Activation of the grp78 and grp94 Promoters by Hepatitis C Virus E2 Envelope Protein (original) (raw)
Abstract
The hepatitis C virus E1 and E2 envelope proteins are targeted to the endoplasmic reticulum, but instead of being secreted, they are retained in a pre-Golgi compartment, at least partly in a misfolded state. Since secretory proteins which are retained in the endoplasmic reticulum frequently can activate the transcription of intraluminal chaperone proteins, we measured the effect of the E1 and E2 proteins on the promoters of two such chaperones, GRP78 (BiP) and GRP94. We found that E2 but not E1 protein activates these two promoters, as assayed by a reporter gene system. Furthermore, E2 but not E1 protein induces the synthesis of GRP78 from the endogenous cellular gene. We also found that E2 but not E1 protein expressed in mammalian cells is bound tightly to GRP78. This association may explain the ability of E2 protein to activate transcription, since GRP78 has been postulated to be a sensor of stress in the endoplasmic reticulum. Since overexpression of GRP78 has been shown to decrease the sensitivity of cells to killing by cytotoxic T lymphocytes and to increase tumorigenicity and resistance to antitumor drugs, this activity of E2 protein may be involved in the pathogenesis of hepatitis C virus-induced diseases.
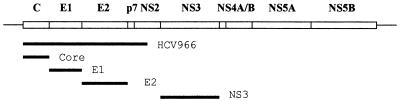
Hepatitis C virus (HCV) is a member of the Flavivididae family and causes the majority of non-A, non-B viral hepatitis in developed nations (10). In contrast to most RNA viruses, HCV causes chronic infection in the majority of those infected. In addition to being at risk for chronic hepatitis and cirrhosis, these chronically infected people also show an increased susceptibility to hepatocellular carcinoma. The viral genome is a single-stranded RNA approximately 9.5 kb in length (10, 23). There is a single open reading frame just over 9 kb in length, which codes for a polyprotein that is proteolytically processed into at least 10 polypeptides. The structural proteins, comprising the capsid (core) protein, the envelope proteins E1 and E2, and possibly p7, are present in that order at the N-terminal end of the polyprotein (Fig. 1). These proteins are released from each other by the signal peptidase in the endoplasmic reticulum (ER), to which the polyprotein is targeted by a signal sequence in the core domain (14). The final result is that both the E1 and E2 proteins end up as luminal ER proteins, with their C termini anchored in the ER membrane.
FIG. 1.
Map of the HCV genome, with the untranslated regions shown as lines and the open reading frame shown as a box divided into the final protein products. The thick lines indicate the regions included in the various expression plasmids used in this study.
The E1 and E2 proteins can interact with each other to form noncovalently linked dimers, and it is presumed that these dimers form the functional subunit of the viral envelope (8, 12, 21). However, when expressed in cultured cells, E1-E2 dimers are formed both slowly and inefficiently (7–9). Instead, heterogeneous complexes of disulfide-linked E1 and E2 are formed. These complexes appear to be aggregates of misfolded proteins that are probably incompetent for viral morphogenesis. The reason for the formation of these aggregates is unclear. One possibility is that they result from overexpression in cultured cells. However, it is also possible that this represents a mechanism for HCV to downregulate its replication rate so as to avoid immune surveillance and/or cytopathic effects. Indeed, electron microscopy of hepatocytes from HCV-infected people has revealed dilated ER with fluffy material within the lumen, consistent with large protein aggregates (30).
It is known that in mammalian cells the accumulation of misfolded proteins within the ER activates an intracellular signaling pathway known as the ER stress response. The presence of misfolded proteins is sensed by the cell and leads to the transcriptional activation of a subset of genes, including those encoding ER luminal chaperone proteins, such as GRP78 (BiP) and GRP94 (15). This feedback response presumably assists the cell in folding and secreting the misfolded proteins. Since the E1 and E2 proteins can accumulate as misfolded aggregates in the ER, we tested if expression of these proteins would lead to increased transcription from the grp78 and grp94 promoters. Our results show that E2 but not E1 protein activates transcription from these two promoters and that E2 but not E1 protein is stably associated with GRP78.
MATERIALS AND METHODS
Plasmids.
The plasmids used for expressing HCV proteins contain the cytomegalovirus immediate-early promoter and are derived from the plasmid pCMV6 (2). The HCV-derived inserts were amplified by PCR of cDNA clones of HCV-1 (4, 25), using appropriate primers. The HCV966 clone expresses codons 1 through 966, the core clone expresses codons 1 through 191, the NS3 clone expresses codons 1027 through 1711, the E1 clone expresses codons 192 through 382, and the E2 clone expresses codons 383 through 715. The last two plasmids also contain the tissue plasminogen activator signal peptide fused to the HCV sequences at the N terminus (2), to allow direction of the expressed proteins into the ER. The reporter plasmids contain the chloramphenicol acetyltransferase (CAT) gene under the control of either the grp78 or grp94 promoter and were kindly provided by A. Lee, University of Southern California (13, 22). The control CAT reporter plasmid under the control of the herpes simplex virus type 1 thymidine kinase (tk) promoter was kindly provided by B. M. Peterlin, University of California, San Francisco (28a).
Transient and stable transfections.
For transient-expression experiments, HuH-7 human hepatoma cells were transfected by the calcium phosphate method (29). Each 60-mm-diameter plate of cells was transfected with 5 μg of each plasmid. After 2 days, the cells were harvested and assayed for CAT activity by thin-layer chromatography. Triplicate plates were used for each experiment. For stable expression of E1 or E2 protein, CHO cells were cotransfected with the appropriate expression plasmid and a plasmid expressing dihydrofolate reductase, and stable transfectants were selected by growth in medium containing hypoxanthine-aminopterin-thymidine. Clones were evaluated for E1 and E2 protein expression by Western blotting, and one high expresser each was picked for subsequent analysis.
Immunofluorescence, Western blotting, and immunoprecipitation.
For immunofluorescence, transfected cells were fixed, permealized with methanol, and exposed first to goat antibody to GRP78 (1:100 dilution; obtained from Santa Cruz Biochemicals) and then to fluorescein-labeled rabbit antibody to goat immunoglobulin G (1:40 dilution; obtained from Sigma). To localize the transfected cells, E1 or E2 protein was similarly detected by immunofluorescence (with a 1:200 dilution of murine monoclonal antibody 3D5/C3 to E1 protein or monoclonal antibody 3E5-1 to E2 protein and a 1:50 dilution of rhodamine-labeled rabbit antibody to mouse immunoglobulin G [obtained from Dako]) (25).
For immunoprecipitation of E2 protein, CHO cells stably expressing E2 protein were lysed with 1% Triton X-100 in phosphate-buffered saline, pH 7.4. After two rounds of centrifugation at 100,000 × g for 45 min each, the cleared lysates were divided into equal aliquots and incubated at 4°C with one of two monoclonal antibodies against E2 protein (3E5-1, which recognizes a linear epitope, or TE5/H7, which recognizes a conformational epitope), with a monoclonal antibody against GRP78 (SPA827 from StressGen), or with no antibody. After 1 h, protein A-Sepharose 4B beads (Sigma) were added, and the mixture was incubated for another hour with gentle rocking. Following washing four times with 1% Triton X-100 in phosphate-buffered saline (pH 7.4), the beads were resuspended in sodium dodecyl sulfate (SDS) sample buffer and boiled for 3 min. The supernatants were electrophoresed on SDS–10 to 20% polyacrylamide gels and then transferred to a nitrocellulose membrane. The membrane was analyzed by Western blotting with rabbit antibodies to GRP78 (StressGen SPA826), using the alkaline phosphatase (Promega) detection system. Immunoprecipitation of E1 protein was performed similarly with CHO cells stably expressing E1 protein, except that monoclonal antibody 3D5/C3 to E1 protein was used.
Partial purification of E1 and E2 proteins.
E1 protein was extracted from stably transfected CHO cells with 1% Triton X-100 in 50 mM Tris-HCl, pH 8.0. The extract was cleared by a 45-min centrifugation at 10,000 × g and loaded on a Galanthus nivalis agglutinin (GNA) lectin column. After washing with 1 M NaCl in buffer A (0.1% Triton X-100 in 20 mM sodium phosphate, pH 6.8), the E1 protein was eluted with 1 M methyl-α-d-mannopyranoside in buffer A. The E1-containing eluate was dialyzed against buffer A, and E1 protein was further concentrated by passing the dialysate through a Sepharose 4B column (Pharmacia) eluted with 0.5 M NaCl in buffer A. The E1-containing fraction was analyzed by electrophoresis on SDS–10 to 20% polyacrylamide gels, followed by Western blotting for GRP78 as detailed above or for E1 protein with monoclonal antibody 3D5/C3. Purification of E2 protein was performed in a similar manner with stably transfected CHO cells. In addition, one aliquot of the crude total extract was incubated with 10 mM each ATP and MgCl2 (Mg-ATP) for 15 min before being loaded onto the GNA column.
RESULTS
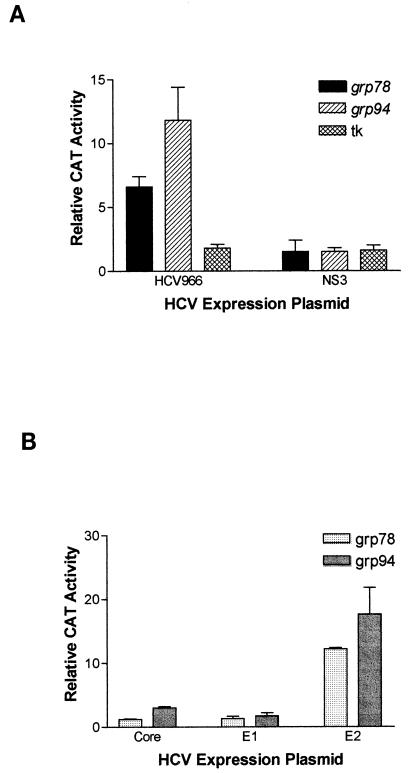
To determine the effect of HCV envelope proteins on the grp78 and grp94 promoters, we used a transient-transfection assay with HuH-7 human hepatoma cells. One plasmid expresses the structural proteins of HCV, including core, E1, E2, and p7, as well as a fragment of the nonstructural protein NS2 (HCV966 in Fig. 1). The other plasmid contains the CAT reporter gene driven by the grp78 or grp94 promoter. As the control plasmid for basal CAT expression, pUC19 was used instead of the expression plasmid. As seen in Fig. 2A, expression of the HCV structural proteins induced a strong induction of CAT expression from both the grp78 and grp94 promoters. This cannot be a nonspecific effect of the expression plasmid we used, since the same vector expressing the HCV NS3 protein did not have any effect on these promoters (Fig. 2A). Furthermore, the HCV structural proteins do not have a global effect on gene expression, since they were not able to activate expression from the herpes simplex virus tk promoter (Fig. 2A).
FIG. 2.
(A) Effects of various HCV proteins on CAT expression driven by the grp78, grp94, or tk promoter in transient transfection into HuH-7 hepatoma cells. The results are normalized to CAT expression in the presence of pUC19 and are shown as the means and standard deviations from three transfections. (B) Effects of the individual HCV structural proteins on CAT expression driven by the grp78 or grp94 promoter in transient transfection into HuH-7 hepatoma cells. The results are normalized to CAT expression in the presence of pUC19 and are shown as the means and standard deviations from three transfections.
To determine which of the HCV structural proteins were affecting the grp78 and grp94 promoters, we used the same vector to drive the separate expression of each the individual proteins except p7. As shown in Fig. 2B, neither the core nor the E1 protein had a significant effect on the grp78 and grp94 promoters. In contrast, the E2 protein was able to induce both promoters >12-fold (Fig. 2B), while having no effect on the tk promoter (data not shown). Note that the p7-coding sequences are not present in this plasmid, thereby ruling out a role for p7. This effect of E2 is neither cell nor species specific, since it was also able to activate the grp78 promoter in HT1080 human fibrosarcoma cells (by 6.0-± 0.9-fold) and in AML12 murine hepatocytes (by 4.6-± 0.2-fold).
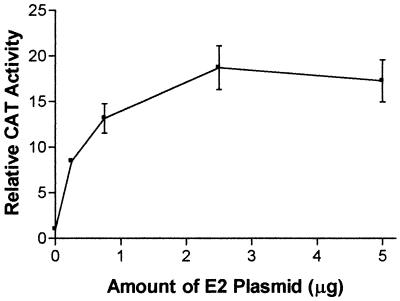
Because the E2 protein is expressed to high levels in the transfected cells, it may be argued that the activation of the grp78 and grp94 promoters seen here may be simply due to overload of the ER. However, the E1 protein is expressed at similarly high levels (24a) (Fig. 3), rendering this possibility unlikely. Furthermore, when a titration of the E2-expressing plasmid is performed, it is observed that even upon transfection of small amounts of this plasmid (0.25 μg/60-mm-diameter dish), there is a substantial activation of the grp94 promoter (Fig. 3). It should be noted that the total amount of transfected DNA was held constant in these experiments to prevent fluctuations in the transfection efficiency. Therefore, even relatively small amounts of E2 protein have the ability to activate transcription.
FIG. 3.
Titration of the amount of E2-expressing plasmid cotransfected with the CAT plasmid driven by the grp94 promoter into HuH-7 cells. The total amount of transfected plasmids was held constant by using pUC19. The results are normalized to CAT expression in the presence of pUC19 only and are shown as the means and standard deviations from three transfections.
In the experiments described above, E2 protein was targeted to the ER lumen. Therefore, it appears that E2 protein activates transcription indirectly by influencing an intracellular signaling pathway rather than acting in the nucleus. Nevertheless, since rare proteins have been described as being localized to both the ER and the nucleus (1, 6, 19), it was conceivable that a small fraction of the E2 protein had somehow escaped from the ER to effect a nuclear function. To rule out this possibility, we deleted the signal sequence from the E2-expressing plasmid. Indeed, this plasmid did not activate CAT expression from the grp78 promoter. Instead, for unknown reasons, it actually decreased expression (0.07 ± 0.01, relative to pUC19). Possibly the ectopic expression of E2 protein in the cytosol led to a cytotoxic effect.
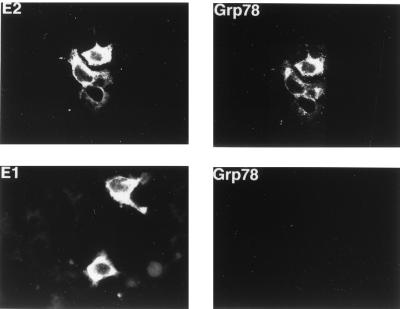
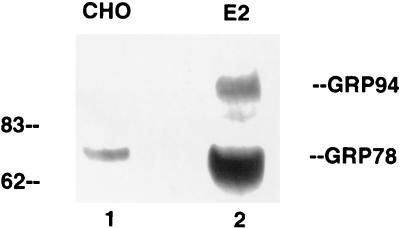
Since the CAT reporter plasmid represents an artificial gene expression system, it was important to determine if E2 protein also affected the expression of the endogenous cellular grp78 and grp94 genes. To answer this question, we first transiently transfected the E2-expressing plasmid into HuH-7 cells and used immunofluorescence to estimate the amount of GRP78 protein in these cells. As exemplified in Fig. 4, top panels, all cells expressing E2 protein also contained high levels of GRP78. In contrast, when the same experiment was performed with cells transfected with the E1-expressing plasmid, the cells that express E1 protein did not show elevated levels of GRP78 (Fig. 4, bottom panels). Therefore, E2 but not E1 protein is capable of inducing the synthesis of endogenous cellular GRP78. We were not able to evaluate GRP94 expression in these cells, since nonmurine antibody to GRP94 is not commercially available. Because immunofluorescence data are not quantitative, we also used Western blotting to assess the effect of E2 protein on GRP78 and GRP94 expression in a CHO cell line stably transfected with E2 protein. As seen in Fig. 5, this cell line expresses substantially more GRP78 and GRP94 than the parental CHO cells, thus confirming the immunofluorescence data.
FIG. 4.
Immunofluorescence detection of GRP78 and E2 or E1 protein expression in HuH-7 cells transiently transfected with either the E2 (top) or E1 (bottom) expression plasmid. A low level of GRP78 is detectable in the untransfected cells, as revealed in the original photographs.
FIG. 5.
Western blot detection of GRP78 and GRP94 in CHO cells (lane 1) or CHO cells stably expressing E2 protein (lane 2). Equal amounts of total cell extracts were separated by SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane for Western blotting against GRP78. The monoclonal antibody against GRP78 (StressGen SPA826) cross-reacts with GRP94. Numbers on the left are molecular weights in thousands.
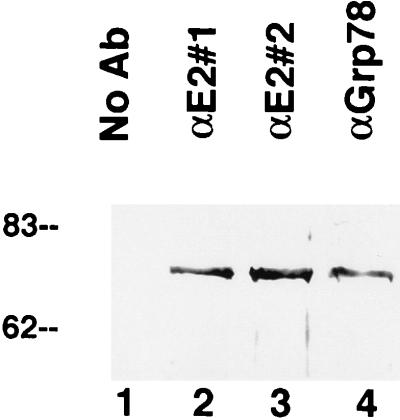
The immunofluorescence studies (Fig. 4) revealed a substantial overlap in the distribution of GRP78 and E2 protein in the cell. This colocalization raised the possibility that these two proteins may be stably bound to each other. This inference is consistent with the observation that GRP78 was coimmunoprecipitated from crude cell extracts when either of two antibodies to E2 protein was used (Fig. 6, lanes 2 and 3) but was not present when the antibody was omitted (Fig. 6, lane 1). The intensity of the band was similar to that of the band when anti-GRP78 antibody was used for immunoprecipitation (Fig. 6, lane 4), indicating that the majority of the GRP78 was associated with E2 protein, since in these experiments the antibodies were present in excess. Interestingly, antibodies to E1 protein did not precipitate GRP78 from E1-expressing cells (data not shown), suggesting that E1 protein does not bind strongly to GRP78.
FIG. 6.
Coimmunoprecipitation of E2 protein and GRP78. Proteins were immunoprecipitated from lysates of E2-expressing CHO cells with either of two different antibodies (Ab) to E2 protein (lanes 2 and 3) or an antibody to GRP78 (αGrp78) (lane 4), electrophoresed on an SDS-polyacrylamide gel, and probed for GRP78 by Western blotting. GRP78 was precipitated by all three antibodies; in contrast, no GRP78 was precipitated when the antibody was omitted (lane 1). Numbers on the left are molecular weights in thousands.
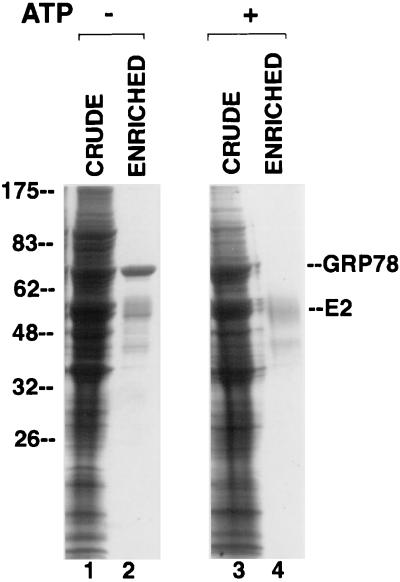
Our conclusion that E2 protein stably binds to GRP78 was strengthened by the observation that a protein migrating at the position expected for GRP78 copurified with E2 protein after lectin affinity chromatography on GNA (Fig. 7, lane 2). Indeed, N-terminal sequencing of this protein revealed the sequence EEEDKK, which corresponds to the known sequence of hamster GRP78 (27). It should be noted that GRP78 is not glycosylated (15, 24) and thus cannot bind directly to GNA. It is known that complexes of GRP78 and misfolded proteins are dissociated by Mg-ATP (18, 28). To determine if this is the case for GRP78 and E2 protein, we added Mg-ATP to lysates of cells expressing E2 protein before purifying the E2 protein on a GNA column. As seen in Fig. 7, lane 4, GRP78 was indeed absent from E2 protein purified in this manner. This absence was not due to protein degradation, since Mg-ATP had no apparent effect on the total proteins present in the crude lysate (Fig. 7, compare lane 3 to lane 1).
FIG. 7.
Copurification of E2 protein and GRP78 in the absence but not the presence of Mg-ATP. Lysates from E2-expressing CHO cells were electrophoresed on an SDS-polyacrylamide gel and stained with Coomassie blue, either before (lanes 1 and 3) or after (lanes 2 and 4) chromatography on a GNA lectin column. In lanes 3 and 4, the lysate was preincubated with Mg-ATP for 15 min. Numbers on the left are molecular weights in thousands.
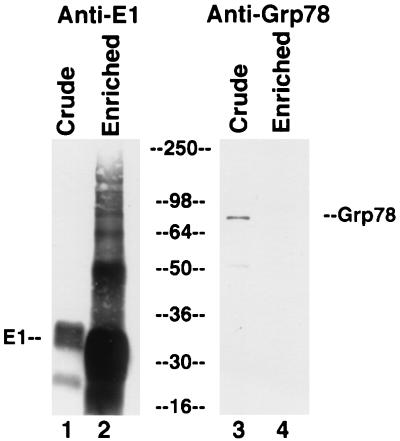
In contrast, GRP78 does not appear to be copurified with E1 protein, as no protein migrating at the expected position was copurified with E1 protein (data not shown). To confirm that GRP78 does not associate with E1 protein, we performed Western blotting for GRP78 with crude and partially purified E1 protein. As seen in Fig. 8, there was only a small amount of GRP78 in the crude lysate (lane 3), and this was lost upon enrichment for E1 protein (lane 4; contrast with the large enrichment for E1 protein in lane 2 compared to lane 1). These results confirm that there is a strong and stable association between GRP78 and E2 protein but not between GRP78 and E1 protein.
FIG. 8.
No evidence for copurification of E1 protein and GRP78. Lysates from E1-expressing CHO cells were electrophoresed on an SDS-polyacrylamide gel, transferred to a membrane, and probed for E1 protein (lanes 1 and 2) or GRP78 (lanes 3 and 4) by Western blotting, either before (lanes 1 and 3) or after (lanes 2 and 4) chromatography on a GNA lectin column. Numbers in the center are molecular weights in thousands.
DISCUSSION
We have shown that the HCV structural proteins can activate CAT gene transcription driven by both the grp78 and grp94 promoters. This property can be ascribed solely to the E2 envelope protein, which must be targeted to the ER lumen to show this effect. Furthermore, E2 protein can also increase expression of the endogenous cellular GRP78. Therefore, E2 protein must activate an intracellular signaling pathway from the ER to the nucleus to influence cellular transcription of ER-resident chaperone proteins.
Because the E1 and E2 proteins can form large disulfide-linked aggregates that accumulate in a pre-Golgi compartment (8, 9), we initially suspected that it is the presence of these presumably misfolded intraluminal proteins that induces grp78 and grp94 transcription. However, while E2 protein can activate transcription, E1 protein cannot. Yet it has been shown that E1 protein actually folds much slower than E2 protein and that its folding is dependent on E2 protein (17). Therefore, the ability to activate transcription may involve a specific property of E2 protein and not be simply related to the presence of large amounts of misfolded proteins in the ER. Thus, even if there is lower expression of E2 protein in the infected hepatocyte, it may be sufficient to increase chaperone protein expression. This inference is strengthened by our observation that even small amounts of the cotransfected E2-expressing plasmid have a _trans_-activation effect.
The mechanism by which E2 protein activates transcription is unknown. Interestingly, however, our data indicate that E2 protein, unlike E1 protein, is stably bound to GRP78. This binding may explain the activation of transcription by E2 protein. It has been hypothesized that GRP78 may itself be a sensor and transducer of ER stress, by binding to unfolded proteins and then undergoing a conformation change that eventuates in signal transduction into the nucleus (15, 20). If so, the stable binding of E2 protein to GRP78 would activate this signaling pathway. During the preparation of this paper, it was reported that both E1 and E2 proteins bind several intraluminal chaperones in the ER, including GRP78 (5). However, those experiments utilized pulse-chase analysis and hence may have detected transient rather than long-term associations. Furthermore, the amount of GRP78 bound to E1 protein appeared to be smaller than that bound to E2 protein (5). Therefore, it is possible that a critical threshold of the amount and duration of bound GRP78 must be reached before a signal is sent to the nucleus to increase chaperone expression.
The physiological and pathological significance of this novel effect of E2 protein is as yet unknown. Interestingly, GRP78 has been shown to protect cells against killing by cytotoxic T lymphocytes, to increase the tumorigenicity of transformed cells, and to increase resistance to antitumor drugs (3, 11, 16, 26). Therefore, it is possible that by increasing GRP78 expression, E2 protein plays a role in the chronicity of HCV infection and in HCV-related carcinogenesis. It is also noteworthy that the hepatitis B virus large surface protein also activates the grp78 and grp94 promoters (29). This common feature may explain how these two disparate viruses cause the identical spectrum of diseases. Further experiments will be needed to clarify the possible role of E2 protein in HCV pathogenesis.
ACKNOWLEDGMENTS
E.L. and Y.-L.F. contributed equally to this work.
We thank Greg Jensen for technical assistance, Jim Ou for critical reading of the manuscript, Amy Lee and Matija Peterlin for the CAT plasmids, and Seishi Murakami for helpful discussions.
This work was supported by NIH grant R01CA55578 to T.S.B.Y. E.L. was partially supported by a UCSF Pathology Department Research Fellowship.
REFERENCES
- 1.Burns K, Duggan B, Atkinson E A, Famulski K S, Nemer M, Bleackley R C, Michalak M. Modulation of gene expression by calreticulin binding to the glucocorticoid receptor. Nature. 1994;367:476–480. doi: 10.1038/367476a0. [DOI] [PubMed] [Google Scholar]
- 2.Chapman B S, Thayer R M, Vincent K A, Haigwood N L. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 1991;19:3979–3986. doi: 10.1093/nar/19.14.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chatterjee S, Cheng M F, Berger S J, Berger N A. Induction of M(r) 78,000 glucose-regulated stress protein in poly(adenosine diphosphate-ribose) polymerase- and nicotinamide adenine dinucleotide-deficient V79 cell lines and its relation to resistance to the topoisomerase II inhibitor etoposide. Cancer Res. 1994;54:4405–4411. [PubMed] [Google Scholar]
- 4.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 5.Choukhi A, Ung S, Wychowski C, Dubuisson J. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J Virol. 1998;72:3851–3858. doi: 10.1128/jvi.72.5.3851-3858.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dedhar S, Rennie P S, Shago M, Hagesteijn C Y, Yang H, Filmus J, Hawley R G, Bruchovsky N, Cheng H, Matusik R J, et al. Inhibition of nuclear hormone receptor activity by calreticulin. Nature. 1994;367:480–483. doi: 10.1038/367480a0. [DOI] [PubMed] [Google Scholar]
- 7.Deleersnyder V, Pillez A, Wychowski C, Blight K, Xu J, Hahn Y S, Rice C M, Dubuisson J. Formation of native hepatitis C virus glycoprotein complexes. J Virol. 1997;71:697–704. doi: 10.1128/jvi.71.1.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubuisson J, Hsu H H, Cheung R C, Greenberg H B, Russell D G, Rice C M. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubuisson J, Rice C M. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J Virol. 1996;70:778–786. doi: 10.1128/jvi.70.2.778-786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houghton M. Hepatitis C viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1035–1058. [Google Scholar]
- 11.Jamora C, Dennert G, Lee A S. Inhibition of tumor progression by suppression of stress protein GRP78/BiP induction in fibrosarcoma B/C10ME. Proc Natl Acad Sci USA. 1996;93:7690–7694. doi: 10.1073/pnas.93.15.7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanford R E, Notvall L, Chavez D, White R, Frenzel G, Simonsen C, Kim J. Analysis of hepatitis C virus capsid, E1, and E2/NS1 proteins expressed in insect cells. Virology. 1993;197:225–235. doi: 10.1006/viro.1993.1583. [DOI] [PubMed] [Google Scholar]
- 13.Li W W, Alexandre S, Cao X, Lee A S. Transactivation of the grp78 promoter by Ca2+ depletion. A comparative analysis with A23187 and the endoplasmic reticulum Ca(2+)-ATPase inhibitor thapsigargin. J Biol Chem. 1993;268:12003–12009. [PubMed] [Google Scholar]
- 14.Lin C, Lindenbach B D, Pragai B M, McCourt D W, Rice C M. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Little E, Ramakrishnan M, Roy B, Gazit G, Lee A S. The glucose-regulated proteins (GRP78 and GRP94): functions, gene regulation, and applications. Crit Rev Eukaryot Gene Expr. 1994;4:1–18. doi: 10.1615/critreveukargeneexpr.v4.i1.10. [DOI] [PubMed] [Google Scholar]
- 16.Menoret A, Meflah K, Le Pendu J. Expression of the 100-kda glucose-regulated protein (GRP100/endoplasmin) is associated with tumorigenicity in a model of rat colon adenocarcinoma. Int J Cancer. 1994;56:400–405. doi: 10.1002/ijc.2910560319. [DOI] [PubMed] [Google Scholar]
- 17.Michalak J P, Wychowski C, Choukhi A, Meunier J C, Ung S, Rice C M, Dubuisson J. Characterization of truncated forms of hepatitis C virus glycoproteins. J Gen Virol. 1997;78:2299–2306. doi: 10.1099/0022-1317-78-9-2299. [DOI] [PubMed] [Google Scholar]
- 18.Munro S, Pelham H R. An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- 19.Ou J H. Molecular biology of hepatitis B virus e antigen. J Gastroenterol Hepatol. 1997;12:S178–S187. doi: 10.1111/j.1440-1746.1997.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 20.Price B D. Signalling across the endoplasmic reticulum membrane: potential mechanisms. Cell Signal. 1992;4:465–470. doi: 10.1016/0898-6568(92)90015-z. [DOI] [PubMed] [Google Scholar]
- 21.Ralston R, Thudium K, Berger K, Kuo C, Gervase B, Hall J, Selby M, Kuo G, Houghton M, Choo Q L. Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J Virol. 1993;67:6753–6761. doi: 10.1128/jvi.67.11.6753-6761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramakrishnan M, Tugizov S, Pereira L, Lee A S. Conformation-defective herpes simplex virus 1 glycoprotein B activates the promoter of the grp94 gene that codes for the 94-kD stress protein in the endoplasmic reticulum. DNA Cell Biol. 1995;14:373–384. doi: 10.1089/dna.1995.14.373. [DOI] [PubMed] [Google Scholar]
- 23.Reed K E, Rice C M. Molecular characterization of hepatitis C virus. Curr Stud Hematol Blood Transfus. 1998;62:1–37. doi: 10.1159/000060472. [DOI] [PubMed] [Google Scholar]
- 24.Rose M D, Misra L M, Vogel J P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 24a.Selby, M. J. Unpublished data.
- 25.Selby M J, Choo Q L, Berger K, Kuo G, Glazer E, Eckart M, Lee C, Chien D, Kuo C, Houghton M. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J Gen Virol. 1993;74:1103–1113. doi: 10.1099/0022-1317-74-6-1103. [DOI] [PubMed] [Google Scholar]
- 26.Sugawara S, Takeda K, Lee A, Dennert G. Suppression of stress protein GRP78 induction in tumor B/C10ME eliminates resistance to cell mediated cytotoxicity. Cancer Res. 1993;53:6001–6005. [PubMed] [Google Scholar]
- 27.Ting J, Wooden S K, Kriz R, Kelleher K, Kaufman R J, Lee A S. The nucleotide sequence encoding the hamster 78-kDa glucose-regulated protein (GRP78) and its conservation between hamster and rat. Gene. 1987;55:147–152. doi: 10.1016/0378-1119(87)90258-7. [DOI] [PubMed] [Google Scholar]
- 28.Toledo H, Carlino A, Vidal V, Redfield B, Nettleton M Y, Kochan J P, Brot N, Weissbach H. Dissociation of glucose-regulated protein GRP78 and GRP78-IgE Fc complexes by ATP. Proc Natl Acad Sci USA. 1993;90:2505–2508. doi: 10.1073/pnas.90.6.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Tong-Starksen S E, Luciw P A, Peterlin B M. Human immunodeficiency virus long terminal repeat responds to T-cell activation signals. Proc Natl Acad Sci USA. 1987;84:6845–6849. doi: 10.1073/pnas.84.19.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Z, Jensen G, Yen T S. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J Virol. 1997;71:7387–7392. doi: 10.1128/jvi.71.10.7387-7392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X J, Zhang T H, Underwood J C. The ultrastructural pathology of chronic hepatitis C. Chung-hua Ping Li Hsueh Tsa Chih. 1993;22:157–159. [PubMed] [Google Scholar]