Madison, WI) in the presence of [35S]methionine (Fritzler et al., 1995; Griffith et al., 1997). TnT reactions were conducted at 30°C for 1.5–2 h. To confirm the presence of translation products, 2- to 5-μl samples were fractionated by SDS-PAGE and analyzed by autoradiography. The in vitro translated products were then used as the substrate in IP reactions, as described above.
Indirect Immunofluorescence
Indirect Immunofluorescence (IIF) analyses used commercially prepared HEp-2 cells (ImmunoConcepts, Sacramento, CA) with fluorescein-conjugated goat anti-human antibodies as previously described (Fritzler et al., 1993). For colocalization studies primary antibodies to the following proteins were used: golgin-97 (murine monoclonal cdf4; Griffith et al., 1997), p58 (murine monoclonal, a gift from Dr. Tom Hobman, University of Alberta), TGN38 (murine monoclonal; Affinity Bioreagents, Golden, CO), clathrin heavy chain (murine monoclonal; Transduction Laboratories, Lexington, KY), rab9 (murine monoclonal, Affinity Bioreagents, Golden, CO), PMP70 (murine monoclonal; Zymed, San Francisco, CA), LAMP1 (murine monoclonal; Drs. August and Hildreth, Pharmacology and Molecular Sciences, John Hopkins University School of Medicine, Baltimore, MD), EEA1 (rabbit polyclonal antibody; Selak et al., 1999), cullin-1 (rabbit polyclonal antibody; NeoMarkers, Fremont, CA), caveolin (rabbit polyclonal antibody, Transduction Laboratories), and p19skp1 (rabbit polyclonal antibody; NeoMarkers). IIF used a Leica confocal microscope fitted with the appropriate filter sets for rhodamine, FITC, and DAPI. Images were taken at 1-μm increments.
Immunogold Electron Microscopy
HeLa cells were grown in log phase, removed from the flask by gentle scraping, and fixed for 1.5 h in ice-cold 4% paraformaldehyde and 0.025% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4. After fixation, free aldehydes were quenched in 50 mM glycine in 0.1 M phosphate buffer. The cells were then resuspended in warm 7.5% gelatin in PBS in microfuge tubes for 10 min and then pelleted at 10,000 rpm for 1 min. The tubes were placed on ice, and the solidified gelatin pellets were removed, transferred to an ice-cold 50 mM glycine solution, cut into small cubes, and then transferred to 2.3 M sucrose in 0.1 M phosphate buffer at 4°C. Each block of tissue was placed on a cryo pin-head, excess sucrose solution was removed, and the pin was immersed and stored in liquid nitrogen. Thin sections (∼100 nm) were cut using glass knives and picked up in wire loops dipped in a mixture of methylcellulose and sucrose. The sections were placed directly onto carbon-coated 200-mesh nickel grids. Each grid was processed on individual droplets of the following solutions at room temperature: 50 mM glycine, 10% fetal calf serum (FCS) in PBS, primary antibody diluted 1/400 in 10% FCS, washed in 0.2% FCS, incubated in protein A tagged with 10 nm gold (Dr. J. Slot, University of Utrecht), washed in PBS, fixed in 1% glutaraldehyde in PBS, washed in double-distilled H2O, and contrasted in uranyl oxalate (pH 7). Individual grids were then picked up in ice-cold uranyl acetate/methyl cellulose (pH 4) mixture using copper wire loops that were then blotted with filter paper to remove excess solution. The grids were then allowed to dry overnight before examination on a Philips CM-100 transmission electron microscope (Mahwah, NJ). Controls included normal incubation with protein A-gold alone, normal human serum, and rabbit antibodies to the Golgi protein, giantin (Lindstedt and Hauri, 1993).
Transfection of HEp-2 Cells with GFP-tagged GW182 cDNA
The _Xho_I/_Sac_II cDNA fragment derived from EST clone KIAA1460 was subcloned into the corresponding sites of the phrGFP-N1 expression vector (Stratagene, La Jolla, CA) and the in-frame fusion of GFP to GW182 fragment was verified by direct nucleotide sequencing. HEp-2 cells were grown in 8-well chamber slides to 50% confluency and transfected with 2 μg GFP-fusion in 100 μl of serum-free media and 6 μl of FuGENE 6 (Roche, Laval, PQ, Canada). After incubation at room temperature for 45 min, this mix was added to HEp-2 cells and incubated for 3 h at 37°C. The media was then replaced with serum containing media and cultured for 41 h at 37°C. The cells were fixed with ice-cold acetone-methanol (3:1 vol/vol), and immunofluorescence was performed as described above. As a control, cells were transfected with the phrGFP-N1 vector alone.
Northern Blot Hybridization
Northern blot analysis was performed using a Multiple Human Tissue blot (catalogue no. 7760–1; CLONTECH). A 1.4-kb_Cla_I/_Eco_RI fragment of clone 5.1 was gel-purified and labeled with [α-32P]dATP using a random 9-mer kit (catalogue no. 300385; Stratagene). β-Actin cDNA (CLONTECH) was used as a control. The probes were purified on size exclusion columns (Stratagene), and the hybridization reactions were carried out according to the manufacturer's instructions.
The GenBank accession number for GW182 is AY035864.
RESULTS
GW182 Is Localized to Cytoplasmic Speckles
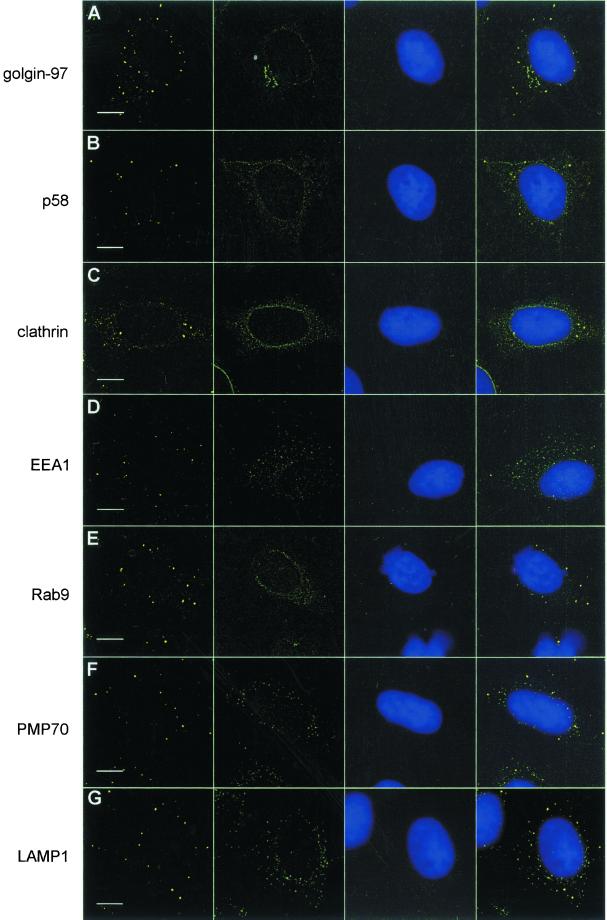
One of the original interests in the prototype serum was its distinctive staining of HEp-2 cytoplasmic bodies. We sought the identity of these structures by using appropriate markers and reagents for cytoplasmic organelles and vesicles in colocalization reactions with the index human serum (Figure 1). The staining obtained with the index human serum did not colocalize with the Golgi compartments, as determined by studies that used antibodies to golgin-97 (Figure 1A; Griffith et al., 1997) and the marker of the ER/Golgi intermediate compartment p58 (Lahtinen_et al._, 1996; Figure 1B) or TGN38, a marker for the trans-Golgi network (TGN38; our unpublished results; Luzio et al., 1990). Brefeldin, a fungal toxin known to rapidly dissociate the Golgi complex (Orci et al., 1991) did not affect the morphology, size, or number of these structures (our unpublished results). The staining did not colocalize with clathrin-coated vesicles (Figure 1C), with the early endosomal protein EEA1 (Figure 1D; Mu_et al._, 1995; Selak et al., 1999; Lawe et al., 2000), with the late endosomal protein rab9 (Lombardi_et al._, 1993; Figure 1E), with the peroxisome membrane protein PMP70 (Figure 1F; Kamijo et al., 1990), with LAMP1 (Figure 1G), a lysosome marker (Fukuda et al., 1988), or with caveolin (our unpublished results; Conrad et al., 1995;Song et al., 1995). Last, antibodies to ubiquitin ligases, cullin-1 (Kipreos et al., 1996; Lisztwan et al., 1998) and p19Skp1 (Zhang et al., 1995;Bai et al., 1996; Connelly and Hieter, 1996), members of the SCF family of ubiquitin ligases (for _S_kp1,_C_dc53/Cullin, _F_-box receptor; Feldman et al., 1997; Skowyra et al., 1997), did not colocalize with these structures either (our unpublished results). To ensure that the method of fixation had not influenced the staining pattern, we compared methanol/acetone-fixed cells with cells fixed in 4% paraformaldehyde/Triton X-100 and found no differences as determined by IIF staining of HEp-2 cells (our unpublished results).
Figure 1.
GW bodies are distinct from known cytoplasmic compartments in HEp-2 cell. The first column represents staining with the index human serum at 1/600 dilution using as a secondary goat anti-human immunoglobulin conjugated to rhodamine. The second column is composed of several different cytoplasmic markers as follows. (A) Mouse monoclonal antigolgin-97 antibody; (B) mouse monoclonal anti-p58 antibody; (C) mouse monoclonal anticlathrin antibody; (D) rabbit anti-EEA1 antibody; (E) mouse monoclonal anti-rab9 antibody; (F) rabbit anti-PMP70 antibody; (G) mouse monoclonal anti-LAMP1 antibody. All these marker antibodies were used at 1/100 dilution. The third column shows corresponding DAPI stained cell nuclei. The fourth column represents images merged from the three columns to the left. The bar lines in the first column represents 10 μm.
Cloning of GW182
The cDNA clone designated clone 5.1 isolated from screening the HeLa cDNA expression library with serum the index human serum was 1.6 kb in length and upon conceptual translation encoded a protein of ∼550 amino acids (Figure 2). Analysis of the coding region using BLASTN revealed several cDNA clones with high similarity to clone 5.1 (Table 1). Interestingly, one human EST clone (KIAA1460; GenBank accession no. BAA95984) had 99% sequence identity to clone 5.1 and represented a longer cDNA in both the 5′ and 3′ ends. However, KIAA1460 also appeared to be incomplete because an upstream methionine start site was absent. Analysis of the sequences obtained from the Human Genome Project using BLASTN identified two clones corresponding to upstream sequences (GenBank accession nos. AC008731 and AW857733; Figure 2). We postulated that these overlapping clones encompassed the entire coding region for GW182.
Table 1.
Representative EST clones highly related to GW182
| Accession no. | Species | Position corresponds to GW182 | % Identity (amino acids) |
|---|---|---|---|
| BAA95984 | Human | 312–1709 | 99 |
| BAA91899 | Human | 1270–1709 | 99 |
| AAH05741 | Mouse | 1270–1702 | 95 |
| Alternative mRNA splicing | ||
|---|---|---|
| BF169182 | Mouse | Missing exon 10 |
| W80996 | Human | Missing exon 10 |
To confirm that the conceptual full-length construct was correct, a primer set was designed for PCR using HeLa cDNA or genomic DNA. The forward primer was designed at the beginning of the coding region, and the reverse primer was derived from clone 5.1 (Figures 2 and3). Our results showed that the PCR generated a predicted fragment of ∼1.5 kb (our unpublished results) that was excised and confirmed by direct DNA sequencing analysis. Thus, the full-length protein has 1709 amino acids with a predicted molecular weight of 182 kDa and a calculated pI of 6.15.
Figure 3.
Human GW182 cDNA and deduced amino acid sequence. The sequence has been submitted to the GenBank under accession no.AY035864. Potential translation start sites (double underlined) and upstream in-frame stop codon (TGA, underlined) are indicated. The GW182 protein consists of 1709 amino acids with a putative nuclear localization signal (NLS; aa 918–937) and RNA recognition motif (RRM; aa 1528–1600) outlined in boxes, respectively. The GW182 protein is rich in tryptophan residues (W, circled), which are often adjacent to glycine (G). Two regions with repeats of GW or WG in tandem are highlighted in bold and double underlined. The region lacking GW is also bracketed. In addition the primers used to amplify the upstream region of the GW182 are shown. Exon–intron junctions and the exact positions of the cDNA clones are indicated.
Characteristics of Clone 5.1 and the GW182 Protein
Using the molecular software tools on the ExPASy website, the full-length protein GW182 had several interesting characteristics, including a potential nuclear localization signal (NLS), an RNA recognition motif (RRM; see boxes in Figure 3), and contained 11.2% glycine and 15.8% serine residues. Throughout the protein there were 60 (3.5%) tryptophan residues (circled in Figure 3), 39 of which are adjacent to glycine residues; exceptions are the RRM and a region labeled as “non-GW” in Figure 3. Many of the tryptophans are in regions that appear to be repetitive and are comprised primarily of glycine/tryptophan (GW) or tryptophan/glycine repeat (WG) amino acid sequences and less often by a variation when tryptophan was followed by another amino acid other than glycine (Figure 3). Of the 39 glycine/tryptophan units, there are 19 GWs, 28 WGs, and 8 GWGs. Although the significance of these repeats is not known, on the basis of this unusual motif and its predicted molecular mass of 182 kDa, we have elected to name the protein GW182 and the associated cytoplasmic structures as GW bodies (GWBs).
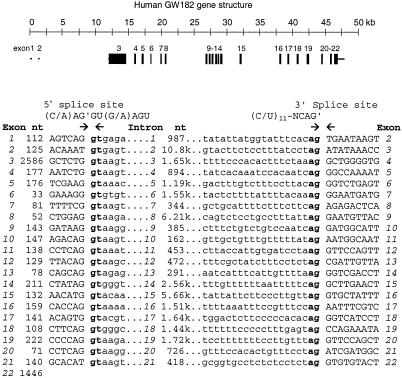
Analysis of the Human Gene for GW182
Analysis of the human genomic sequence (GenBank accession no.AC008731), which was detected by BLASTN search using the cDNA sequence of GW182, revealed the complete gene structure (Figure4). The gene has 22 exons, is ∼46 kb long, and resides on chromosome 16p12. Analysis of human and mouse EST clones from BLASTN search revealed at least two clones that are missing exon 10 at the exact sites predicted for the exon–intron junctions (Table 1). This suggests that alternative products derived from mRNA splicing of GW182 gene exist. The exon–intron junctions of the human GW182 gene are summarized in Figure 4.
Figure 4.
The human GW182 gene is located on chromosome 16p12 and has 22 exons.
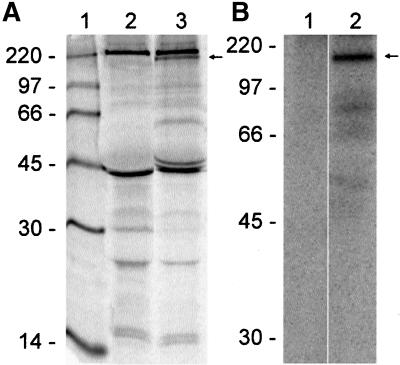
GW182 Protein
The predicted molecular weight of GW182 was verified by IP using extract of [35S]methionine-labeled HeLa cells and the index human serum (Figure 5A). The index human serum immunoprecipitated two predominant proteins of ∼180 and 50 kDa. The 180-kDa protein is also specifically precipitated by other human autoimmune sera with a similar staining pattern on HEp-2 cells (our unpublished results). Whether the 50-kDa protein represents a degradation product or alternative mRNA splicing–derived product of GW182 is unclear and awaits further investigation.
Figure 5.
Immunoprecipitation analysis detected a cellular phosphoprotein of ∼180 kDa specifically recognized by the human index serum. (A) Immunoprecipitation of an extract from [35S]methionine-labeled HeLa cells using normal human serum (lane 2) and the human index serum (lane 3) resolved by SDS-PAGE on a 12.5% gel. Lane 1 contains the 14C-labeled molecular weight standard. B. Immunoprecipitation of an extract of [32P]phosphate-labeled HeLa cells using normal human serum (lane 1) and the index human serum (lanes 2) on 10% gel. The specific signals at ∼180 kDa are indicated by arrow. The nonspecific bands at 220 and 45 kDa detected in both lanes 1 and 2 of A are most likely nonmuscle myosin and actin, respectively.
GW182 Is Phosphorylated
Throughout the GW182 protein there were multiple predicted serine-threonine and tyrosine phosphorylation sites. To determine whether the GW protein was phosphorylated, HeLa cells were labeled with [32P]orthophosphate, and the cell extracts were immunoprecipitated using the index human serum (Figure 5B). A 180-kDa phosphorylated protein was immunoprecipitated with the index serum (lane 2), and normal human serum did not immunoprecipitate any phosphoproteins (lane 1). In contrast to the results obtained from [35S]methionine-labeled cells (Figure 5A), no protein product of 50 kDa was observed.
GFP-tagged Expression of GW182
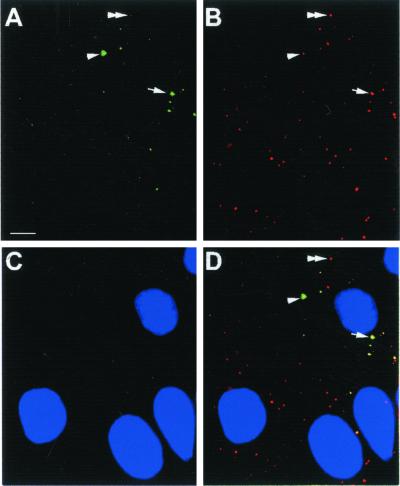
To determine whether GW182 is able to localize to GWBs, a green fluorescent protein (GFP) fusion construct pGFP-GWaa313–1709 was prepared and transfected into HEp-2 cells as described in MATERIALS AND METHODS. The GFP vector showed both nuclear and cytoplasmic staining in transfected cells alone (our unpublished results). In contrast, the GFP-tagged partial protein of GW182 shows distinct cytoplasmic domains (Figure 6A) that are recognized and stained with the index human serum, as shown in Figure 6D. It is noted that different GWBs incorporated varying amounts of GFP-fusion protein relatively to endogenous GW182 detected by the index human serum (Figure 6).
Figure 6.
Transfection of HEp-2 cells with a construct of GFP fused to a GW182 cDNA fragment. The transfected GFP-tagged protein produced a staining pattern in transfected cells (A) highly similar to the GWBs recognized by the index human serum (B). When the images in panels A and B were merged with the DAPI stained nuclei of C, colocalization of the GFP product was observed in the cytoplasmic GWBs (D). Three populations of GWBs can be distinguished by the difference in green to red signal. Some GWBs have apparently equal intensity of green and red signals (arrow), whereas others have more green (arrowhead) or more red signal (double arrowhead). Bar, 10 μm. Original magnification, ×600.
Antibodies to Recombinant Protein Derived from Clone 5.1 Recognize GW Bodies
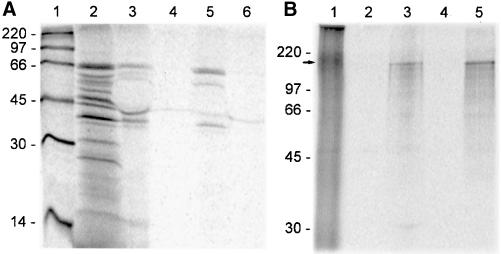
Rabbit antibodies to the recombinant protein encoded by clone 5.1 (rabbit anti-GW182p) were raised to verify that the GW182 resided in the cytoplasmic bodies observed with the index human serum (see below). To ensure that the rabbit antibodies did indeed recognize the protein product encoded by clone 5.1, in vitro transcription and translation (TNT) was used in IP with the index human serum and with rabbit antibodies to the recombinant protein (Figure7A). The recombinant protein was immunoprecipitated by both the index human serum (lane 3) and the rabbit antiserum (lane 5) but not by the rabbit preimmune serum (lane 4) or the NHS (lane 6). The same IP specificity was observed with the in vitro–derived full-length GW182 protein, which migrated at ∼180 kDa (Figure 7B).
Figure 7.
Analysis of in vitro–translated GW182 proteins specifically recognized by the index serum and rabbit antibodies raised to recombinant protein. (A) Immunoprecipitation of translation products derived from clone 5.1. Lane 1 shows the14C-labeled molecular weight standard. Lane 2 shows the proteins produced from in vitro transcription and translation with the largest polypeptide migrating at ∼66 kDa. Immunoprecipitation of the in vitro–translated protein product was performed using the index human serum (lane 3), rabbit preimmune serum (lane 4), postimmune rabbit serum (lane 5), and normal human serum (lane 6). (B) Immunoprecipitation of translation product derived from the full-length construct (lane 1) with the major protein migrating at ∼180 kDa. The latter was immunoprecipitated by the index human serum (lane 2) and rabbit antiserum (lane 5) but not by a normal human serum control (lane 2) and the rabbit preimmune serum (lane 4).
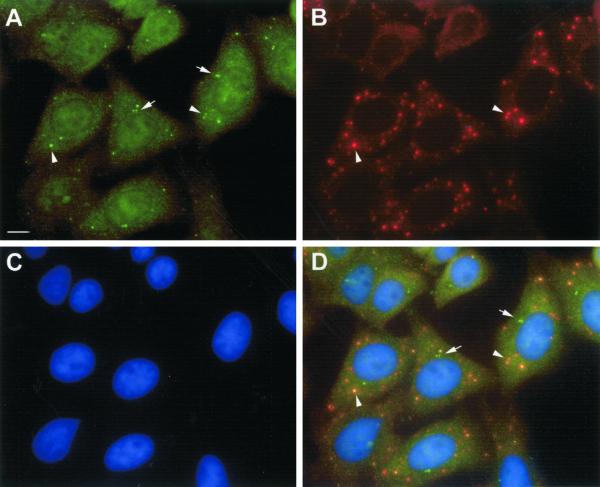
The most striking feature of IIF using the index human serum was cytoplasmic bodies numbering from 0 in mitotic cells to 30 in some interphase cells (Figure 8B). In addition, finer cytoplasmic speckling was observed as well. IIF studies with the index human serum did not show any nuclear staining of HEp-2 cells. On average, ∼20–30 of the cytoplasmic bodies are clearly seen as larger bodies but higher magnification revealed polymorphic GWBs varying in size from ∼0.2 to 1 μm. The cytoplasmic staining observed with the immune rabbit serum was similar to that observed with the human serum, and in colocalization studies, the rabbit and the index human serum reacted with the same cytoplasmic domains (Figure 8).
Figure 8.
Immunofluorescence analysis of HeLa cell GWBs shows costaining of rabbit anti-GW182 antibody and the index human serum. (A) HeLa cell staining with rabbit anti-GW182 antibody at 1/100 dilution. (B) Staining with the index human serum at 1/600 dilution. (C) Nuclei counterstained with DAPI. (D) Merged image with GWBs (arrowheads) recognized by both antibodies. Note that the rabbit anti-GW182 serum also shows unrelated staining of centrioles (arrows) that is also evident in preimmune rabbit serum (our unpublished results). Bar, 10 μm. Original magnification, ×600.
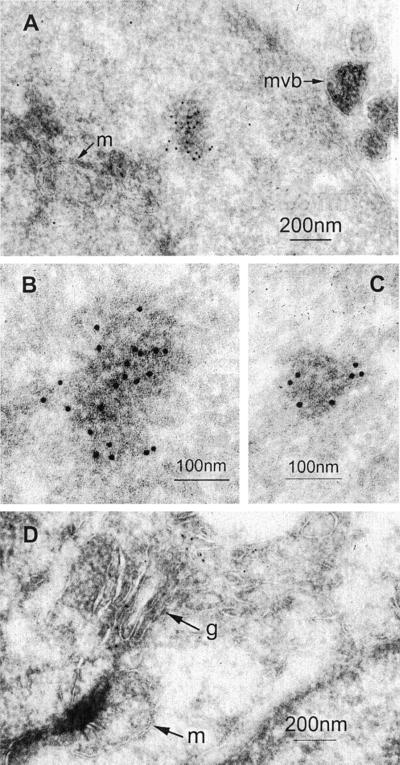
The observation that GWBs are distinct from endosomes, lysosomes, mitochondria, and vesicular elements of the Golgi complex is supported by immunogold electron microscopy (IEM; Figure9). The structures postimmunolabeled with protein A-gold and anti-GW182 are granular, electron dense bodies that are devoid of membranous structures (Figure 9, A–C). This is distinct from multivesicular bodies (Figure 9A), mitochondria (Figure9, A and D), and the Golgi complex (Figure 9D). No immunogold staining was seen with normal or preimmune serum or with immunogold in the absence of primary antibodies (our unpublished results). By comparison, antibodies to giantin bound to the Golgi membrane stack and vesicles in the lateral aspect of the membrane stack (Figure 9D). In addition, in agreement with size estimated by confocal microscopy, the immunogold-labeled GWBs vary in size from 100 to 200 nm (Figure 9, A–C).
Figure 9.
Immunogold electron microscopy (IEM) localization of GW182 in HeLa cells. Frozen sections of fixed and gelatin-embedded HeLa cells were incubated with the index human serum or rabbit antigiantin diluted 1/400 and then postimmunolabeled with protein A-gold. Examples of the structures labeled by anti-GW182 are shown in A–C and by antigiantin in D. Antibodies to GW182 labeled an electron dense cytoplasmic body that is not bound by a membrane and is distinct from mitochondria (m) and multivesicular bodies (mvb). (B) High-power view of the electron dense body in A. (C) A high-power view of another representative immunogold labeled cytoplasmic electron dense body. Antibodies to giantin label the lateral aspects of the Golgi membrane stack (g). Scale bars are shown in the lower right.
GW182 Binds to mRNA
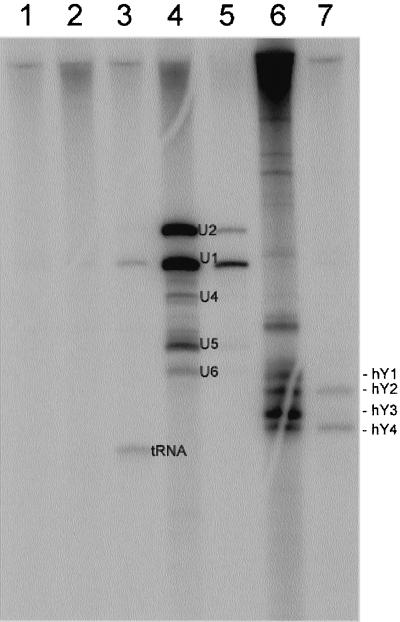
Because GW182 contained an RRM, we questioned whether this protein could bind small RNA species that are targets of other autoimmune diseases. The prototype serum that immunoprecipitated GW182 did not immunoprecipitate tRNA, snRNA, or hYRNA (Figure10). By contrast, human anti–Jo-1 (antihistidyl tRNA synthetase) precipitated tRNA, human anti-Sm serum precipitated U1, U2, U4, U5, and U6, human anti-U1 serum precipitated U1 RNA, human anti–SS-A/B precipitated hYRNA, and human anti–SS-A/Ro antibodies precipitated hY2/4 RNA (Figure 10).
Figure 10.
Immunoprecipitation of RNA from32P-labeled HeLa cell extracts. IP reactions were performed using 100 μl of the cell extract. Normal human serum (lane 1) and the prototype human serum (lane 2) did not precipitate small RNAs. However, prototype anti-tRNA synthetase (lane 3), anti-Sm serum (lane 4), anti-U1RNP serum (lane 5), anti–SS-A/Ro and anti–SS-B/La serum (lane 6), and anti–SS-A/Ro serum (lane 7) all precipitated expected RNA molecules.
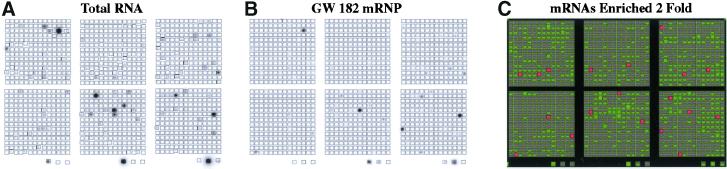
Although anti-GW182 did not immunoprecipitate tRNA, snRNA, or hYRNA, HeLa cell mRNA was recovered in IP pellets and was used to probe a 1200-probe set human cDNA array on which many candidate mRNA targets were identified (Figure 11). Table2 identifies the genes that showed high levels (fold enrichment > 2) of reactivity with the mRNA immunoprecipitated by the prototype human anti-GW182. They included glycoprotein hormone alpha subunit precursor, 60S ribosomal protein L6 (RPL6), metalloproteinase inhibitor 1 precursor (TIMP1), activated RNA polymerase II transcriptional coactivator p15, cAMP-dependent transcription factor ATF-4, serum- and glucocorticoid-regulated serine/threonine protein kinase (SGK), PTPCAAX1 nuclear tyrosine phosphatase, and neuroleukin (NLK). Sequence analysis of these genes and their related mRNAs did not disclose any obvious common sequence motif in the translated or untranslated regions that might account for binding to GW182. The specificity of these reactions was confirmed and validated as illustrated before using normal human serum and sera from patients with systemic lupus erythematosus and paraneoplastic syndromes that react with proteins associated with known mRNAs (Tenenbaum_et al._, 2000; Brown et al., 2001).
Figure 11.
Identification of mRNAs associated with GW182-mRNP complexes using cDNA microarray analysis. As described in MATERIALS AND METHODS, RNA was extracted from immunoprecipitated GW182-mRNPs or total cell lysate and used to make reverse-transcribed radiolabeled probes for Atlas Human 1.2 Arrays containing 1188 singly spotted cDNA segments (CLONTECH). Phosphorimages were scanned and stored as gel files and then analyzed using ATLASIMAGE 1.01 software (CLONTECH). A default external background setting was used in conjunction with a background-based signal threshold to determine gene signal significance. Comparison of cDNA array images was performed using an average of all of the gene signals on the array (global normalization) to normalize the signal intensity between arrays. Messenger RNAs associated with GW182–mRNP complexes were considered significant if they were twofold or greater enriched over total cellular RNA. (A) Total cellular RNA; (B) mRNAs associated with GW182–mRNP complexes; (C) computer-generated overlay comparison of representative arrays A and B. Red bars represent species of mRNAs that were enriched twofold or greater as compared with total cellular RNA (genes listed in Table 2). Green bars indicate mRNAs that were present on one or both of the arrays but were not enriched at least twofold.
Table 2.
mRNA targets enriched in the GW182 mRNP
| Fold enrichment | Gene name | GenBank |
|---|---|---|
| 17.50 | Glycoprotein hormone alpha subunit precursor | V00518 |
| 16.00 | 60S ribosomal protein L6 (RPL6); TAX-responsive enhancer element binding protein 107 (TAXREB107); neoplasm-related protein C140 | X69391 |
| 12.00 | Metalloproteinase inhibitor 1 precursor (TIMP1); erythroid potentiating activity (EPA); fibroblast collagenase inhibitor | X03124 |
| 8.00 | Activated RNA polymerase II transcriptional coactivator p15; PC4 | U12979 |
| 8.00 | cAMP-dependent transcription factor ATF-4; DNA-binding protein TAXREB67; cAMP-response element binding protein (CREB2) | D90209 |
| 5.00 | Serum- and glucocorticoid-regulated serine/threonine protein kinase (SGK) | AJ000512 |
| 5.00 | PTPCAAX1 nuclear tyrosine phosphatase (PRL-1) | U48296 |
| 5.00 | Neuroleukin (NLK); glucose-6-phosphate isomerase (GPI); phosphoglucose isomerase (PGI); phosphohexose isomerase (PHI) | K03515 |
| 4.00 | Proliferating cyclic nuclear antigen (PCNA); cyclin | M15796 |
| 3.00 | Prothymosin alpha (ProT-alpha; PTMA) | M26708 |
| 3.00 | 40S ribosomal protein S19 (RPS19) | M81757 |
| 3.00 | Ubiquitin-conjugating enzyme E2 H10; ubiquitin-protein ligase; ubiquitin carrier protein | U73379 |
| 3.00 | Serine/threonine-protein kinase PCTAIRE 1 (PCTK1) | X66363 |
| 3.00 | ALG-2 calcium-binding protein | AF035606 |
| 3.00 | Mitochondrial matrix protein P1 precursor; p60 lymphocyte protein; chaperonin homolog; HUCHA60; heat shock protein 60 (HSP-60); HSPD1 | M34664 |
| 2.75 | Alpha1 catenin (CTNNA1); cadherin-associated protein; alpha E-catenin | D13866 |
| 2.50 | Transcription factor AP-2 (TFAP2; AP2TF) | M36711 |
| 2.33 | Acyl-CoA-binding protein (ACBP); diazepam binding inhibitor (DBI); endozepine (EP) | M14200 |
Northern Blot Analysis
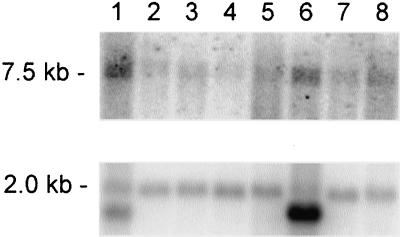
Northern blot analysis using a human multiple tissue blot probed with the 1.4 kb labeled fragment derived from clone KIAA1460, showed highest expression in heart (Figure12). In addition, expression of the GW182 gene was observed in all tissues suggesting that this protein is ubiquitously expressed. The actin control shows that mRNA loading is quite comparable among the lanes. IIF studies of a number of tissues and cell lines (HeLa, 3T3, MOLT-4, human chondrocyte and osteoblast, chicken fibroblast) suggest that GWBs are variably but widely expressed in species as well as tissues and cell types (our unpublished results).
Figure 12.
Northern blot analysis of GW182 mRNA. The order of polyA+ RNA from human tissues was as follows (lanes 1–8): heart, whole brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas. The membrane was probed with a_Cla_I/_Eco_RI fragment from the clone KIAA1460 (top panel) and reprobed using actin β-actin cDNA (bottom panel).
DISCUSSION
We have identified a novel protein that we have named GW182 because of its multiple glycine-tryptophan repeats found throughout the protein and the predicted and observed molecular mass of 182 kDa. By IIF, antibodies to native and recombinant GW182 appear to identify a cytoplasmic domain that we have tentatively named GWBs. The contention that GW182 resides in a distinct cytoplasmic domain is supported by the observation that antibodies to GW182 did not colocalize with markers of the Golgi complex, endosomes, lysosomes, or peroxisomes. Although there may appear to be a few overlaps with of some of these cytoplasmic markers, the overall IIF patterns observed with these markers and the IIF staining by the prototype human serum or rabbit anti-GW182p are clearly very different. Another possibility is that the GWBs might represent aggresomes or sites of protein degradation. However, our experiments using proteosome inhibitors or a target of the SCF ubiquitin ligase–mediated pathway for protein degradation suggests that GWBs are not aggresomes (Garcia-Mata et al., 1999;Johnston et al., 2000). Last, the observation that GWBs are not colocalized to sites marked by anticlathrin suggests that these bodies are probably not involved in clathrin-dependent vesicle transport (Hirst and Robinson, 1998). These conclusions are supported by IEM data showing that anti-GW182 identifies a granular, electron-dense body in the cytoplasm of HeLa cells that is the same size as that identified by IIF and colocalization studies.
The data showing that the GW182 protein binds to several different mRNA species is quite intriguing. The lack of colocalization of GWBs with various known cytoplasmic organelles may not be surprising considering the interaction of GW182 with mRNA. Perhaps the GW182 protein is involved in stabilizing and/or regulating translation and/or storing mRNAs. Several mRNAs as members of mRNP complexes have been identified using antibodies to RNA proteins such as ELAV/Hu, elf-4E, and poly(A) binding proteins (Tenenbaum et al., 2000; Keene, 2001). ELAV/Hu proteins are involved in stabilizing and/or regulating translation of early response gene transcripts expressed primarily in neurons (Keene, 1999; Brennan and Steitz, 2001). Analysis of the transcripts revealed some limited sequence similarity, suggesting that a common theme exists among these transcripts that allows recognition by ELAV/Hu proteins. Interestingly, ELAV/Hu proteins along with a fraction of polyadenylated mRNA have been observed in discrete clusters within the cytoplasm of medulloblastoma cells (Antic and Keene, 1998). It is possible that transcripts may be clustered in vivo with similar fates and/or functions. For example, upon retinoic acid treatment of P19 cells to induce neuronal differentiation, the population of mRNAs identified in the ELAV/Hu protein complex changed; additional mRNAs with AU-rich elements known to be upregulated in neurons were found (Tenenbaum et al., 2000).
At the present time, we have not identified common functional or structural features among the mRNAs that are immunoprecipitated by the anti-GW182 antibodies. Some of the proteins are known to be key components of the cell cycle, whereas others have no immediately apparent relationship with one other. New approaches to elucidate unique structural features of mRNA are being developed and may shed light on the role of GW182 and related proteins. We are pursuing in situ hybridization using the mRNAs identified in this study and appropriate controls to determine if these mRNAs are located in GWBs.
In general the various functions of the cell are highly coordinated and regulated, and among these functions, gene transcription and translation are also highly regulated. The movement of mRNA transcripts into the cytoplasm and subsequent coordinated translation after appropriate endogenous or exogenous signals would necessarily involve highly sophisticated levels of control (Keene, 1999; Tenenbaum et al., 2000; Keene, 2001). For example, some mRNAs may be required for rapid protein production and may be stored or protected from degradation. Because the cytoplasmic domains observed by IIF using the index human sera and rabbit anti-GW182 antibody failed to colocalize with markers of several cytoplasmic organelles and because of our observation that specific mRNAs are bound by GW182, it seems reasonable to conclude that GW182 is involved in mRNA expression and that this occurs in a defined cytoplasmic domain that we identify as GWBs. It might also be that GW182 is involved in degradation of mRNA but GWBs do not colocalize with the SCF ubiquitin ligase-mediated pathway for protein degradation.
Along these lines of discussion, the GWBs described in this study may be related to mRNA-associated particles described in other systems, particularly neuronal cells (Triedge et al., 1991; Miyashiro_et al._, 1994; Knowles et al., 1996; Martone_et al._, 1996; Gazzaley et al., 1997; Racca_et al._, 1997; Bassell et al., 1998; Steward_et al._, 1998). In oligodendrocytes, injected fluorescent-labeled myelin basic protein mRNA localized to granules that had a radius of 0.6–0.8 μm, contained elongation factors, rRNA and other mRNAs. It was concluded that these granules may represent supramolecular complexes of a translational unit (Barbarese et al., 1995; Ainger et al., 1997). These observations are consistent with the size of HeLa and HEp-2 cell granules observed in our study and suggest that GW182 marks a subset of these cytoplasmic bodies that contain a distinct subset of mRNAs. RNA granules containing mRNAs have also been described in fibroblasts (Ross et al., 1997) and mast cells (Dvorak and Morgan, 2000; Dvorak and Morgan, 2001). In other organisms, similar observations have been made for the 3′ UTR of ASH1 mRNA in budding yeast (Hazelrigg, 1998) and the bcd and_PROSPERO_ mRNA in Drosophila oocytes (Wang and Hazelrigg, 1994; Oleynikov and Singer, 1998; Bassell and Oleynikov, 1999). In studies of fibroblasts, the β-actin mRNA is localized at the leading edge of the cell, and the 3′UTR binds a protein called zipcode-binding protein 1 (ZBP-1; Ross et al., 1997). GW182 does not have significant sequence similarity to ZBP-1, but they both have a RRM and NLS in common. Although the putative NLS motif suggests that ZBP-1 and GW182 proteins may be able to enter the nucleus, both proteins are predominantly found in the cytoplasm. In the case of GW182, the evidence showed that cytoplasmic staining was observed using both the index human serum and the rabbit antibodies to the GW182 recombinant protein. In addition, the partial protein of GW182 tagged with GFP was localized to the cytoplasm, specifically to GWBs, and no expression was detected in the nucleus. Last, no differences were observed in the IIF staining pattern with formaldehyde or other fixation was used (our unpublished data compared methanol/acetone fixation with paraformaldehyde/Triton X-100 fixation). Interestingly, HuB, HuC, and HuD isoforms are mainly cytoplasmic in neurons with a small amount of protein observed with the nucleus as well (Gao and Keene, 1996). Therefore, we believe that the potential NLS motif may either be nonfunctional or is blocked in cells grown under our conditions and requires a stimulus.
It is likely that GW182 protein is one member of a family of proteins residing in GWBs. As observed with many organelles and vesicles in the cell, it is simplistic to think that one protein comprises these complex structures. It is possible that more than one alternatively spliced protein product from the GW gene may reside in GWBs. First, the existence of two EST clones missing exon 10 (accession nos.BF169182 and W80996, from mouse and human, respectively) supports this possibility (Table 1). Second, IP of extracts from radiolabeled HeLa cells by the index human serum reveals the presence of two proteins of 180 and 50 kDa. This could either be due to cross-reactivity or the association of these two proteins in a complex. We are currently pursuing isolating other proteins that are associated with GWBs.
Considering the sequence characteristics of GW182, it is not surprising that we were able to show that GW182 is likely a phosphoprotein. GWBs are heterogeneous in size and vary in number in individual cells. The variation in size and number may be related to the physiological state of the cell and the stage of the cell cycle. Detailed studies are currently underway to confirm this preliminary observation.
The significance of the GW repeats in GW182 is unclear. Although ESTs that contain this motif are in the database, to date no other mammalian proteins with this motif have been reported. A clue to the function of GW repeats may come from studies of bacteria such as Listeria monocytogenes, Staphylococcus caprae, and_Erysipelothrix rhusiopathiae,_ where it has been suggested that proteins bearing these repeats play an important role in anchoring bacterial proteins to the surface of the cell (Makino et al., 1998) and anchoring bacteria to target cells (Braun et al., 1997; Milohanic et al., 2001). The mode of binding is not clearly understood, but is thought to occur via interaction with lipoteichoic acid or with specific cell surface proteins. For example, the protein internalin B (InIB) produced by L. monocytogenes, is a key protein in promoting adherence and entry of the bacteria to host cells during infection. InIB has a C-terminal cell wall–anchoring domain containing 80-amino-acid GW repeats (Braun_et al._, 1997). Deletion of this domain impaired adherence to host cells, whereas addition of GW repeats improved the binding to the cell surface. Similarly, the autolysin Ami in L. monocytogenes contains a N-terminal catalytic domain and a C-terminal domain that is homologous to the GW domain in InIB but contains 8 GW modules arranged in tandem (Braun et al., 1997; Milohanic et al., 2001). Similar six to eight tandem repeat motifs have been described in the S. caprae atlC gene product (Alligent et al., 2001) and the surface protective antigen (SpaA) of E. rhusiopathiae (Makino et al., 1998). Of interest, the 6 GW repeats in _S. caprae_occur in the fibronectin-binding domain of the atlC protein. This may have relevance to the GW182 protein in mammalian cells that might bind to cytoskeletal elements to promote stabilization or movement of certain RNA species to their physiological target. The role of GW182 in binding or adherence of RNA or other proteins to cytoskeletal components or membrane moieties requires further study.
Summary
We have identified a novel protein GW182 that contains multiple glycine/tryptophan repeats and a classical RNA-binding domain at the C terminus. GW182 appears to react with a subset of mRNAs and localizes to cytoplasmic domains that do not belong to any of the known conventional organelles in the cytoplasm. Hence, we tentatively identified the cytoplasmic domain marked by antibodies directed against GW182 as GW bodies or GWBs.
ACKNOWLEDGMENTS
The authors acknowledge the assistance of Joan Miller, Carol L. Peebles, and Dr. Doug Zochodne (University of Calgary) for arranging the collection of the serum samples. Malcolm R. Wood provided valuable assistance with immunogold electron microscopy in The Scripps Research Institute Core Microscopy Unit. This work was supported by in part by the Canadian Institutes for Health Research grant MOP-38034 and the National Institutes of Health (NIH) grants CA56956 and AI39645, and CA79907 and AI46451. This work was also supported in part by the Sam and Rose Stein Charitable Trust and NIH grant M01RR00833 to the General Clinical Research Center of the Scripps Research Institute. M.J.F. hold the Arthritis Society Chair at the University of Calgary. This is publication 14253-MEM from the Scripps Research Institute.
Abbreviations used:
EEA1
early endosome antigen 1
IIF
indirect immunofluorescence
IP
immunoprecipitation
NLS
nuclear localization signal
ORF
open reading frame
RRM
RNA recognition motif
snRNP
small nuclear ribonucleoprotein
TnT
in vitro transcription and translation
Footnotes
REFERENCES
- Ainger K, Avossa D, Diana AS, Barry C, Barbarese E, Carson JH. Transport and localization elements in myelin basic protein mRNA. J Cell Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alligent J, Aubert S, Dyke KGH, Solh NE. Staphylocoocus caprae strains carry determinants known to be involved in pathogneicity: a gene encoding an autolysin-binding fibronectin and the icaoperon involved in biofilm formation. Infect Immun. 2001;69:712–718. doi: 10.1128/IAI.69.2.712-718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Antic D, Keene JD. Messenger ribonucleoprotein complexes containing human ELAV proteins: interactions with cytoskeleton and translational apparatus. J Cell Sci. 1998;111:183–197. doi: 10.1242/jcs.111.2.183. [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Barbarese E, Koppel DE, Deutscher MP, Smith CL, Ainger K, Morgan F, Carson JH. Protein translation components are colocalized in granules in oligodendrocytes. J Cell Sci. 1995;108:2781–2790. doi: 10.1242/jcs.108.8.2781. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Zhang HL, Byrd AL, Femino AM, Singer RH, Taneja KL, Liftshitz LM, Herman IM, Kosik KS. Sorting of beta actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell G, Oleynikov Y. The travels of mRNAs through all cells large and small. FASEB J. 1999;13:447–454. doi: 10.1096/fasebj.13.3.447. [DOI] [PubMed] [Google Scholar]
- Bestagno M, Cerino A, Riva S, Ricotti GCBA. Improvements of Western blotting to detect monoclonal antibodies. Biochem Biophys Res Comm. 1987;146:1509–1514. doi: 10.1016/0006-291x(87)90820-5. [DOI] [PubMed] [Google Scholar]
- Braun L, Dramsi S, Dehoux P, Bierne H, Lindahl G, Cossart P. InIB: an invasion protein of Listeria monocytogeneswith a novel type of surface association. Mol Microbiol. 1997;25:285–294. doi: 10.1046/j.1365-2958.1997.4621825.x. [DOI] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JD, O'Donnell WT, Tenenbaum ST, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of Fragile X-Mental Retardation protein (FMRP)-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Chan EKL, Francoeur A-M, Tan EM. Epitopes, structural domains, and asymmetry of amino acid residues in SS-B/La nuclear protein. J Immunol. 1986;136:3744–3749. [PubMed] [Google Scholar]
- Chan, E.K.L., and Fritzler, M.J. (1998). Golgins: coiled-coil-rich proteins associated with the Golgi complex. Electronic J. Biotechnol. 1,http://ejb.ucv.cl/content/vol1/issue2/full/1.
- Chan EKL, Tan EM. Human autoantibody-reactive epitopes of SS-B/La are highly conserved in comparison with epitopes recognized by murine monoclonal antibodies. J Exp Med. 1987;166:1627–1640. doi: 10.1084/jem.166.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad PA, Smart EJ, Ying YS, Anderson RGW, Bloom GS. Caveolin cycles between plasma membrane caveolae and the Golgi complex by microtubule-dependent and microtubule-independent steps. J Cell Biol. 1995;131:1421–1433. doi: 10.1083/jcb.131.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak AM, Morgan ES. Ultrastructural immunogold cytochemistry with autoimmune human sera and an antibody to uridine implicate human mast cell granules in RNA biology. Histochem J. 2000;32:685–696. doi: 10.1023/a:1004119500801. [DOI] [PubMed] [Google Scholar]
- Dvorak AM, Morgan ES. Ribosomes and secretory granules in human mast cells: close associations demonstrated by staining with a chelating agent. Immunol Rev. 2001;179:94–101. doi: 10.1034/j.1600-065x.2001.790110.x. [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan K B, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphoylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Fritzler MJ, Hamel JC, Ochs RL, Chan EKL. Molecular characterization of two human autoantigens: unique cDNAs encoding 95- and 160-kD proteins of a putative family in the Golgi complex. J Exp Med. 1993;178:49–62. doi: 10.1084/jem.178.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzler MJ, Lung C-C, Hamel JC, Griffith K, Chan EKL. Molecular characterization of golgin-245: a novel Golgi complex protein containing a granin signature. J Biol Chem. 1995;270:31262–31268. doi: 10.1074/jbc.270.52.31262. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Vitalla J, Matteson J, Carlsson SR. Cloning of cDNAs encoding human lysosomal membrane glycoproteins, h-lamp-1 and h-lamp-2. Comparison of their deduced amino acid sequences. J Biol Chem. 1988;263:18920–18928. [PubMed] [Google Scholar]
- Gao FB, Keene JD. Hel-N1/Hel-N2 proteins are bound to poly(A)+mRNA in granular RNP structures and are implicated in neuronal differentiation. J Cell Sci. 1996;109:579–589. doi: 10.1242/jcs.109.3.579. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata R, Bebok Z, Sorscher EJ, Sztul ES. Characterization and dynamics of aggresome formation by a cytososlic GFP-chimera. J Cell Biol. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzaley A, Benson D, Huntley G, Morrison J. Differential subcellular regulation of NMDAR1 protein and mRNA in dendrites of dentate. Neuron. 1997;14:433–445. doi: 10.1523/JNEUROSCI.17-06-02006.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith K, Chan EKL, Hamel JC, Miyach K, Fritz MJ. Molecular characterization of a novel 97 kDa Golgi complex autoantigen recognized by autoimmune antibodies from patients with Sjögren's syndrome. Arthritis Rheum. 1997;40:1693–1702. doi: 10.1002/art.1780400920. [DOI] [PubMed] [Google Scholar]
- Hazelrigg T. The destinies and destinations of RNAs. Cell. 1998;95:451–460. doi: 10.1016/s0092-8674(00)81613-x. [DOI] [PubMed] [Google Scholar]
- Hirst J, Robinson MS. Clathrin and adaptors. Biochim Biophys Acta. 1998;1404:173–193. doi: 10.1016/s0167-4889(98)00056-1. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes. A cellular response to misfolded proteins. J Cell Biol. 2000;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo K, Taketani S, Yokota S, Osumi T, Hashimoto T. The 70-kDa peroxisomal membrane protein is a member of the Mdr (P-glycoprotein)-related ATP-binding protein superfamily. J Biol Chem. 1990;265:4534–4540. [PubMed] [Google Scholar]
- Keene JD. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci USA. 1999;96:14085–14090. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and proteome. Proc Natl Acad Sci USA. 2001;98:7018–7024. doi: 10.1073/pnas.111145598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C. elegansand identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Basell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lahtinen U, Hellman U, Wernstedt C, Saraste J, Pettersson RF. Molecular cloning and expression of a 58-kDa cis-Golgi and intermediate compartment protein. J Biol Chem. 1996;271:4031–4037. doi: 10.1074/jbc.271.8.4031. [DOI] [PubMed] [Google Scholar]
- Lawe DC, Patki V, Heller-Harrison R, Lambright D, Corvera S. The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3-phosphate and Rab5 binding—critical role of this dual interaction for endosomal localization. J Biol Chem. 2000;275:3699–3705. doi: 10.1074/jbc.275.5.3699. [DOI] [PubMed] [Google Scholar]
- Lerner MR, Steitz JA. Snurps and scyrps. Cell. 1981;25:298–300. doi: 10.1016/0092-8674(81)90047-7. [DOI] [PubMed] [Google Scholar]
- Lindstedt AD, Hauri HP. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol Biol Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan J, Marti A, Sutterluty H, Gstaiger M, Wirbelauer C, Krek W. Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45(SKP2): evidence for evolutionarily conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 1998;17:368–383. doi: 10.1093/emboj/17.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi D, Soldati T, Riederer MA, Goda Y, Zerial M, Pfeffer SR. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio JP, Brake B, Banting G, Howell KE, Braghetta P, Stanley KK. Identification, sequencing and expression of an integral membrane protein of the trans Golgi network (TGN38) Biochem J. 1990;270:97–102. doi: 10.1042/bj2700097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Yamamoto K, Murakami S, Shirahata T, Uemura K, Sawada T, Wakamoto H, Morita T. Properties of repeat domain found in a novel protective antigen, SaA, of Erysipelothrix rhusiopathiae. Microbiol Pathogen. 1998;25:101–109. doi: 10.1006/mpat.1998.0216. [DOI] [PubMed] [Google Scholar]
- Martone ME, Pollock JA, Jones YZ, Ellisman MH. Ultrastructural localization of dendrite messenger RNA in adult rat hippocampus. J Neurosci. 1996;16:7437–7446. doi: 10.1523/JNEUROSCI.16-23-07437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milohanic E, Jonquières R, Cossart P, Berche P, Gaillard J-L. The autolysin Ami contributes to the adhesion of Listeria monocytogenesto eukaryotic cells via its cell wall anchor. Mol Microbiol. 2001;39:1212–1224. doi: 10.1111/j.1365-2958.2001.02208.x. [DOI] [PubMed] [Google Scholar]
- Miyashiro K, Dichter M, Eberwine J. On the nature and differential distribution of mRNAs in hippocampal neurites: implications for neuronal functioning. Proc Natl Acad Sci USA. 1994;91:10800–10804. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu FT, Callaghan JM, Steele-Mortimer HS, Parton RG, Campbell PL, McCluskey J, Yeo JP, Tock EPC, Toh BH. EEA1, an early endosomal protein. J Biol Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Darnell RB. Paraneoplastic neurologic disease antigens: RNA-binding proteins and signaling proteins in neuronal degeneration. Annu Rev Neurosci. 2001;24:239–262. doi: 10.1146/annurev.neuro.24.1.239. [DOI] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH. RNA localization: different zipcodes, same postman? Trends Cell Biol. 1998;8:381–383. doi: 10.1016/s0962-8924(98)01348-8. [DOI] [PubMed] [Google Scholar]
- Orci L, Tagaya M, Amherdt A, Perrelett A, Donaldson JG, Lippincott-Schwartz J, Klausner RD, Rothman JE. Brefeldin A, a drug that blocks secretion, prevents the assembly of non-clathrin-coated buds on Golgi cisternae. Cell. 1991;64:1183–1195. doi: 10.1016/0092-8674(91)90273-2. [DOI] [PubMed] [Google Scholar]
- Racca C, Gardiol A, Triller A. Dendritic and postsynaptic localization of glycine receptor RNA. J Neurosci. 1997;17:1691–1700. doi: 10.1523/JNEUROSCI.17-05-01691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer G, Raska I, Tan EM, Scheer U. Human autoantibodies: probes for nucleolus structure and function. Virch Arch B Cell Pathol. 1987;54:131–143. doi: 10.1007/BF02899205. [DOI] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RL. Characterization of a β-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak S, Scheonroth L, Senecal J-L, Fritzler MJ. Early endosome antigen 1: an autoantigen associated with neurological diseases. J Invest Med. 1999;47:311–318. [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Song KS, Tang Z, Li S, Lisanti MP. Mutational analysis of the properties of caveolin-1. A novel role for the C-terminal domain in mediating homo-typic caveolin-caveolin. J Biol Chem. 1995;272:4398–4403. doi: 10.1074/jbc.272.7.4398. [DOI] [PubMed] [Google Scholar]
- Steward O, Wallaca CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:1–20. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Tan EM. Autoantibodies in pathology and cell biology. Cell. 1991;67:841–842. doi: 10.1016/0092-8674(91)90356-4. [DOI] [PubMed] [Google Scholar]
- Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci USA. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triedge H, Fremeau J, Weinstock PH, Arancio O. Dendritic location of neural BC1 RNA. Proc Natl Acad Sci USA. 1991;88:2093–2097. doi: 10.1073/pnas.88.6.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Muhlen CA, Tan EM. Autoantibodies in the diagnosis of systemic rheumatic disease. Semin Arth Rheum. 1995;24:323–358. doi: 10.1016/s0049-0172(95)80004-2. [DOI] [PubMed] [Google Scholar]
- Waite RL, Sentry JW, Stenmark H, Toh BH. Autoantibodies to a novel early endosome antigen 1. Clin Immunol Immunopath. 1998;86:81–87. doi: 10.1006/clin.1997.4455. [DOI] [PubMed] [Google Scholar]
- Wang S, Hazelrigg T. Implications for bcd mRNA localization from spatial distribution of exu protein in Drosophilaoogenesis. Nature. 1994;369:400–403. doi: 10.1038/369400a0. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kobayashi R, Galaktionov K, Beach D. p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S phase kinase. Cell. 1995;82:915–922. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]