Nucleoporins Nup98 and Nup214 Participate in Nuclear Export of Human Immunodeficiency Virus Type 1 Rev (original) (raw)
Abstract
Human immunodeficiency virus type 1 (HIV-1) Rev contains a leucine-rich nuclear export signal that is essential for its nucleocytoplasmic export mediated by hCRM1. We examined the role of selected nucleoporins, which are located in peripheral structures of the nuclear pore complex and are thought to be involved in export, in Rev function in human cells. First, we found that upon actinomycin D treatment, Nup98, but not Nup214 or Nup153, is able to translocate to the cytoplasm of HeLa cells, demonstrating that Nup98 may act as a soluble factor. We further showed that Rev can recruit Nup98 and Nup214, but not Nup153, to the nucleolus. We also found that the isolated FG-containing repeat domains of Nup98 and Nup214, but not those of Nup153, competitively inhibit the Rev/RRE-mediated expression of HIV. Taken together, the recruitment of Nup98 and Nup214 by Rev and the competitive inhibition exhibited by their NP domains demonstrate direct participation of Nup98 and Nup214 in the Rev-hCRM1-mediated export.
The active export of macromolecules from the nucleus is mediated by short peptide signals that act as ligands for nuclear export factors. The best-studied example is human immunodeficiency virus type 1 (HIV-1) Rev, which is responsible for the export of the Rev-responsive element containing viral RNA. The export of Rev of HIV-1 and other lentiviruses is mediated via its leucine-rich nuclear export signals (NES) (13, 29). Functional Rev-like NES have also been identified in several other proteins (for a recent review, see reference 23). Several NES-containing proteins such as Rev have been shown to interact directly with the human protein hCRM1 (14, 18, 42). This interaction is inhibited by the antibiotic leptomycin B (LMB) (51). hCRM1 belongs to the importin β protein superfamily (15, 21). Some of the importin β-like proteins have been shown to be involved in the nucleocytoplasmic transport. The known and predicted characteristics of the family include (i) preferential binding to Ran GTPase in its GTP-bound form and (ii) binding to repeated FG-containing motifs characteristic of some nucleoporins (NP repeats). hCRM1 and a related protein, CAS (26), have been implicated as direct receptors mediating the nuclear export of NES-containing proteins and importin α, respectively, and hence define a subset of proteins in the importin β superfamily termed exportins. The key feature of exportins is their ability to cooperatively bind their export substrate and GTP-bound Ran. The ternary complex is then believed to interact with some components of the nuclear pore complex (NPC) via an exportin, thus delivering the export cargo to the site of translocation. Upon translocation of the ternary complex through the NPC, GTP hydrolysis occurs on Ran, resulting in the dissociation of the complex and release of the export substrate into the cytoplasm. In this model, exportins act as transient linkers between the export cargo and the NPC, which are regulated by the nucleotide-bound status of Ran (14, 26).
The NPC possesses extensive peripheral structures extending into the nuclear interior (for recent reviews, see references 32 and 33) including filamentous basket-like structures that have been proposed to provide docking sites for nuclear export intermediates (2). Vertebrate NP-repeat-containing nucleoporins Nup98, Nup153, and Nup214 have been assigned to peripheral NPC structures (4, 8, 37) and have been implicated in the nuclear export (2, 3, 35, 36, 48). In the Saccharomyces cerevisiae two-hybrid system, it has been shown that FG-containing repeat domains of different nucleoporins interact with the Rev NES via CRM1 with various efficiencies, suggesting that nucleoporins play a role in NES-mediated export (5, 16, 17, 31, 45).
In this report, we addressed the roles of Nup98, Nup153, and Nup214 for Rev function in human cells. We show that Nup98 and Nup214 are not solely stationary components of the NPC since (i) the presence of actinomycin D leads to cytoplasmic translocation of Nup98 and (ii) Rev can associate with Nup98 and Nup214, but not Nup153, outside the NPC. The codistribution of Rev with Nup98 and Nup214 as well as with hCRM1 and the competitive inhibition of Rev function by the isolated nucleoporin repeat domains of Nup98 and Nup214 suggest that Nup98 and Nup214, but not Nup153, are the major downstream partners of hCRM1 and play a direct role in the export of Rev from the nucleolus to the cytoplasm.
MATERIALS AND METHODS
Recombinant DNA.
The molecular clones of HIV-1 pRev(−)HIV (pNL4-3fB [12, 40]) and pRev(−)RRE(−).CTE [previously named pNL43Rev(−)RRE(−).S (53)] as well as the expression vectors for the tagged green (green fluorescent protein [GFP]) and blue (blue variant of GFP [BFP]) variants of Rev and NES(−)Rev (43) have been described elsewhere. The plasmids expressing GFP-tagged proteins were prepared by insertion of the PCR-amplified coding sequences (CDS) into the _Nhe_I site of pCMV-GFPsg25 (44). The hybrid genes encode the N-terminal tripeptide Met-Ala-Ser followed by the respective protein starting at its second amino acid residue and the complete GFP. The untagged versions of these proteins were produced by providing a stop codon after their CDS in the above-described constructs, generating proteins with authentic C termini. The GFP-tagged and untagged NP domains of Nup98 and Nup153 contain the CDS from amino acids (aa) 2 to 494 of the human Nup98 protein (6), and aa 894 to 1475 of Nup153. The hemagglutinin (HA)-tagged Nup98 plasmid was based on p37R vector (46, 52) and included an N-terminal HA epitope (Tyr-Pro-Tyr-Asp-Val-Pro-Asp-Tyr-Ala) inserted between aa 1 and 2 of these proteins. HA-tagged Nup214-NP expression plasmid was a gift from J. Hauber. HA-tagged Nup153 plasmid (2) was obtained from B. Burke. To construct the expression plasmid for hCRM1, its CDS was PCR amplified using hCRM1 cDNA (15) and inserted into p37R vector as described above for HA-tagged plasmids. The GFP-NES plasmid encodes the GFP protein fused to the Rev NES and was a gift from E. Afonina and G. N. Pavlakis. The authenticity of the constructs was verified by double-strand sequencing.
Cell culture and transfections.
HLtat is a HeLa-derived cell line that constitutively produces HIV-1 Tat protein (39). Human 293 is an embryonic kidney cell line. Cells were maintained in Dulbecco modified Eagle medium (DMEM) containing 10% fetal calf serum plus penicillin and streptomycin (53). For microscopy of living cells, 60-mm-diameter plastic transfection plates were seeded in DMEM without phenol red. Cells were transfected by the calcium phosphate coprecipitation technique, using DNA purified on QIAGEN columns, and total protein was extracted (53). L3luc luciferase expression plasmid was included in the transfection mixtures, and the luciferase activity was measured (41). GFP fluorescence was measured in cell lysates with a CytoFluor fluorimeter (43). HIV-1 p24_gag_ antigen was measured by using commercial antigen capture assay kits (Cellular Products) (53). LMB was a generous gift from B. Wolff (51). Stock solutions of LMB were made at 0.5 mM in 1:1 dimethyl sulfoxide-isopropanol and stored at −70°C. Working solutions of 0.5 μM LMB in DMEM were kept at −20°C. For 293 and HLtat cells, LMB treatment was performed with 10 nM LMB for 4 h at 37°C.
Antibodies, immunofluorescence, and microscopy.
The rabbit polyclonal antibodies for Rev (10) and GFP (44) have been described elsewhere. The monoclonal antibody for the influenza virus HA epitope was obtained from BAbCo (Berkeley, Calif.). Rabbit antisera for hCRM1 were obtained from J. van Deursen and G. Grosveld. Affinity-purified rabbit antibodies for Xenopus Nup98 have been described elsewhere (36) and were a generous gift from M. Powers and D. Forbes. For indirect immunofluorescence, cells grown on plastic plates were fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature, made permeable by treatment with 0.1% Nonidet P-40 or 0.1% Triton X-100 in PBS for 10 min, and labeled with the appropriate antibodies as described previously (43). Integrity of the nuclei was confirmed by using goat polyclonal anti-lamin A antibodies (Santa Cruz Biotechnology) or, in living cells, a DNA-specific live fluorescent dye (Hoechst 33342). Cells were observed under appropriate illumination with a Zeiss Axiovert 135TV microscope. Digital images were acquired with a SenSys charge-coupled device camera (Photometrics) and processed with IPLab Spectrum software. Digital deconvolution was performed using the HazeBuster extension of IPLab Spectrum software. Measurements of GFP fusion proteins in cell lysates were performed as described previously (52).
RESULTS
Nup98 can translocate to the cytoplasm.
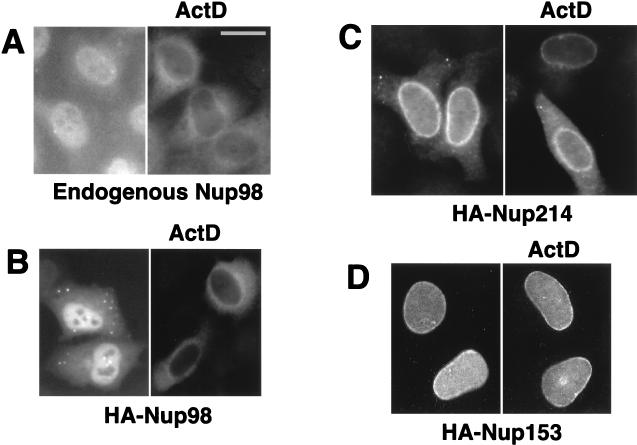
We studied the localization of Nup98, Nup153, and Nup214 in the human HeLa-derived HLtat cell line by using indirect immunofluorescence. We used an affinity-purified antibody raised against Xenopus Nup98 which has been shown to react with human Nup98 (36) to visualize endogenous Nup98 (Fig. 1A), and we used an anti-HA mouse monoclonal antibody to visualize the HA epitope-tagged transfected Nup98 (Fig. 1B). Both endogenous Nup98 and HA-tagged Nup98 show similar subcellular localization. They are found predominantly in the nucleoplasm, at the nuclear rim, and as dots in the cytoplasm and over the nucleus but are excluded from the nucleoli (Fig. 1A, left panel). The nuclear rim association is more prominent upon digital deconvolution microscopy (data not shown). Similar localization of endogenous Nup98 was found in cultured Xenopus cell line A6 with the same antibody, in agreement with published data (36).
FIG. 1.
Nup98, but not Nup153 and Nup214, is able to translocate to the cytoplasm. Endogenous Nup98 was visualized by using affinity-purified anti-Xenopus Nup98 serum (A), whereas HA-tagged Nup98, Nup214, and Nup153 proteins were detected with anti-HA antibody (B to D). HLtat cells were transiently transfected with 2 μg of the indicated HA-tagged nucleoporin expression plasmids (B to D) or not transfected (A). Some cells were treated with 2 μg of actinomycin D (ActD) per ml for 2 h. The indirect immunofluorescence and fluorescence microscopy were performed as described in Materials and Methods, and the images were filtered with a linear filter (3 × 3 kernel), using IPLab Spectrum software. Bar, 20 μm.
To examine the possible ability of Nup98 to shuttle between the nuclear and cytoplasmic compartments, we asked whether the presence of an inhibitor of transcription affects the localization of Nup98, as has been reported for several other shuttle proteins such as Rev (29). Interestingly, we found that the endogenous Nup98 (Fig. 1A) and HA-Nup98 (Fig. 1B) completely translocate from the nucleus to the cytoplasm of the HeLa cells upon treatment with actinomycin D in the absence or presence of the translation inhibitor cycloheximide. This translocalization was most prominent in confluent monolayers of HLtat cells, whereas we did not find this effect in A6 cells (not shown). These data demonstrate that Nup98 can be exported from the nucleus of HeLa cells.
Upon transient transfection of HLtat cells, HA-Nup214 predominantly localizes to the nuclear rim and can also be detected in the cytoplasm (Fig. 1B, left panel), in agreement with published data (25). The HA-tagged Nup153 (Fig. 1C, left panel) localizes to the nuclear rim and the nucleoplasm, as previously described (2). In contrast to Nup98, treatment with actinomycin D did not affect the localization of either Nup214 or Nup153 (Fig. 1B and C, right panels). These findings indicate that Nup98, in contrast to Nup153 and Nup214, is a dynamic factor rather than a stationary component of the NPC.
Rev recruits Nup98 and Nup214 to the nucleoli.
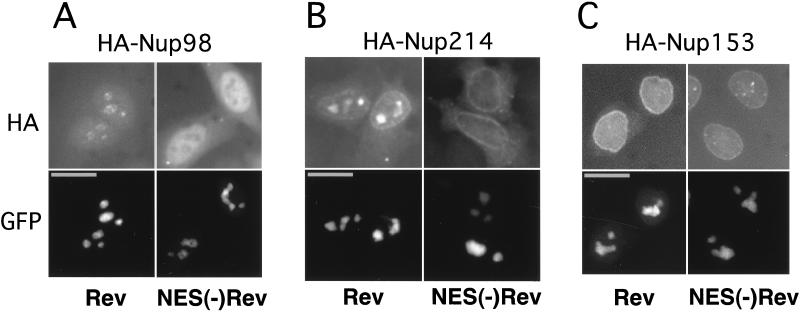
Next, we asked whether Rev can associate with intact nucleoporins Nup98, Nup153, and Nup214 in mammalian cells and thereby affect their subcellular localization. We expressed these HA-tagged nucleoporins in HLtat cells in the presence of GFP-tagged Rev or, as a control, the Rev mutant that lacks the NES [GFP-NES(−)Rev]. As expected from the previous studies, both Rev and NES(−)Rev localized predominantly to the nucleoli (Fig. 2, bottom panels). As shown in Fig. 2A, the presence of Rev resulted in the redistribution of Nup98 to the nucleoli. In contrast, the presence of NES(−)Rev had no effect on the localization of Nup98, which remained excluded from the nucleoli (Fig. 2A). Similarly, we found that Rev, but not NES(−)Rev, can recruit Nup214 to the nucleoli (Fig. 2B). In contrast to Nup98 and Nup214, Rev did not affect the localization of Nup153 (Fig. 2C). Western immunoblots and fluorescence measurements of GFP-tagged proteins confirmed the presence of comparable amounts of Rev and NES(−)Rev proteins, and similar results were obtained with untagged Rev proteins, which were visualized by indirect immunofluorescence (data not shown).
FIG. 2.
Rev recruits Nup98 and Nup214 to the nucleoli. HLtat cells were transfected with 0.5 μg of the HA-tagged nucleoporin expression plasmids in the presence of 1 μg of GFP-Rev or 1 μg of GFP-NES(−)Rev expression plasmid as indicated at the bottom of the figure. The GFP fluorescence and HA indirect immunofluorescence were detected in the same cells, as indicated to the left. Fluorescence microscopy was performed as described in the legend to Fig. 1. Bars, 20 μm.
We further found that in the presence of Rev, the isolated NP domains of Nup98 and Nup153 are also recruited to the nucleoli and that this localization is mediated by the NES of Rev and hCRM1, since it is interrupted by LMB (data not shown). The recruitment of NP domain of Nup153 is in contrast to our finding of the lack of intact Nup153 to codistribute with Rev (Fig. 2) and is most likely due to the generic ability of various NP domains to interact with hCRM1/Rev, as suggested by Neville et al. (31).
Taken together, these experiments demonstrate that Rev can recruit intact Nup98 and Nup214, but not Nup153, to the nucleoli. This finding demonstrates that both Nup98 and Nup214 are dynamic factors and, importantly, are able to associate with Rev outside the NPC. These results further suggest a role of Nup98 and Nup214 in Rev function.
Rev recruits hCRM1 to the nucleoli.
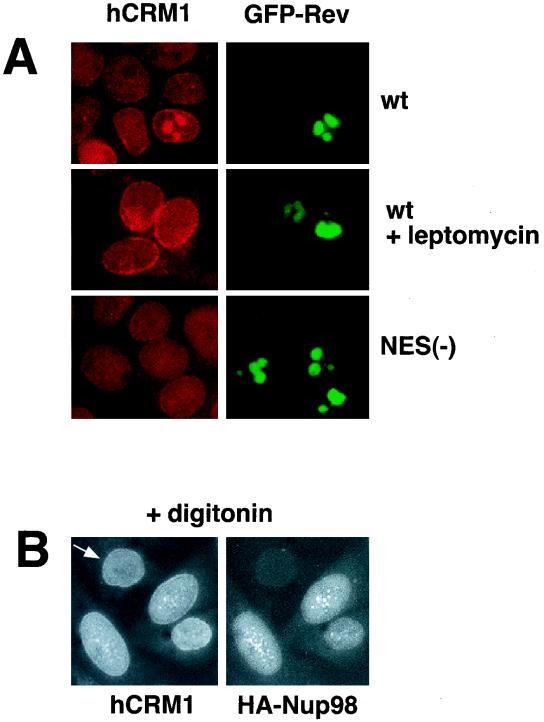
To examine the association of hCRM1 and HIV-1 Rev in mammalian cells, we studied whether the presence of Rev affects the localization of hCRM1. We transfected human 293 cells with the BFP-tagged Rev protein (44) or, as a control, the BFP-tagged NES(−)Rev protein. Endogenous hCRM1 was mostly found at the nuclear rim as well as within the nucleus (Fig. 3A), in agreement with published data (15), while both Rev and NES(−)Rev localized predominantly to the nucleoli (Fig. 3A, right panels). Importantly, in the cells that expressed Rev, but not NES(−)Rev, a significant fraction of hCRM1 was found in the Rev-containing nucleoli (left top panel). Such colocalization was observed in all Rev-expressing cells. Upon treatment with LMB, the effect of Rev on hCRM1 localization was completely abolished, whereas no effect of LMB on hCRM1 distribution was observed in cells that did not produce detectable amounts of Rev or that expressed the NES(−)Rev. Similar results were obtained with untagged Rev and NES(−)Rev, which were detected by indirect immunofluorescence as well as in studies using another cell line such as HLtat (data not shown). Taken together, these findings suggest that Rev associates with hCRM1 (Fig. 3A), which further allows the recruitment of Nup98 and Nup214 (Fig. 2) to the nucleoli.
FIG. 3.
hCRM1 codistributes with Rev and Nup98. (A) Human 293 cells were transfected with 1 μg of BFP-Rev (wild type [wt]) or BFP-NES(−)Rev [NES(−)] expression plasmid as indicated to the right, and treated with 6 nM LMB for 4 h. The endogenous hCRM1 was visualized by indirect immunofluorescence with rabbit anti-hCRM1 serum (15) and rhodamine red-conjugated goat anti-rabbit secondary antibodies as previously described (43). BFP fluorescence is shown pseudocolored in green. (B) HLtat cells were transfected with 0.5 μg of HA-Nup98 and subjected to double indirect immunofluorescence for HA-Nup98 and the endogenous hCRM1, as indicated. Before immunodetection, cells were extracted in situ with 0.004% digitonin in 0.3× PBS for 10 min at 4°C. Fluorescence microscopy was performed as described in the legend to Fig. 1.
We next addressed the question whether HA-Nup98 and hCRM1 codistribute in the absence of Rev. We found that these proteins colocalize in nucleoplasmic foci in transfected HLtat cells (Fig. 3B). Since both proteins showed diffuse nucleoplasmic localization (Fig. 1B and 3A), we performed in situ extraction with digitonin prior to immunostaining in order to remove the fraction of proteins that is loosely bound to nuclear structures. This treatment revealed an extensive dot-like pattern in which HA-Nup98 and hCRM1 colocalized, as shown in Fig. 3B. This dot-like pattern of hCRM1 was not observed in the absence of HA-Nup98 (Fig. 3B, indicated by an arrow). A similar but less-extensive colocalization pattern was obtained without digitonin treatment (data not shown). This result shows that Nup98 is able to colocalize with hCRM1 independent of Rev outside the NPC.
Competitive inhibition of Rev function by the isolated nucleoporin repeat domains of Nup98 and Nup214.
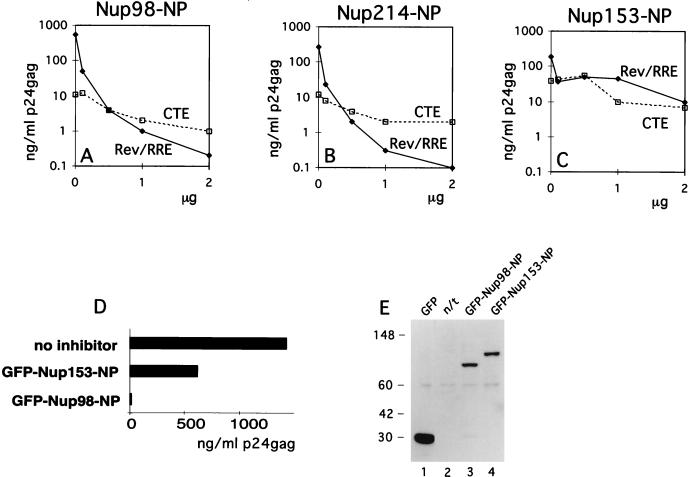
We then explored the effects of the isolated NP domains of Nup98, Nup214, and Nup153 on Rev function. We reasoned that exogenously expressed, isolated NP domains may be able to titrate soluble factors that interact with the respective endogenous nucleoporins and that this depletion might affect Rev-mediated HIV-1 expression, resulting in competitive inhibition. Therefore, a Rev-deficient molecular clone was cotransfected into human 293 cells in the presence of sufficient amounts of Rev necessary to achieve maximal HIV expression together with increasing amounts of plasmids expressing the NP domains of Nup98, Nup214, or Nup153. One day later, the cells were harvested and cell extracts were analyzed for Gag production as a measure of Rev-mediated HIV-1 expression. Rev function was potently inhibited by the NP domains of Nup98 (Fig. 4A) and Nup214 (Fig. 4B). The inhibition by these NP domains occurred to similar degrees and in a dose-dependent manner. In contrast, Nup153-NP inhibited Rev function ∼100-fold less (Fig. 4C). In another experiment, we obtained a similar ∼100-fold difference (Fig. 4D) in the inhibition of HIV-1 by Nup98-NP and Nup153-NP and we also confirmed that similar amounts of GFP-tagged proteins were present (Fig. 4E). Using the yeast two-hybrid system, Fritz and Green (16) showed strong interaction of all three NP domains with Rev. Interestingly, we found a differential effect of these three NP domains on Rev function reflecting intrinsic properties of these properties in mammalian cells. Despite the differences in size and the number of nucleoporin repeats (19 FG repeats within the 227-aa Nup214-NP versus 39 repeats within the 492-aa Nup98-NP), the extent of Rev inhibition was very similar for these two NP domains. The NP domain of Nup153 contains 25 FG repeats, which is comparable to those of Nup98 and Nup214. The different numbers of FG repeats probably do not account for the observed differential effects on Rev function. We conclude that the observed inhibition by Nup153-NP may reflect the ability of generic NP repeats to interfere with Rev (45), while the strong differential effect of the NP domains of Nup98 and Nup214 likely indicates the direct participation of these nucleoporins in the Rev pathway.
FIG. 4.
NP domains of Nup98 and Nup214 specifically affect the function of Rev. (A to C) Human 293 cells were transfected with 1 μg of Rev(−)HIV in the presence of 25 ng of the pBsRev expression plasmid (Rev/RRE) or 1 μg of the Rev(−)RRE(−).CTE Rev-independent HIV-1 (CTE). All transfections included 0.1 μg of L3luc, a luciferase expression vector. Increasing amounts of expression plasmid Nup98-NP (A), Nup214-NP (B), or Nup153-NP (C) were cotransfected (plotted on the x axis in micrograms). Gag production was measured in the lysates at one day posttransfection (p24_gag_ measured in nanograms per milliliter) and is plotted on the y axis. Results from a representative experiment are shown. Similar results were obtained in three independent transfection experiments. (D and E) 293 cells were cotransfected with 1 μg of Rev(−)HIV in the presence of 25 ng of pBsRev expression plasmid and in the absence or presence of the indicated GFP-tagged NP expression plasmids. HIV expression is determined by measuring Gag production and expressed as a percentage of the value obtained in the absence of the NP domains (D). Western immunoblot analysis (E) of the extracts shown in panel D. GFP-tagged proteins were detected by using rabbit anti-GFP serum for the experiment shown in panel D. The positions of protein markers (in kilodaltons) are shown to the left. The expected molecular masses of GFP fusion proteins were 27 kDa (GFP), 77 kDa (GFP-Nup98-NP), and 86 kDa (GFP-Nup153-NP). n/t, nontransfected cells.
As a control for the specificity of the inhibition, we used an otherwise isogenic Rev-RRE-deficient molecular clone of HIV-1 that contains the type D retrovirus posttranscriptional control element (CTE) necessary for expression (53). CTE has been shown to utilize an export pathway distinct from that of Rev-RRE (34, 38, 52) and involves the cellular TAP protein (22). We also used a luciferase mRNA transcribed from the HIV-1 long terminal repeat promoter, representing a transcript that does not depend on posttranscriptional regulation. The NP domains of all three nucleoporins tested affected the CTE-mediated expression of HIV-1 (Fig. 4A to C) and the luciferase expression (not shown) similarly but to a much lesser extent than the Rev-mediated HIV expression. Based on this comparison, we concluded that the observed dose-dependent inhibition of Rev-mediated expression by Nup98-NP and Nup214-NP was due to a preferential inhibition of Rev function.
The inhibition of Rev function induced by the NP domain of Nup98 can be counteracted by excess Rev.
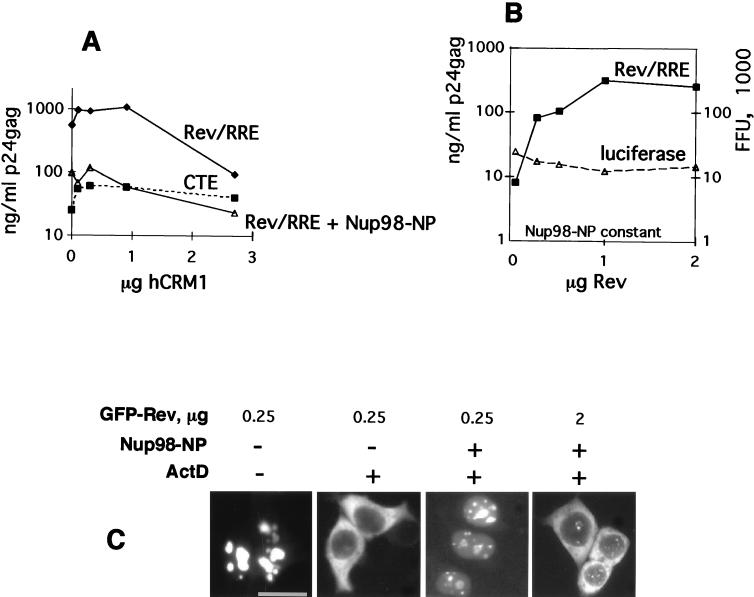
Since hCRM1 is the likely molecular link between Rev and the nucleoporins, we asked whether hCRM1 can outcompete the inhibition mediated by the NP domain of Nup98. We found that cotransfection of more than 1 μg of the hCRM1-producing plasmid preferentially inhibited the Rev-mediated HIV expression compared to expression of isogenic HIV-1 mRNAs that are controlled by the CTE (Fig. 5A). It is plausible that CRM1 is present at the optimal amounts required for nucleocytoplasmic export and that excess CRM1 disturbs this balance and negatively interferes with processes involving CRM1-mediated export. In addition, we found that the presence of excess hCRM1 did not counteract the Nup98-NP-induced inhibition of Rev-mediated expression (Fig. 5A). One explanation for the observed lack of complementation is that the amount of exogenous hCRM1 required for the titration of Nup98-NP interferes with cellular CRM1-mediated export and thereby with HIV-1 expression per se. Alternatively, Nup98-NP may bind more strongly to the hCRM1-Rev complex than to free hCRM1. In this model, hCRM1-Rev complexes, but not free hCRM1, will preferentially compete for Nup98-NP domain, and the amount of Rev, but not the amount of CRM1, will determine the outcome of the experiment. To test this model, we cotransfected constant amounts of the Rev(−)HIV-1 clone and of the Nup98-NP expression plasmid necessary to achieve ∼100-fold inhibition. The transfection mixtures also contained increasing amounts of a Rev expression vector. The amount of Rev used has been shown to allow maximal HIV expression in the absence of inhibitor (data not shown). As shown in Fig. 5B, the increasing amounts of Rev led to a dose-dependent rescue of HIV-1 expression. Similar results were obtained when a wide range of Rev and Nup98-NP concentrations was used (data not shown). The increase in HIV expression is specific, since increasing amounts of Rev did not affect the CTE-mediated expression of HIV-1 (data not shown) or luciferase expression (Fig. 5B). Therefore, our data favor the model in which Nup98-NP interacts better with Rev-CRM1 than with CRM1 alone.
FIG. 5.
Rev counteracts the inhibitory effect of the NP of Nup98. (A) 293 cells were transfected with 1 μg of Rev(−)HIV and 0.25 μg of pBsRev in the absence or presence of 0.25 μg of Nup98-NP. In parallel, transfections were done with 1 μg of Rev(−)RRE(−).CTE in the absence of Nup98-NP (CTE). Increasing amounts of hCRM1 expression plasmid were cotransfected (plotted on the x axis in micrograms). p24_gag_ expression was measured and is plotted on the y axis as raw values (in nanograms per milliliter of lysate). Similar results were obtained in three independent transfection experiments. Panels A to C show results from representative experiments. (B) Rev counteracts the effect of Nup98-NP on HIV-1 expression. Human 293 cells were transfected with 1 μg of Rev(−)HIV (Rev/RRE) in the presence of 0.2 μg of Nup98-NP. Transfections included the luciferase plasmid. Increasing amounts of pBsRev plasmid, starting at 25 ng, were cotransfected (plotted on the x axis in micrograms). p24_gag_ expression (in nanograms per milliliter of lysate) is plotted on the left y axis as raw values. Luciferase values are plotted on the right y axis as multiples of 103 firefly units (FFU). (C) 293 cells were cotransfected with 0.25 or 2 μg of GFP-Rev expression plasmid in the presence or absence of 0.2 μg of Nup98-NP plasmid, as indicated. At day 1 posttransfection, some cells were treated with actinomycin D (+) in the presence of cycloheximide as indicated at the top, and GFP fluorescence was visualized as described previously (43).
We further demonstrated that Nup98’s NP domain directly affects the nuclear export of Rev by studying the subcellular localization of a fluorescent GFP-tagged Rev protein in living cells. A constant amount of the Nup98-NP expression vector necessary to inhibit HIV expression was cotransfected with either small or large amounts of a GFP-tagged Rev expression plasmid. One day later, the cells were treated with actinomycin D and the nuclear export of GFP-tagged Rev protein was visualized directly. Figure 5C shows that when Rev is present at small amounts, its export is abolished by Nup98-NP. In contrast, when present at large amounts, Rev counteracts this inhibition and regains its ability to translocate to the cytoplasm. The same results were obtained by using untagged Rev proteins and indirect immunofluorescence (not shown). To further support the specificity of the effect of Nup98-NP, we also studied the export of a GFP hybrid containing the NES of Rev as well as the export of RanBP1, a protein we previously showed to contain a Rev-like NES which determines its cytoplasmic accumulation (52). Nup98-NP inhibited the export of both NES-GFP and RanBP1, demonstrating that this inhibition reflects the interference of the NP domain with CRM1 function in general, and this interference is independent of the nature of its ligand.
Under the conditions leading to the inhibition of Rev’s nuclear export by Nup98-NP, we did not observe reduced nuclear accumulation of Rev or NES(−)Rev or inhibition of the nuclear import of human glucocorticoid receptor and human hnRNP A1 protein (not shown). Therefore, the effect of the isolated Nup98-NP domain is clearly on export but not import, in agreement with published data (2, 35, 36). Taken together, the data provided in Fig. 5 show that competitive inhibition of Rev’s function by Nup98-NP is due to the competitive inhibition of Rev’s nuclear export.
DISCUSSION
In this study, we show that in HeLa cells Nup98 can translocate to the cytoplasm upon actinomycin D treatment and that Rev is able to recruit both Nup98 and Nup214, but not Nup153, to the nucleolus. We further provided evidence that the isolated NP domains of Nup98 and Nup214 are able to competitively inhibit Rev-mediated expression by interfering with Rev’s nuclear export. Taken together, these data suggest that Nup98 and Nup214, but not Nup153, are dedicated downstream partners of hCRM1 in human cells. The fact that the interaction of Rev and these nucleoporins is mediated via hCRM1 and our finding that hCRM1 associates with Rev via its nuclear export signal in the nucleoli further point to the nucleolus as a integral part necessary for the Rev function. hCRM1 has previously been shown to be a dynamic factor, able to relocalize from the nuclear envelope to the nucleoplasm in response to expression of Nup214-NP (15). The nucleolus has been implicated as part of normal hCRM1 route, based on the presence of hCRM in the nucleoli of Nup214-depleted embryos and based on its nucleolar relocalization upon treatment with actinomycin D in HtTA-1 cells (15). Since we did not observe actinomycin D-induced translocation of hCRM1 in 293 and HLtat cell lines (data not shown), the presence of the NES of Rev thus appears to be necessary and sufficient for hCRM1’s nucleolar recruitment. This observation indicates that hCRM1 associates with Rev on the normal route of both proteins.
The above findings led us to propose a model of participation of Nup98 and Nup214 in the hCRM1-mediated nuclear export of Rev. We speculate that transport intermediates assemble in the nucleoli or the nucleoplasm and that these intermediates include Rev, hCRM1, and Nup98 or Nup214. In this model, Nup98 and Nup214 act as soluble factors by targeting the preassembled Rev-hCRM1-nucleoporin complexes to the NPC.
Nup98 and Nup214 are proto-oncogenes that have been implicated in human myeloid leukemias. Chromosomal translocations leading to acute myeloid leukemia (AML) produced fusions of Nup98’s NP domain with HOXA9, a transcription factor potentially involved in myeloid differentiation (6, 30), or with a putative RNA helicase, DDX10 (1). The Nup98-HOXA9 fusion was predicted to act through its HOXA9 moiety by inhibition of HOXA9-mediated differentiation. Based on the predicted properties of DDX10, the Nup98-DDX10 protein was not expected to act on the level of transcription. Therefore, the presence of a NP moiety of Nup98 on both fusions indicates that it may contribute to AML in a transcription-independent manner. Nup214 is also activated by fusions of its NP domain to different genes unrelated to transcription initiation factors (49), leading to myeloid leukemias. This led us to speculate that Nup98 and Nup214 oncogenic fusions may contribute to these malignancies by the same underlying mechanism, mediated through their NP domains.
The NES-containing proteins IκBα and inhibitor of protein kinase A (PKI) regulate the activity and the nucleocytoplasmic transport of NF-κB and protein kinase A, respectively. IκBα and PKI have been suggested to negatively regulate signal transduction by clearing their respective substrates from the nucleus to ensure timely signal termination (11, 16, 50). Mitogen-activated protein kinase kinase (MAPKK) has been shown to control the nuclear accumulation of mitogen-activated protein kinase (MAPK) by a similar mechanism (20, 24). The nuclear export of MAPKK is mediated by a Rev-type NES that has been recently shown to interact with hCRM1 (18). Disruption of the NES within constitutively active MAPKK has been shown to increase its ability to induce transformation and led to a dramatic increase of activated MAPK in the nucleus (19). Therefore, the inhibition of NES-mediated export may be predicted to result in enhanced or constitutive signals through the above transducers. In our experimental model, the nucleoporin repeat domains of Nup98 and Nup214 alone or as a fusion to a neutral moiety, GFP, resulted in extremely strong and specific inhibition of NES-mediated export, by hCRM1-bridged titration of a low-abundance NES substrate. The configuration of the Nup98-NP within GFP hybrid protein used in this report directly mimics the above-described Nup98 oncogenic fusion proteins. Therefore, the phenotypes of NP proteins described here may reflect the contribution of the NP moiety to the phenotypes of Nup98 and Nup214 oncogenic fusion proteins. If there is an analogy between our experimental system and primary leukemia cells, these oncoproteins may cause unregulated signal transduction through NF-κB, protein kinase A, or MAPK, contributing to AML. In support of this model, constitutive activation and MAPK and MAPKK is sufficient for cell transformation (7, 9, 27, 28) and has been implicated in AML (47).
ACKNOWLEDGMENTS
We thank J. Hauber, E. Afonina, G. N. Pavlakis, D. Forbes, M. Powers, G. Grosveld, and J. van Deursen for reagents and J. Bear for technical assistance. We are grateful to A. Gragerov, G. N. Pavlakis, and E. Izaurralde for critically reading the manuscript.
This research was sponsored by the National Cancer Institute, DHHS, under contract with ABL.
ADDENDUM IN PROOF
We showed by a cell fusion assay that both Nup98 and Nup153 were able to shuttle between nuclei. Nevertheless, Nup153 could not be recruited to the nucleoi by Rev (Fig. 2). Recently, Bogerd et al. (H.P. Bogerd, A. Echarria, T. M. Ross, and B. R. Cullen, J. Virol. **72:**8627–8635, 1998) also found that NP-Nup214 has a differential effect on Rev/RRE- versus CTE-controlled expression.
REFERENCES
- 1.Arai Y, Hosoda F, Kobayashi H, Arai K, Hayashi Y, Kamada N, Kaneko Y, Ohki M. The inv(11)(p15q22) chromosome translocation of de novo and therapy-related myeloid malignancies results in fusion of the nucleoporin gene, NUP98, with the putative RNA helicase gene, DDX10. Blood. 1997;89:3936–3944. [PubMed] [Google Scholar]
- 2.Bastos R, Lin A, Enarson M, Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boer J, Bonten-Surtel J, Grosveld G. Overexpression of the nucleoporin CAN/NUP214 induces growth arrest, nucleocytoplasmic transport defects, and apoptosis. Mol Cell Biol. 1998;18:1236–1247. doi: 10.1128/mcb.18.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boer J M, van Deursen J M, Croes H J, Fransen J A, Grosveld G C. The nucleoporin CAN/Nup214 binds to both the cytoplasmic and the nucleoplasmic sides of the nuclear pore complex in overexpressing cells. Exp Cell Res. 1997;232:182–185. doi: 10.1006/excr.1997.3502. [DOI] [PubMed] [Google Scholar]
- 5.Bogerd H P, Fridell R A, Madore S, Cullen B R. A novel cellular co-factor for HIV-1 Rev. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 6.Borrow J, Shearman A M, Stanton V P, Jr, Becher R, Collins T, Williams A J, Dube I, Katz F, Kwong Y L, Morris C, Ohyashiki K, Toyama K, Rowley J, Housman D E. The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat Genet. 1996;12:159–167. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- 7.Brunet A, Pages G, Pouyssegur J. Constitutively active mutants of MAP kinase kinase (MEK1) induce growth factor-relaxation and oncogenicity when expressed in fibroblasts. Oncogene. 1994;9:3379–3387. [PubMed] [Google Scholar]
- 8.Cordes V C, Reidenbach S, Kohler A, Stuurman N, van Driel R, Franke W W. Intranuclear filaments containing a nuclear pore complex protein. J Cell Biol. 1993;123:1333–1344. doi: 10.1083/jcb.123.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowley S, Paterson H, Kemp P, Marshall C J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 10.D’Agostino D M, Ciminale V, Pavlakis G P, Chieco-Bianchi L. Intracellular trafficking of the human immunodeficiency virus type 1 Rev protein: involvement of continued rRNA synthesis in nuclear retention. AIDS Res Hum Retroviruses. 1995;11:1063–1072. doi: 10.1089/aid.1995.11.1063. [DOI] [PubMed] [Google Scholar]
- 11.Fantozzi D A, Harootunian A T, Wen W, Taylor S S, Feramisco J R, Tsien R Y, Meinkoth J L. Thermostable inhibitor of cAMP-dependent protein kinase enhances the rate of export of the kinase catalytic subunit from the nucleus. J Biol Chem. 1994;269:2676–2686. [PubMed] [Google Scholar]
- 12.Feinberg M B, Jarrett R F, Aldovini A, Gallo R C, Wong-Staal F. HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell. 1986;46:807–817. doi: 10.1016/0092-8674(86)90062-0. [DOI] [PubMed] [Google Scholar]
- 13.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 14.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 15.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fritz C C, Green M R. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr Biol. 1996;6:848–854. doi: 10.1016/s0960-9822(02)00608-5. [DOI] [PubMed] [Google Scholar]
- 17.Fritz C C, Zapp M L, Green M R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda M, Gotoh I, Adachi M, Gotoh Y, Nishida E. A novel regulatory mechanism in the mitogen-activated protein (MAP) kinase cascade. Role of nuclear export signal of map kinase kinase. J Biol Chem. 1997;272:32642–32648. doi: 10.1074/jbc.272.51.32642. [DOI] [PubMed] [Google Scholar]
- 20.Fukuda M, Gotoh Y, Nishida E. Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J. 1997;16:1901–1908. doi: 10.1093/emboj/16.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 23.Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- 24.Jaaro H, Rubinfeld H, Hanoch T, Seger R. Nuclear translocation of mitogen-activated protein kinase kinase (MEK1) in response to mitogenic stimulation. Proc Natl Acad Sci USA. 1997;94:3742–3747. doi: 10.1073/pnas.94.8.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraemer D, Wozniak R W, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutay U, Bischoff F R, Kostka S, Kraft R, Gorlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 27.Mansour S J, Candia J M, Gloor K K, Ahn N G. Constitutively active mitogen-activated protein kinase kinase 1 (MAPKK1) and MAPKK2 mediate similar transcriptional and morphological responses. Cell Growth Differ. 1996;7:243–250. [PubMed] [Google Scholar]
- 28.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Vande Woude G F, Ahn N G. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 29.Meyer B E, Meinkoth J L, Malim M H. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals. J Virol. 1996;70:2350–2359. doi: 10.1128/jvi.70.4.2350-2359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura T, Largaespada D A, Lee M P, Johnson L A, Ohyashiki K, Toyama K, Chen S J, Willman C L, Chen I M, Feinberg A P, Jenkins N A, Copeland N G, Shaughnessy J D., Jr Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12:154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 31.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 32.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 33.Pante N, Aebi U. Molecular dissection of the nuclear pore complex. Crit Rev Biochem Mol Biol. 1996;31:153–199. doi: 10.3109/10409239609106583. [DOI] [PubMed] [Google Scholar]
- 34.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjöld M-L, Dahlberg J E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powers M A, Forbes D J, Dahlberg J E, Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powers M A, Macaulay C, Masiarz F R, Forbes D J. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol. 1995;128:721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radu A, Moore M S, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 38.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz S, Felber B K, Benko D M, Fenyö E M, Pavlakis G N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sodroski J, Goh W C, Rosen C, Dayton A, Terwilliger E, Haseltine W. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature. 1986;321:412–417. doi: 10.1038/321412a0. [DOI] [PubMed] [Google Scholar]
- 41.Solomin L, Felber B K, Pavlakis G N. Different sites of interaction for Rev, Tev, and Rex proteins within the Rev-responsive element of human immunodeficiency virus type 1. J Virol. 1990;64:6010–6017. doi: 10.1128/jvi.64.12.6010-6017.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 43.Stauber R, Gaitanaris G A, Pavlakis G N. Analysis of trafficking of Rev and transdominant Rev proteins in living cells using green fluorescent protein fusions: transdominant Rev blocks the export of Rev from the nucleus to the cytoplasm. Virology. 1995;213:439–454. doi: 10.1006/viro.1995.0016. [DOI] [PubMed] [Google Scholar]
- 44.Stauber R H, Carney P, Horie K, Hudson E A, Tarasova N I, Gaitanaris G A, Pavlakis G N. Development and applications of enhanced autofluorescent protein mutants. BioTechniques. 1998;24:462–471. doi: 10.2144/98243rr01. [DOI] [PubMed] [Google Scholar]
- 45.Stutz F, Izaurralde E, Mattaj I W, Rosbash M. A role for nucleoporin FG repeat domains in export of human immunodeficiency virus type 1 Rev protein and RNA from the nucleus. Mol Cell Biol. 1996;16:7144–7150. doi: 10.1128/mcb.16.12.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tabernero C, Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J Virol. 1996;70:5998–6011. doi: 10.1128/jvi.70.9.5998-6011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Towatari M, Iida H, Tanimoto M, Iwata H, Hamaguchi M, Saito H. Constitutive activation of mitogen-activated protein kinase pathway in acute leukemia cells. Leukemia. 1997;11:479–484. doi: 10.1038/sj.leu.2400617. [DOI] [PubMed] [Google Scholar]
- 48.van Deursen J, Boer J, Kasper L, Grosveld G. G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J. 1996;15:5574–5583. [PMC free article] [PubMed] [Google Scholar]
- 49.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol Cell Biol. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wen W, Harootunian A T, Adams S R, Feramisco J, Tsien R Y, Meinkoth J L, Taylor S S. Heat-stable inhibitors of cAMP-dependent protein kinase carry a nuclear export signal. J Biol Chem. 1994;269:32214–32220. [PubMed] [Google Scholar]
- 51.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleocytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 52.Zolotukhin A S, Felber B K. Mutations in the nuclear export signal of human ran-binding protein RanBP1 block the Rev-mediated posttranscriptional regulation of human immunodeficiency virus type 1. J Biol Chem. 1997;272:11356–11360. doi: 10.1074/jbc.272.17.11356. [DOI] [PubMed] [Google Scholar]
- 53.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]