Hepatitis B Virus X Protein Is both a Substrate and a Potential Inhibitor of the Proteasome Complex (original) (raw)
Abstract
The hepatitis B virus X protein (HBX) is essential for the establishment of HBV infection in vivo and exerts a pleiotropic effect on diverse cellular functions. The yeast two-hybrid system had indicated that HBX could interact with two subunits of the 26S proteasome. Here we demonstrate an association in vivo of HBX with the 26S proteasome complex by coimmunoprecipitation and colocalization upon sucrose gradient centrifugation. Expression of HBX in HepG2 cells caused a modest decrease in the proteasome’s chymotrypsin- and trypsin-like activities and in hydrolysis of ubiquitinated lysozyme, suggesting that HBX functions as an inhibitor of proteasome. In these cells, HBX is degraded with a half-life of 30 min. Proteasome inhibitors retarded this rapid degradation and caused a marked increase in the level of HBX and an accumulation of HBX in polyubiquitinated form. Thus, the low intracellular level of HBX is due to rapid proteolysis by the ubiquitin-proteasome pathway. Surprisingly, the proteasome inhibitors blocked the transactivation by HBX, and this effect was not a result of a squelching phenomenon due to HBX accumulation. Therefore, proteasome function is possibly required for the transactivation function of HBX. The inhibition of protein breakdown by proteasomes may account for the multiple actions of HBX and may be an important feature of HBV infection, possibly in helping stabilize viral gene products and suppressing antigen presentation.
Hepatitis B virus (HBV) has a unique fourth open reading frame (ORF) coding for a protein known as hepatitis B virus X (HBX) (for reviews, see references 3, 10and 55). The HBX gene is well conserved among the mammalian hepadnaviruses and codes for a 16.5-kDa protein, which has been detected in both the nucleus and cytoplasm. HBX mRNA (0.7 kb) has been detected in infected liver, but the protein has not been easy to detect. However, this protein must be expressed in vivo because antibodies against HBX have been detected in infected individuals. The HBX gene has been shown to be essential for the establishment of HBV infection in vivo (4, 57). Its gene product also activates a variety of viral and cellular promoters in diverse cell types. Recently, HBX has been shown to participate in signal transduction pathways, in particular the activation of the Ras/Raf pathway (1,9). Furthermore, components of the basal transcription complex such as CREB/ATF2 (31) and TATA binding protein (36,37), p53 (51, 52), ERCC3 (a general transcription factor involved in nucleotide excision repair) (53), RPB5 (a common subunit of RNA polymerases) (5), XAP-1/UVDDB (a DNA repair protein) (26, 27), and a cell senescence-associated protein (47) have been reported to be potential cellular targets of HBX. Although many of these findings may explain the biological functions of HBX, definitive functional evidence supporting these claims is lacking. Recently HBX expression has been linked to the induction of apoptosis (6, 46). Finally, the demonstration of the oncogenic potential of the HBX gene in a transgenic mouse model suggests that HBX may contribute to the pathogenesis of HBV-associated hepatocellular carcinoma (23).
Using the yeast two-hybrid system, we previously identified a putative target of HBX as the α subunit of the 20S proteasome (PSMA7; initially referred to as XAPC7) and demonstrated that this interaction is functionally important for the transactivation function of HBX (22). This interaction has also been independently reported by two other groups (12, 45). In addition, during further analyses of the HBX-interacting clones from the two-hybrid screen, we found that the PSMC1 subunit of the 19S proteasome complex also interacts specifically with HBX (56). The proteasome complex is responsible for the majority of nonlysosomal protein degradation in eukaryotic cells (for reviews, see references 8 and21). The 26S proteasome is an ∼2,000-kDa, multisubunit, ATP-dependent proteolytic complex. It consists of the 19S cap complex, which is required for the recognition and degradation of ubiquitinated proteins, and the ∼700-kDa 20S proteasome core, where proteins are degraded (for reviews, see references 8and 28). The 20S particle can also interact with another multisubunit complex, PA28, which is induced by gamma interferon and functions to activate peptide hydrolysis by the 20S proteasome (18, 30). The proteasome functions in diverse cellular processes ranging from cell differentiation, cell cycle control, signal transduction, stress response, transcriptional activation, DNA repair, apoptosis, and antigen presentation (for reviews, see references 8 and21).
In view of the pleiotropic effects of HBX on signal transduction, transcription, cell proliferation, and transformation, we reasoned that the interaction between HBX and the proteasome may possibly result in alterations in proteasome function and contribute to the observed effects of HBX on cells. In this study, we further demonstrated the structural and functional interactions between HBX and proteasome in vivo. HBX appeared to be not only a substrate but also a potential inhibitor of the protease activities of cellular proteasomes. In addition, our data suggested that proteasome function may be required for the transactivation function of HBX. Our results indicate that these structural and functional interactions between HBX and the proteasome may represent a major mechanism underlying the biological effects of HBX.
MATERIALS AND METHODS
Reagents, antibodies, and plasmids.
The inhibitors of calpain_N_-acetyl-l-leucinyl-l-leucinal-l-norleucinal (aLLN) and_N_ - acetyl - l - leucinyl - l - leucinal -methional (aLLM) were obtained from Sigma Chemical Co. (St. Louis, Mo.). The proteasome inhibitors lactacystin and MG132 were obtained from E. J. Corey (Harvard University, Cambridge, Mass.) and Peptide Institute Inc. (Osaka, Japan), respectively. The peptide substrates of the proteasomes Suc-Leu-Leu-Val-Tyr-AMC (LLVY), Boc-Leu-Arg-Arg-AMC (LRR), and Z-Leu-Leu-Glu-βNA (LLE) were obtained from Bachem (Torrance, Calif.). Plasmids pCDNA1, pHook2, and pHooKLacZ were obtained from Invitrogen (San Diego, Calif.). pHookHBX was constructed by inserting the HBX gene (_Hin_dIII-_Bam_HI fragment) from pCDHBX (22) into the expression vector pHook2. HBX cDNA fused to the Flag epitope sequence (Eastman Kodak, New Haven, Conn.) at its 3′ end and cloned into the expression vector pCEP4 (pCEPHBXFlag), and recombinant adenoviruses containing Flag-tagged HBX or HBX0, where the start codon of the HBX gene was mutated to a stop codon (AdHBx and AdHBX0), were provided by Robert Schneider (9). pCMVUb, which contains a human ubiquitin cDNA fused to the influenza virus hemagglutinin (HA) epitope at the 5′ end driven by a cytomegalovirus (CMV) expression vector, was generously provided by Mathias Treier (50). pHookUb-R-βgal was generated by inserting the ubiquitin–Arg–β-galactosidase fusion construct (17), provided by Alexander Varshavsky, into plasmid pHook2. Plasmids RSV-Luc, AP1-CAT, and SP1-Luc have been described previously (22). The mouse monoclonal antibody M2 against the Flag epitope was obtained from Eastman Kodak, antibody 12CA5 against the HA epitope was obtained from Boehringer Mannheim (Indianapolis, Ind.), and rabbit polyclonal antiubiquitin serum was obtained from Sigma. Mouse monoclonal antibodies MCP34 and MCP168 against human proteasome subunits PSMA7 (α subunit) and PSMB7 (β subunit), respectively, were kindly provided by Klavs Hendil (19). Mouse monoclonal antibodies 2-17 against the 20S α subunit PSMA1 and against the 19S subunits TBP1 (PSMC3) and S4 (PSMC1) were gifts from Keiji Tanaka (24, 25). Chloramphenicol acetyltransferase (CAT) and luciferase assays were performed by using kits from Boehringer Mannheim and Promega (Madison, Wis.), respectively.
Plasmid transfection and virus infection.
The HepG2 cell line was maintained in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum. DNA transfection was performed by the standard calcium phosphate method. Recombinant adenoviruses were diluted in Dulbecco’s modified Eagle medium at a concentration of 108PFU/ml. The cells were washed twice with phosphate-buffered saline (PBS) and then infected with 1 ml of the virus preparation per 10-cm-diameter dish at a multiplicity of infection of 10 for 1 h with intermittent mixing. This multiplicity of infection gave a high level of expression at 24 h postinfection. The cells were then placed in regular medium and cultured for 24 h before harvesting; this early time point was chosen to minimize the effect of adenovirus replication and late gene expression on the cells.
Immunoprecipitation and Western immunoblotting.
Plasmid-transfected or virus-infected cells were treated with the various protease or proteasome inhibitors for 6 h before harvest. Cells were lysed directly in a 10-cm-diameter dish with 1 ml of cold standard lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% Triton X-100, 1 μg of leupeptin per ml, 1 μg of aprotinin per ml, 1 mM phenylmethylsulfonyl fluoride, and 1 μM MG132. The lysed cells were centrifuged at 13,000 × g for 15 min at 4°C to remove the nuclei and other insoluble cell debris. Immunoprecipitation was performed by incubating the cell lysates with 20 μl of agarose beads conjugated with antibody M2 (3 mg per ml of beads; Eastman Kodak). The immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed with a PhosphorImager (Storm; Molecular Dynamics, Sunnyvale, Calif.). Western immunoblotting was performed with a chemiluminescence kit from Amersham (Arlington Heights, Ill.).
Isolation of pHook-transfected cells and purification of proteasomes.
pHook-transfected cells were isolated by Capture-tec beads as instructed by the manufacturer (Invitrogen). Briefly, 48 h after transfection, the cells from 10-cm-diameter dishes were washed twice with PBS and detached in 1 ml of PBS–3 mM EDTA. The cells were gently dispersed to single-cell suspension, which was then mixed with the Capture-tec beads (20 μl beads per ml of cell suspension) and incubated at 37°C with continuous gentle mixing. The transfected cells were then isolated by using a magnetic stand. The purity of the selected cells was monitored by cell staining with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal).
Cells for proteasome preparation were homogenized on ice in a Dounce homogenizer in buffer A containing 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM ATP, and 250 mM sucrose. A proteasome-rich fraction was isolated by differential centrifugation at 5,000 × g for 10 min, 1,000 × g for 30 min, 100,000 × g for 1 h, and then 200,000 × g for 5 h (14,15). The final pellet containing proteasomes was resuspended in buffer A with 10% glycerol. In some experiments, the samples prepared by differential centrifugation were further purified and analyzed by sucrose gradient centrifugation (54). The proteasome preparations were layered on top of a 5-ml 10 to 40% sucrose gradient containing 20 mM Tris-HCl, 5 mM MgCl2, 1 mM DTT, and 1 mM ATP and centrifuged in SW50.1 rotor for 12 h at 29,000 rpm. Fractions were collected and used for assay of peptidase activity, Western blotting, and immunoprecipitation. Yeast proteasomes were purified to near homogeneity as described elsewhere (40).
Analyses of proteasome activities.
The peptidase activities of the proteasomes were measured with various fluorogenic peptide substrates as described previously (14, 15). In brief, 3 μg of the proteasome preparation was incubated with 0.1 mM LLVY, LRR, or LLE in a 50-μl total volume of buffer A at 37°C for 20 min. The reactions were stopped by adding 1 ml of 1% SDS. The resulting fluorescence was measured with a spectrofluorometer (Packard Instrument, Downers Grove, Ill.). The ubiquitin-dependent proteinase activity of the proteasomes was measured by incubating the proteasome with 125I-ubiquitin-lysozyme, which was prepared as described previously (40).125I-ubiquitin-lysozyme was incubated with the proteasome preparation at 37°C for 20 min in the presence of 1 mM of ATP and then precipitated with 10% trichloroacetic acid (TCA). The TCA-soluble fraction reflecting degraded 125I-ubiquitin-lysozyme was measured with a γ counter.
For activity gel analysis, partially purified proteasomes were electrophoresed on a 4% native acrylamide gel in 90 mM Tris-borate–5 mM MgCl2–1 mM ATP–0.5 mM DTT–0.1 mM EDTA. After electrophoresis, the gel was incubated with 0.2 mM LLVY at 37°C for 15 min and visualized with a UV transilluminator. The bands representing 26S and 20S proteasome activities were photographed and quantitated by using ImageQuant software (Molecular Dynamics).
RESULTS
Inhibition of HBX degradation by proteasome inhibitors.
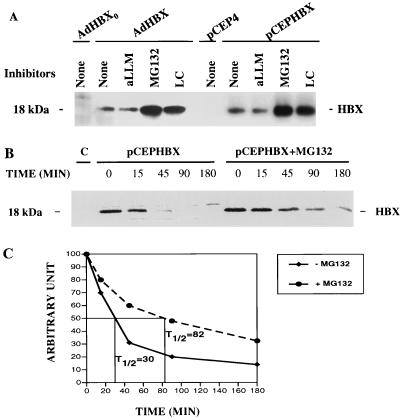
HBX was expressed in HepG2 cells by infection with a recombinant adenovirus vector or through transient transfection with CMV-driven plasmid. For ease of detection, the HBX ORF was fused to the Flag epitope at its C terminus. This construct was shown to be active in the transactivation function of HBX (data not shown); a similar construct had also been used successfully to study the functions of HBX (9). Using immunoprecipitation followed by Western immunoblotting with the anti-Flag antibody M2, we identified a single 18-kDa protein representing HBX in the cell lysates of both expression systems (Fig.1A). The effects of various protease inhibitors on HBX expression were tested. Treatment of cells with specific proteasome inhibitors MG132 and lactacystin for 6 h led to an increase of greater than 10-fold in the steady-state level of HBX protein, whereas aLLM (inhibitor of calpains and lysosomal cysteine proteases) had no effect on the level of HBX. Cells treated with these inhibitors appeared healthy and similar to control cells for at least 16 h. This accumulation of HBX in the presence of MG132 and lactacystin suggests that this protein is rapidly degraded by the proteasome.
FIG. 1.
Effects of proteasome inhibitors on HBX expression. (A) HBX expression. HBX tagged with a Flag epitope was expressed by an adenovirus vector via infection or by a CMV-driven expression plasmid via transient transfection. AdHBX0 and plasmid pCEP4 were used as controls; 108 PFU of virus or 15 μg of the plasmid was used per 10-cm-diameter dish of cells. At 24 h after infection or 42 h after transfection, cells were treated with various inhibitors (20 μM each) for 6 h and then lysed and subjected to immunoprecipitation with antibody M2. The immunoprecipitates were subjected to SDS-PAGE (15% gel) and immunoblotted with antibody M2. (B) HBX degradation. Three 10-cm-diameter dishes of HepG2 cells were transiently transfected with pCEPHBXFlag (15 μg per dish); 18 h later, the cells were evenly distributed among 10 60-mm-diameter dishes. Cells transfected with plasmid pCEP4 were used as a control. After another 24 h, cells were incubated with methionine-free medium for 30 min, pulse-labeled with [35S]methionine (100 μCi/ml) for 20 min, and then chased with medium containing excessive unlabeled methionine (50 μg/ml). Cells were lysed at the end of the labeling and various times after chase. One set of cells was exposed to MG132 (20 μM) starting 1 h before the pulse-labeling and continuing during the chase. The cell lysates were immunoprecipitated with antibody M2. The control represents time zero lysate of pCEPHBX-transfected cells incubated with mouse immunoglobulin G. The immunoprecipitates were electrophoresed on an SDS–15% polyacrylamide gel. The gel was dried, and the levels of HBX expression were analyzed with a PhosphorImager (B). The values of HBX signals at time zero were arbitrarily set at 100, and the signal intensities of other time points were adjusted accordingly. The half-lives (T1/2) are indicated in panel C.
To clarify the mechanism of the increased HBX expression by proteasome inhibitors, a pulse-chase experiment was performed to measure the rate of HBX synthesis and degradation. HBX-expressing cells were pulse-labeled with [35S]methionine and chased for various time points with excess of unlabeled methionine in the presence or absence of the proteasome inhibitor MG132 (20 μM). Labeled HBX was immunoprecipitated with the M2 antibody, analyzed by SDS-PAGE, and quantitated with a PhosphorImager (Fig. 1B). The rate of synthesis as represented by time zero was not altered by treatment of cells with proteasome inhibitor. In the control cells, HBX protein was quickly degraded with a half-life of about 30 min. In MG132 treated cells, the degradation of HBX was much slower, with a half-life of 82 min. A similar reduction in HBX degradation was also seen with lactacystin (data not shown). These results indicate that HBX is degraded by the proteasome.
Polyubiquitination of HBX.
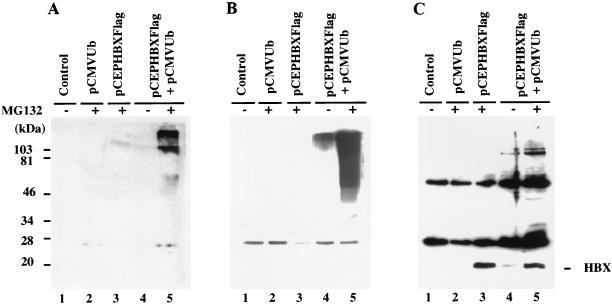
Most cellular proteins degraded by the proteasome are first covalently modified by conjugation to multiple molecules of ubiquitins, and attachment of five or more ubiquitin moieties leads to rapid hydrolysis by the 26S proteasome. To test whether HBX is ubiquitinated before its degradation, an expression construct containing the ubiquitin ORF fused to the HA epitope (pCMVUb) was cotransfected with the HBX expression construct into HepG2 cells. The transfected cells were incubated with or without MG132 (20 μM) for 6 h, lysed, and subjected to immunoprecipitation with antibody M2. The immunoprecipitates were electrophoresed, and Western blotting was performed with an antibody against ubiquitin, HA (12CA5), or Flag (M2) (Fig. 2). In the blot probed with the antiubiquitin antibody, high-molecular-weight smears representing polyubiquitinated HBX proteins were observed in cells cotransfected with pCMVUb and pCDHBX (lanes 4 and 5). Typically ubiquitinated proteins tend to vary in length depending on the number of ubiquitin molecules attached to the substrate and as a result migrate as a ladder or smear on SDS-PAGE. When proteasome function was inhibited by MG132 treatment, there was a much larger accumulation of ubiquitinated proteins (lane 5). This result indicates degradation of the ubiquitinated HBX by the proteasome. By contrast, cells transfected with vector plasmid (lane 1) or pCMVUb alone (lane 2) showed no such pattern. Sample from cells expressing HBX alone showed a slightly positive signal with antiubiquitin antibody, presumably representing HBX linked to the endogenous ubiquitin (lane 3). In the pCMVUb- and HBX-cotransfected cells, the M2 immunoprecipitates probed with antibody against HA or Flag epitope showed clear ladders of high-molecular-weight proteins. Since they withstood denaturation in the presence of SDS, exposure to reducing agent, and boiling prior to electrophoresis, these structures must represent polyubiquitinated HBX (lanes 4 and 5). It is not clear why the patterns of polyubiquitinated bands appeared dissimilar in assays using three different antibodies (antiubiquitin, 12CA5, and M2), all of which should recognize polyubiquitinated HBX. It is possible that proteins with polyubiquitinated side chains are differentially accessible to antibodies recognizing disparate epitopes.
FIG. 2.
Ubiquitin conjugation to HBX. HepG2 cells were plated in 10-cm-diameter dishes and transiently transfected with control plasmid pCEP4 (lane 1), pCMVUb (15 μg; lane 2), pCEPHBXFlag (5 μg; lane 3), or pCMVUb (15 μg) and pCEPHBXFlag (5 μg) (lanes 4 and 5), respectively. The control plasmid pCEP4 was used to equalize the total amount of transfected plasmid at 20 μg; 42 h after transfection, the cells were treated with or without MG132 (20 μM) as indicated for 6 h. The cells were then lysed and denatured in the presence of 1% SDS and 1% β-mercaptoethanol followed by boiling for 5 min. The denatured lysate was then diluted 1:3 with standard lysis buffer and subjected to immunoprecipitation with anti-Flag antibody M2. The immunoprecipitates were divided equally into three aliquots, analyzed by SDS-PAGE (15% gel), and Western blotted with antibodies against ubiquitin (A), the HA epitope (12CA5) (B), and the Flag epitope (M2) (C).
Interaction of HBX and proteasome complex in vivo.
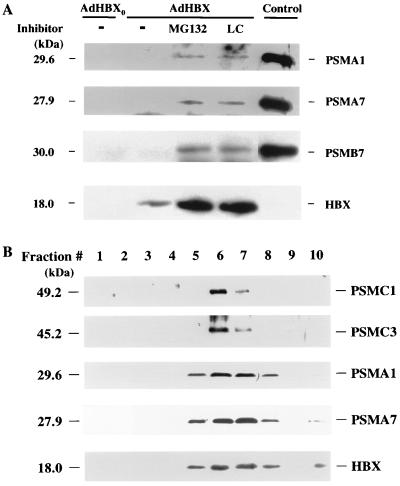
We have previously demonstrated the specific association of HBX and proteasome subunit PSMA7 in vitro. To demonstrate an interaction in vivo between HBX and proteasomes, we attempted to coimmunoprecipitate HBX and the proteasome complex from transfected HepG2 cells. We previously were not able to demonstrate such an interaction by coimmunoprecipitation in various cell lines. However, in light of the finding that HBX was rapidly degraded and that its level could be substantially increased in the presence of proteasome inhibitors, we performed the coimmunoprecipitation experiment in cells treated with MG132 or lactacystin. HepG2 cells were infected with the recombinant AdHBX and control adenovirus, and the HBX was immunoprecipitated from cell lysates with antibody M2. The immunoprecipitates were then subjected to SDS-PAGE and Western immunoblotting with antibodies against various proteasome subunits, including two α subunits (PSMA1 and PSMA7) and one β subunit (PSMB7) (Fig. 3A). In the presence of proteasome inhibitors, several proteasome subunits were coimmunoprecipitated with HBX, suggesting an in vivo association of HBX and the proteasome complex. The specificity of coimmunoprecipitation was verified by the absence of any signals, using an irrelevant antibody for immunoprecipitation (not shown).
FIG. 3.
Interaction of HBX and proteasome complex in vivo. (A) Coimmunoprecipitation of HBX and proteasome in the presence of proteasome inhibitors. HepG2 cells were plated in 10-cm-diameter dishes and infected with AdHBX or AdHBX0 (108 PFU per dish); 24 h after infection, cells were treated with or without proteasome inhibitor MG132 or lactacystin (20 μM) for 6 h. The cells were then lysed and subjected to immunoprecipitation with antibody M2. The immunoprecipitates were divided equally into four aliquots, subjected to SDS-PAGE on a 15% gel, and probed with antibodies against three different proteasome subunits as indicated and antibody M2. Partially purified proteasomes from uninfected HepG2 cells was used as positive controls for immunoblotting of various proteasome subunits. (B) Colocalization of HBX and proteasome by sucrose gradient centrifugation. Partially purified proteasome from HBX-expressing cell lysate as described above was subjected to sucrose gradient centrifugation as described in Materials and Methods; 0.5-ml fractions were collected from the top (fractions 1 to 10) and used for Western immunoblotting with antibodies against two 20S proteasome subunits (PSMA1 and PSMA7) and two 19S subunits (PSMC1 and PSMC3) and immunoprecipitation followed by Western blotting with antibody M2.
The 20S and 26S proteasome complexes can be separated by sucrose velocity gradient centrifugation (54); we therefore fractionated the proteasomes prepared from cells infected with AdHBX and treated with MG132 by this method. HBX was found to colocalize with both the 20S and 26S peaks by Western immunoblotting for various subunits of the 20S (PSMA1 and PSMA7 subunits) and 19S (PSMC1 and PSMC3 subunits) proteasome components and HBX (antibody M2) (Fig. 3B). Peptidase activities were assayed as markers of the 20S and 26S particles (see Fig. 8; note that the fractions were collected in half the volume for Fig. 8). HBX, along with the proteasome, was also present in the last fraction of the gradient, which possibly represents large proteasome-HBX aggregates formed during the purification procedures. The predominant form of HBX in association with the proteasome complex in the above experiments was not ubiquitinated. This result suggests a direct interaction of HBX and proteasome rather than ubiquitin-mediated association of ubiquitinated HBX with proteasome.
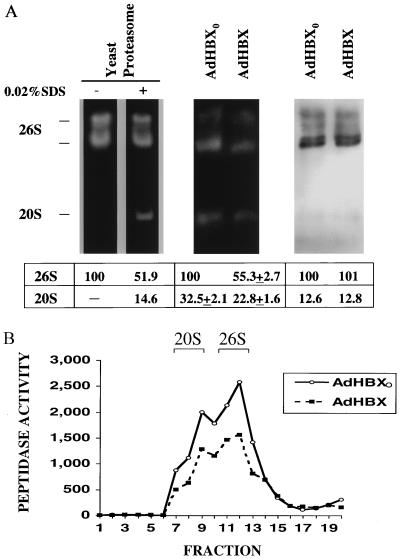
FIG. 8.
HBX inhibits the peptidase activities of both 20S and 26S proteasome complexes. HepG2 cells were plated in 10-cm-diameter dishes and infected with recombinant AdHBX or AdHBX0(108 PFU per dish); 24 h after infection, proteasomes were partially purified as described above and subjected to activity gel analysis (A) or sucrose gradient centrifugation (B) followed by analysis of peptidase activities of each fraction (0.25 ml). In panel A, purified yeast proteasomes treated with or without 0.02% SDS were analyzed on an activity gel (left). The activity gel of proteasome preparation from HBX-expressing or control cells is shown in the middle, and Western immunoblot analysis using anti-PSMA1 antibody is shown on the right. The activity gel and sucrose gradient centrifugation are described in Materials and Methods. The locations of 20S and 26S bands in the sucrose gradient are indicated. Signal intensities of the 20S and 26S bands on the activity gel and Western blot were quantitated by using the ImageQuant software, and the value of the 26S band of the control (AdHBX0) in each gel is arbitrarily set at 100. The data are shown below the gel as means ± standard deviations from three separate experiments.
Inhibition of HBX transactivation by proteasome inhibitors.
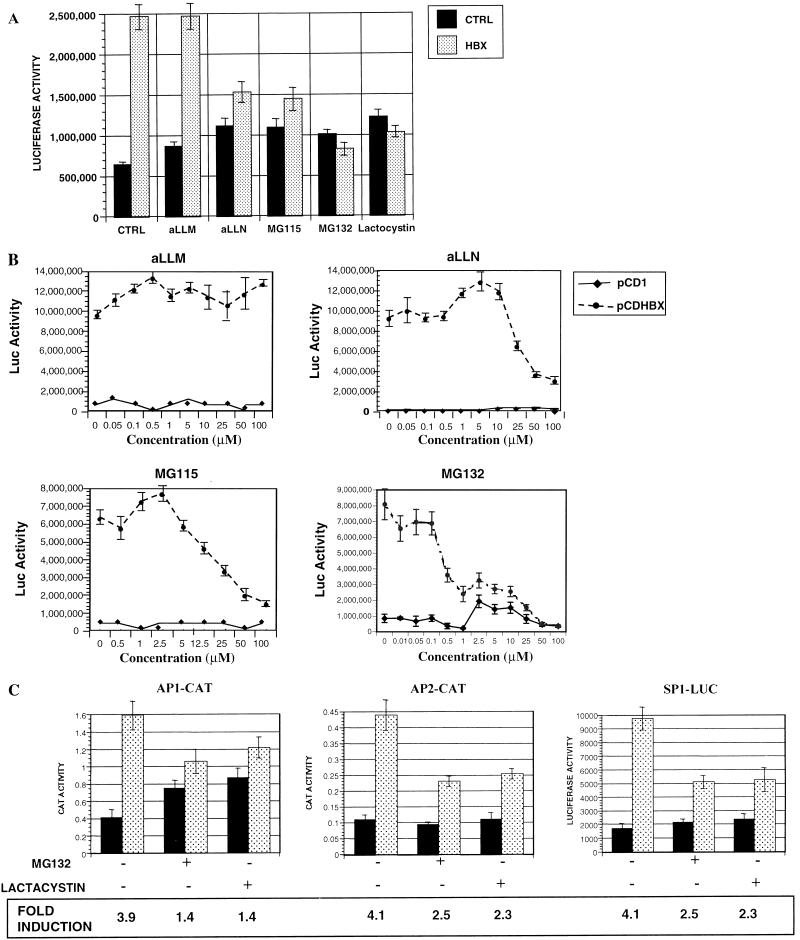
To understand the functional significance of the interaction between HBX and the proteasome complex, we examined whether the proteolytic activity of the proteasome complex might be linked to the function of HBX. The function of HBX was evaluated by testing the transactivation activity of HBX. HepG2 cells were cotransfected with RSV-Luc and pCDHBX or pCD1 and then exposed to the proteasome inhibitors (MG115, MG132, and lactacystin) or the calpain protease inhibitors (aLLM and aLLN) for 16 h prior to determination of luciferase activities. aLLN has been shown to exhibit moderate inhibitory activity against proteasomes, whereas aLLM has little or no effect on the proteasome (38). The transactivation by HBX was reduced significantly, in some cases to the baseline, when the proteasome inhibitors or aLLN was present, whereas aLLM had no effect (Fig. 4A). With these different agents, the half-maximal concentrations of the various inhibitors needed for in vivo inhibition of HBX function paralleled their potencies in inhibiting the chymotryptic activity in vitro (Table 1). These trends strongly suggest that the inhibitors affect HBX transactivation by inhibiting the proteasome. Furthermore, the inhibition of HBX transactivation by MG115, MG132, and aLLn was dose dependent (Fig. 4B) and was seen at concentrations known to inhibit proteasome activities in vivo (38).
FIG. 4.
Effects of proteasome inhibitors on HBX transactivation. (A and B) Effects on complex promoter. HepG2 cells were cotransfected with RSV-Luc and pCDHBX or pCD1 as a control at a ratio of 1 to 5 with a total DNA of 0.6 μg per well in a six-well plate; 20 h after transfection, the cells were treated with various inhibitors at a concentration of 20 μM (A) or at different concentrations (B); 12 h later, the cells were lysed and luciferase (Luc) activities were measured. Data presented are means ± standard deviations in triplicates, and the results are representative of three separate experiments. (C) Effect on simple promoter. HepG2 cells were cotransfected with reporter plasmids AP1-CAT, AP2-CAT, and SP1-Luc as indicated and pCDHBX (░⃞) or pCD1 (control; ■) at a ratio of 1 to 5 with a total DNA of 0.6 μg per well in a six-well plate. At 20 h after transfection, the cells were treated for 12 h in the presence of 20 μM MG132 or 30 μM lactacystin. The cells were lysed, and luciferase or CAT activities were measured. Data presented are means ± standard deviations in triplicates, and the results are representative of three separate experiments.
TABLE 1.
Potency of inhibitors in blocking HBX transactivation correlates with potency against pure 20S proteasomes
| Inhibitor | K i (μM) for inhibition of: | |
|---|---|---|
| HBX function in vivoa | LLVY hydrolysisb | |
| aLLM | None | >1 |
| aLLN | 40–50 | 0.14 |
| MG115 | 10–25 | 0.021 |
| MG132 | 2.5–10 | 0.011 |
To further demonstrate that the inhibition of HBX transactivation by proteasome inhibitors was not limited to a particular promoter, we expanded the experiment to measure several other promoters including basal promoters linked to AP1, AP2, or SP1 motif sequences, all of which were previously shown to respond to HBX transactivation (22). Similarly, HBX transactivations (fold induction) of AP1, AP2, and SP1 were significantly inhibited by both MG132 and lactacystin (Fig. 4C). It is interesting that the proteasome-specific inhibitors caused higher reporter activities in cells transfected with the reporters only, which in the case of AP1 reporter resulted in a nearly twofold higher activity. This finding can be explained by the general increased levels of many transcriptional factors, including Jun/Fos (represented by AP1 reporter activity), as a result of inhibition of their degradation by proteasome inhibitors.
MG132 and lactacystin act via different mechanisms: MG132 is a peptide aldehyde that inhibits the chymotrypsin-like and post-acidic peptidase activities reversibly by forming a transition-state complex with the active sites of the β subunits in the proteasome complex. Lactacystin is an irreversible inhibitor and after converting to a β-lactone derivative reacts covalently with the active sites threonine residues of multiple β subunits (11). To confirm that the inhibition of HBX transactivation by these inhibitors is due to compromised proteasome activities instead of some other mechanism, we tested whether this inhibition of the transactivation function of HBX is reversible by the removal of MG132 or lactacystin. As shown in Fig.5, the transactivation activity of HBX was reduced significantly after 6 h of treatment with MG132 or lactacystin. Six hours after removal of MG132, the transactivation activity returned to the same high level as for the untreated samples. In contrast, HBX transactivation remained substantially suppressed after removal of lactacystin. Thus, HBX transactivation parallels closely changes in proteasome function.
FIG. 5.
Reversibility of inhibition of HBX transactivation by proteasome inhibitors. HepG2 cells were cotransfected with RSV-Luc and pCDHBX or pCD1 as a control (CTRL) at a ratio of 1 to 5 with a total DNA of 0.6 μg per well in a six-well plate; 20 h after transfection, the cells were treated for 6 h with 10 μM MG132, a reversible inhibitor of the proteasome, or 20 μM lactacystin, an irreversible proteasome inhibitor. At the end of 6 h, one set of cells treated with or without inhibitors was harvested for luciferase determination. The second set of cells was incubated for another 6 h; the third set of cells was washed with PBS and exposed to fresh medium without inhibitors for another 6 h. At the end of 6 h, both sets of cells were harvested for luciferase determination. Data presented are the means ± standard deviations in triplicates, and the results are representative of three separate experiments.
HBX transactivation is dose dependent and does not exhibit a squelching phenomenon.
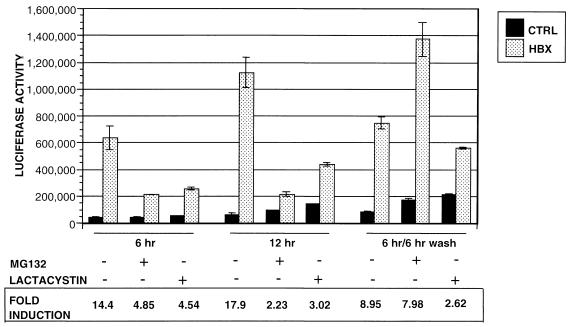
In transient transfection experiments, many transcription factors at high doses exhibit the phenomenon of squelching in which there is a paradoxical reduction of transcription of reporter plasmids (32, 35). Because the proteasome inhibitors markedly increased the level of HBX expression, it is possible that the inhibition of HBX transactivation by MG132 and lactacystin results from a squelching of transcription by high levels of HBX. To test this possibility, RSV-Luc was cotransfected with increasing doses of the HBX expression plasmid. The extent of HBX transactivation increased progressively with incremental amounts of pCDHBX transfected (Fig. 6A). MG132 inhibited HBX transactivation at all doses of pCDHBX examined (Fig.6B). In parallel, we performed immunoprecipitation and Western blotting using antibody M2 to analyze the levels of HBX protein expression with different transfected doses of plasmid pCDHBX. The level of HBX expression correlated well with the amount of plasmid transfected, and the proteasome inhibitor caused a further accumulation of HBX at each dose of the plasmid transfected. These observations suggest that the high level of HBX transactivation with high doses of transfected pCDHBX was the result of increased HBX expression and that HBX does not exhibit a squelching effect in this assay. Therefore, the inhibition of HBX transactivation by proteasome inhibitors cannot be attributed to such an anomalous effect.
FIG. 6.
HBX transactivation is dose dependent and does not exhibit a squelching phenomenon. (A) HepG2 cells were grown in a six-well plate and cotransfected with 0.1 μg of RSV-Luc and increasing amounts (0.02, 0.05, 0.1, 0.5, 0.8, and 1 μg) of pCDHBXFlag. pCD1 was added to make the total transfected DNA 1.1 μg per well. The cells were lysed 48 h later, and luciferase activities were measured. (B) HepG2 cells were cotransfected with RSV-Luc and three different amounts of pCDHBXFlag as indicated; 20 h after transfection, the cells were treated with proteasome inhibitor MG132 at concentrations of 10 and 20 μM for the transactivation assay (top) or 20 μM for measurement of HBX levels (bottom). The cells were lysed and assayed for either luciferase activities or HBX levels by Western blot analysis using antibody M2. The results are shown as fold induction, calculated by dividing the luciferase activity of HBX transfected cells by that of control. Data presented are means ± standard deviations in triplicates, and the results are representative of three separate experiments.
Inhibition of protease activities of proteasome by HBX.
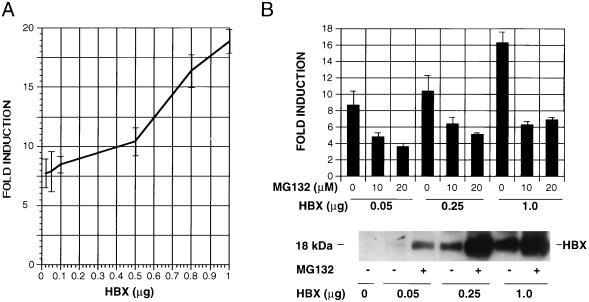
Since HBX binds to proteasomes and the transactivation by HBX requires proteasome function, we further investigated whether the interaction of HBX with proteasomes leads to alterations in the proteasome function. The 20S proteasome has three distinct peptidase sites that are capable of cleaving peptides on the carboxyl side of hydrophobic, basic, and acidic residues. These peptidase activities are referred to as the chymotrypsin-like, trypsin-like, and post-acidic activities, which can be assayed with the fluorogenic substrates LLVY, LRR, and LLE, respectively. HepG2 cells were transfected with pHookHBX or control plasmid pHook LacZ, and the transfected cells were isolated by using magnetic beads. The proteasome-rich fraction was isolated from the selected cells, and activities against these three peptide substrates were assayed. These activities were due to proteasome because they could be reduced more than 90% by 1 μM MG132 (Fig.7A). In HBX-expressing cells, the hydrolysis of the three substrates was consistently reduced below control levels but to various degrees (Fig. 7A). The inhibition of the chymotrypsin-like activity was the greatest (∼50%). Similar reduction of these three peptidase activities was also observed in cells in which HBX was expressed by recombinant AdHBX or HBX-vaccinia virus infection (data not shown).
FIG. 7.
Inhibition of proteasome activities by HBX. HepG2 cells were transiently transfected with pHookHBX or control plasmid pHookLacZ0. Transfected cells were isolated by pHook magnetic beads as described in Materials and Methods. The proteasomes were partially purified by differential centrifugation from the isolated cells. The proteasome contents of the preparations were shown to be similar by immunoblotting with anti-PSMA1 antibody. (A) Peptidase activities. The proteasome preparations (3 μg) were incubated with the three fluorogenic peptide substrates specific for each active site (LLVY for chymotrysin-like activity, LRR for trypsin-like activity, and LLE for post-acidic activity) with or without MG132 (1 μM). The resulting fluorescence reflecting the peptidase activity of proteasomes was measured with a spectrofluorometer. The fluorescence signal from the proteasome of pHookLacZ0-transfected cells without MG132 treatment was standardized as 100% activity, and all the other fluorescence activities were calculated as percent activities. (B) Degradation of 125I-ubiquitin-lysozyme. The partially purified proteasome (3 μg) was incubated with125I-ubiquitin-lysozyme with or without MG132 at 37°C for 20 min and then precipitated with 10% TCA. The TCA-soluble fraction reflecting the degraded 125I-ubiquitin-lysozyme was measured with a γ counter. The soluble counts per minute from the pHookLacZ-transfected cells was standardized as 100% activity, and all other counts were calculated as percent activity. As controls, the partially purified proteasomes from pHookLacZ-transfected cells (A) and from both HBX- and LacZ-transfected cells (B) were treated with 1 μM MG132 to confirm the proteasome’s involvement in these reactions.
We also tested the capacity of the proteasomes from pHook-selected cells to degrade the ubiquitinated substrate,125I-ubiquitin-lysozyme. The degradation of ubiquitinated proteins is an ATP-dependent process catalyzed by the 26S proteasome. As was found for peptidase activities, the rate of degradation of125I-ubiquitin-lysozyme by proteasome from the HBX-expressing cells was approximately half of that in the control-transfected cells (Fig. 7B). Since HBX was expressed at a high level in these experimental conditions, it is conceivable that HBX, as a rapidly degraded protein, competitively inhibits degradation by proteasome of these substrates. To examine this possibility, we studied the effect of expressing high levels of a ubiquitin–Arg–β-galactosidase fusion protein, a very rapidly degraded substrate of the ubiquitin-proteasome pathway (17), on proteasome activities. No effect of overexpression of this short-lived protein on the peptidase assays was seen (not shown).
HBX inhibits both 20S and 26S proteasome activities.
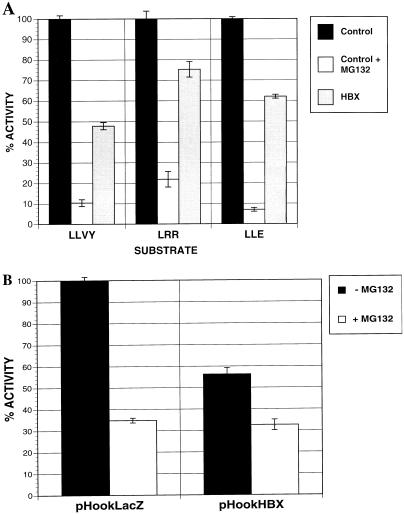
Since HBX inhibited the overall peptidase activities of the proteasome preparation which contained both the 20S and 26S complexes, it is not clear whether the 20S and 26S forms are affected equally or differentially. To address this issue, we performed activity gel analysis of the partially purified proteasomes from control or HBX-expressing cells (Fig. 8A). In a native low-percentage polyacrylamide gel, one can separate the 20S from the 26S complexes as well as analyze their activities by incubating the gel with a fluorogenic peptide substrate (44). For the leftmost panel, purified yeast proteasomes were used as controls. Consistent with previous observations (48), the activity of 20S proteasomes against the LLVY substrate was activated by 0.02% SDS treatment, whereas that of 26S forms was modestly inhibited. The peptidase activities of 20S and 26S proteasomes were similarly inhibited by HBX (Fig. 8A, middle), whereas the quantities of proteasomes as determined by Western blotting for the PSMA1 subunit (Fig. 8A, right) were comparable in the two samples. This observation was also confirmed by analyses of peptidase activities of the sucrose fractions corresponding to the 20S and 26S complexes in sucrose gradient centrifugation of the proteasome preparations (Fig. 8B).
DISCUSSION
The roles of HBX protein in the viral life cycle and in the pathogenesis of HBV infection have been studied extensively. It is commonly accepted that HBX exhibits a promiscuous transactivation function, activating a variety of viral and cellular promoters (3,55), although transactivation may not be essential for viral replication and may not even reflect the primary role of HBX in natural infection. Studies in the woodchuck model of HBV demonstrated that HBX is required for the establishment of infection in vivo (4,57). However, inactivation of HBX in a tissue culture transfection system suggested that HBX plays only a minor role in viral replication (2, 57). Speculations about the molecular actions of HBX have generated much debate and widely divergent hypotheses. In addition, many cellular proteins with rather diverse functions have been shown by a variety of techniques to interact physically with HBX (5, 22, 26, 31, 36, 47, 51–53). Therefore, it is possible that HBX has multiple effector pathways leading to its pleiotropic effects. In the interest of space, we will not discuss in detail all possible pathways of HBX function but instead cite several recent reviews on HBX (3, 10, 55).
Using the yeast two-hybrid system, our group has demonstrated a specific interaction of HBX with an α subunit of the 20S proteasome complex, PSMA7 (22), as well as the PSMC1 subunit of the 19S regulatory component (56). The ATPase-rich 19S particles are found at either end of the 20S proteasome and must bind directly to the α subunits which comprise the outer rings of the 20S particle. Thus, HBX has the inherent capacity to associate with two proteins that may themselves interact within the 26S proteasome.
In agreement with a previous study (42), HBX was shown to have a very short half-life, in the order of 30 min. In the presence of the proteasome inhibitors MG132 and lactacystin, this half-life was prolonged to more than 80 min. Additional experiments revealed that HBX is ubiquitinated and in the presence of proteasome inhibitors accumulates in polyubiquitinated form. Thus, it behaves as a typical substrate of the ubiquitin-proteasome pathway. This rapid degradation probably explains the extreme difficulties encountered by previous workers in detecting HBX in vivo. Using the proteasome inhibitors, we were also able to demonstrate that HBX associates with the proteasome complex in vivo. Formation of this complex appears to be independent of HBX ubiquitination, because this association was first revealed by the two-hybrid analysis and then demonstrated through an in vitro binding experiment (22). Our findings suggest that HBX during its brief half-life interacts directly with the proteasome complex and may become ubiquitinated independently, leading to rapid hydrolysis, as occurs with many other cellular regulatory proteins such as p53, c-Jun, cyclins, and cyclin-dependent kinases (7, 16, 33, 50).
Because the proteasome inhibitors retarded the degradation of HBX and are also known to stabilize many other transcription factors, we anticipated that the transactivation function of HBX would be markedly augmented by these agents. On the contrary, transactivation by HBX was markedly inhibited by MG132 and lactacystin, and the less potent proteasome inhibitor MG115 was also quite effective in suppressing the transactivation. In addition, aLLN, which was originally identified as a calpain inhibitor but was recently shown to also cause an inhibition of proteasomes (38), similarly diminished transactivation by HBX. By contrast, aLLM, a potent calpain inhibitor that has only a very weak activity against proteasome, had no effect on HBX transactivation at the concentration of 100 μM. It is interesting that in control-transfected cells, proteasome inhibitors enhanced expression of the reporter genes up to twofold, suggesting that inhibition of proteasomes generally causes higher levels of many transcription factors and increased transcription of reporter genes. Analysis of the concentration dependence of all these inhibitors indicated that their relative potencies in blocking HBX transactivation correlated with their potencies against the peptidase activities of purified 20S proteasomes. These observations, together with the reversible block in transactivation seen with MG132 but not with the irreversible inhibitor lactacystin, are further evidence that proteasome function may be necessary for transactivation by HBX. Finally, this unexpected ability of proteasome inhibitors to block HBX transactivation was not an artifact due to a squelching phenomenon (32, 35), because there is no demonstrable squelching effect at high levels of HBX expression and because the proteasome inhibitors had similar effects at all levels of HBX expression. Although we cannot eliminate the possibility that the inhibition by HBX of proteasome function prevents transactivation through some indirect or nonspecific effect, this possibility seems unlikely because in the presence of these inhibitors, the control-transfected cells were perfectly capable of expressing the reporter genes. In addition, the cells appeared healthy and morphologically normal after exposure to proteasome inhibitors for the duration of the experiment (∼12 h).
How these observations translate into a mechanism of HBX transactivation is not entirely clear. One possible mechanism is that HBX is processed by the 26S proteasome into an intermediate form that is active in transactivation, similar to the generation of p50 subunit of NF-κB from its p105 precursor (34). Perhaps this HBX species then modifies the proteasome directly, leading to an alteration in the general degradation of cellular proteins. Alternatively, it may induce the selective degradation of a general inhibitor which negatively regulates transcription (transcription corepressor). These models are in line with the effects of proteasome inhibitors on HBX transactivation.
In addition to its transactivation whose biological importance in vivo remains uncertain, HBX may be important in natural infection because of its inhibitory effects on cellular proteasomes. Our data suggested that HBX expression reduced all three peptidase activities of proteasome, especially the chymotrypsin-like activity, which appears to be the most important active site in protein breakdown (8). This reduction in peptide hydrolysis in the 20S particles presumably accounts for the reduced capacity of the 26S proteasome to degrade its natural substrate, ubiquitinated proteins, as shown here with125I-ubiquitin-lysozyme. This inhibition of proteasome function was found to be reproducible with a variety of methods for HBX expression, including transient transfection (Fig. 7), adenovirus transduction (Fig. 8), and vaccinia virus infection (data not shown). Future biochemical studies will be necessary to clarify exactly how, at the molecular level, HBX binding reduces the proteolytic activities of the 20S and 26S proteasomes. Possibly, HBX can act as a reversible inhibitor of the proteasome; for example, the binding of HBX may lead to conformational changes in the particle leading to diminished peptidase activity or reduced substrate entry into the particle. It is well known that minor distortions of the conformation of the 20S particle (with SDS or Mg2+) can lead to large changes in peptide and protein hydrolysis (8). It is noteworthy that other viral proteins with transactivation functions, such as human immunodeficiency virus type 1 Tat and human T-cell leukemia virus type 3 Tax proteins, have also been shown to physically interact with and functionally alter the proteolytic activities of the proteasome (39, 44).
The biological consequences of this inhibition of cellular proteolysis are of interest. We propose that HBX exists to counteract the increased proteolytic function of the cells. Such a viral strategy would ensure the proper processing and assembly of viral proteins during viral replication. Interestingly, certain bacteriophages (e.g., T4) have been found to encode a specific inhibitor of cellular ATP-dependent protease (20). One cellular response to viral infection is induction of heat shock proteins including ubiquitin; this response enhances the host cell’s capacity to degrade proteins with abnormal conformations, such as viral proteins (41). A recent finding indicates that proteasome plays an active antiviral role in degrading incoming human immunodeficiency virus type 1 proteins following viral infection (43). In addition, one major function of the ubiquitin-proteasome pathway is to provide the peptides presented on the cell surface to the immune system (8, 38). Gamma interferon, which is induced early during viral infections, has been shown to stimulate the synthesis of certain proteasome subunits and induction of PA28 activator resulting in enhanced peptide hydrolysis by the proteasome (13, 14, 18, 29). HBX, by inhibiting proteasome-mediated proteolysis, may interfere with antigen presentation of viral proteins, leading to viral evasion of host immune response. There are ample examples of viral gene products whose functions are to interfere with the process of antigen presentation at various levels (49). It is intriguing to think that HBV, an important viral pathogen causing chronic hepatitis, may have evolved a novel mechanism to alter proteasome function in order to guarantee its persistence in infected individuals.
ACKNOWLEDGMENTS
We thank Keiji Tanaka, Klavs Hendil, Mathias Treier, Robert Schneider, and Alexander Varshavsky for their generosity in providing various reagents for this study, and we thank Hucheng Bei and Miranda Fang for excellent technical assistance. We are also grateful for helpful discussions with Kenneth Rock, Jay Hoofnagle, Margaret Koziel, and Reed Wickner.
REFERENCES
- 1.Benn J, Schneider R J. HBV HBx protein establishes a Ras, Raf, MAP kinase signalling cascade and stimulates cell proliferation through activation of p34cdc2 kinase-cyclin B complexes. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum H E, Zhang Z-S, Galun E, von Weizsacker F, Garner B, Liang T J, Wands J R. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J Virol. 1992;66:1223–1227. doi: 10.1128/jvi.66.2.1223-1227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caselmann W H. Trans-activation of cellular genes by hepatitis B virus proteins: a possible mechanism of hepatocarcinogenesis. Adv Virus Res. 1996;47:253–302. doi: 10.1016/s0065-3527(08)60737-x. [DOI] [PubMed] [Google Scholar]
- 4.Chen H-S, Kaneko S, Girones R, Anderson R W, Hornbuckle W E, Tennant B C, Cote P J, Gerin J L, Purcell R H, Miller R H. The woodchuck hepatitis X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheong J, Yi M, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chirillo P, Pagano S, Natoli G, Puri P L, Burgio V L, Balsano C, Levrero M. The hepatitis B virus X gene induces p53-mediated programmed cell death. Proc Natl Acad Sci USA. 1997;94:8162–8167. doi: 10.1073/pnas.94.15.8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciechanover A, DiGiuseppe J A, Bercovich B, Orlan A, Richter J D, Schwartz A L, Brodeur G M. Degradation of nuclear oncoproteins by ubiquitin proteolysis system in vitro. Proc Natl Acad Sci USA. 1991;88:139–143. doi: 10.1073/pnas.88.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 9.Doria M, Klein N, Lucito R, Schneider R J. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feitelson M A, Duan L-X. Hepatitis B virus X antigen in the pathogenesis of chronic infections and the development of hepatocellular carcinoma. Am J Pathol. 1997;150:1141–1157. [PMC free article] [PubMed] [Google Scholar]
- 11.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 12.Fischer M, Runkel L, Schaller H. HBx protein of hepatitis B virus interacts with the C-terminal portion of a novel human proteasome alpha-subunit. Virus Genes. 1995;10:99–102. doi: 10.1007/BF01724303. [DOI] [PubMed] [Google Scholar]
- 13.Fruh K, Gossen M, Wang K, Bujard H, Peterson P A, Yang Y. Displacement of housekeeping proteasome subunits by MHC-encoded LMPs: a newly discovered mechanism for modulating the multicatalytic proteinase complex. EMBO J. 1994;13:3236–3244. doi: 10.1002/j.1460-2075.1994.tb06625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaczynska M, Rock K, Goldberg A. γ-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature. 1993;365:264–266. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 15.Gaczynska M, Rock K L, Spies T, Goldberg A L. Peptidase activities of proteasomes are differentially regulated by the major histocompatibility complex-encoded genes for LMP2 and LMP7. Proc Natl Acad Sci USA. 1994;91:9213–9217. doi: 10.1073/pnas.91.20.9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glotzer M, Murray A W, Kirschner M W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 17.Gonda D K, Bachmair A, Wunning I, Tobias J W, Lane W S, Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- 18.Groettrup M, Soza A, Eggers M, Kuehn L, Dick T P, Schild H, Rammensee H, Koszinowski U H, Kloetzel P M. A role for the proteasome regulator PA28 alpha in antigen presentation. Nature. 1996;381:166–168. doi: 10.1038/381166a0. [DOI] [PubMed] [Google Scholar]
- 19.Hendil K B, Kristensen P, Uerkvitz W. Human proteasomes analysed with monoclonal antibodies. Biochem J. 1995;305:245–252. doi: 10.1042/bj3050245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilliard J J, Simon L D, Van Melderen L, Maurizi M R. PinA inhibits ATP hydrolysis and energy-dependent protein degradation by Lon protease. J Biol Chem. 1998;273:524–527. doi: 10.1074/jbc.273.1.524. [DOI] [PubMed] [Google Scholar]
- 21.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, Kwong J, Sun E C, Liang T J. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J Virol. 1996;70:5582–5591. doi: 10.1128/jvi.70.8.5582-5591.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim C M, Koike K, Saito I, Miyamura T, Jay G. HBX gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;315:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 24.Kominami K, DeMartino G N, Moomaw C R, Slaughter C A, Shimbara N, Fujimuro M, Yokosawa H, Hisamatsu H, Tanahashi N, Shimizu Y, et al. Nin1p, a regulatory subunit of the 26S proteasome, is necessary for activation of Cdc28p kinase of Saccharomyces cerevisiae. EMBO J. 1995;14:3105–3115. doi: 10.1002/j.1460-2075.1995.tb07313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumatori A, Tanaka K, Inamura N, Sone S, Ogura T, Matsumoto T, Tachikawa T, Shin S, Ichihara A. Abnormally high expression of proteasomes in human leukemic cells. Proc Natl Acad Sci USA. 1990;87:7071–7075. doi: 10.1073/pnas.87.18.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee T H, Elledge S J, Butel J S. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Y, Nomura T, Cheong J, Dorjsuren D, Iida K, Murakami S. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcription factor IIB and the RNA polymerase II subunit 5. J Biol Chem. 1997;272:7132–7139. doi: 10.1074/jbc.272.11.7132. [DOI] [PubMed] [Google Scholar]
- 28.Lupas A, Flanagan J M, Tamura T, Baumeister W. Self-compartmentalizing proteases. Trends Biochem Sci. 1997;22:399–404. doi: 10.1016/s0968-0004(97)01117-1. [DOI] [PubMed] [Google Scholar]
- 29.Ma C-P, Slaughter C A, Demartino G N. Identification, purification, and characterization of a protein activator (PA28) of the 20S proteasome (macropain) J Biol Chem. 1992;267:10515–10523. [PubMed] [Google Scholar]
- 30.Ma C-P, Vu J H, Proske R J, Slaughter C A, DeMartino G N. Identification, purification, and characterization of high molecular weight, ATP-dependent activator (PA700) of the 20S proteasome. J Biol Chem. 1994;269:3539–3547. [PubMed] [Google Scholar]
- 31.Maguire H F, Hoeffler J P, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 32.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 33.Pagano M, Tam S W, Theodoras A M, Beer-Romero P, Del Sal G, Chau V, Yew P R, Draetta G F, Rolfe M. Role of the ubiquitin proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 34.Palombella V J, Rando O, Goldberg A L, Maniatis T. Processing of the NF-kB1 (p105) precursor protein and the degradation of IkB-a require the proteasome. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 35.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 36.Qadri I, Conaway J W, Conaway R C, Schaack J, Siddiqui A. Hepatitis B virus transactivator protein, HBx, associates with the components of TFIIH and stimulates the DNA helicase activity of TFIIH. Proc Natl Acad Sci USA. 1996;93:10578–10583. doi: 10.1073/pnas.93.20.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qadri I, Maguire H F, Siddiqui A. Hepatitis B virus transactivator protein X interacts with TATA-binding protein. Proc Natl Acad Sci USA. 1995;92:1003–1007. doi: 10.1073/pnas.92.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Inhibitors of the proteasome block the degradation of most cell protein and generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 39.Rousset R, Desbois C, Bantignies F, Jalinot P. Effects on NF-kB1/p105 processing of the interaction between the HTLV-1 transactivator Tax and the proteasome. Nature. 1996;381:328–331. doi: 10.1038/381328a0. [DOI] [PubMed] [Google Scholar]
- 40.Rubin D M, Coux O, Wefes I, Hengartner C, Young R A, Goldberg A L, Finley D. Identification of the gal14 suppressor Sug1 as a subunit of the yeast 26S proteasome. Nature. 1996;379:655–658. doi: 10.1038/379655a0. [DOI] [PubMed] [Google Scholar]
- 41.Santoro M G. Viral infection. In: Feige V, Morimoto R I, Yahara I, Polla B S, editors. Stress inducible cellular responses. Boston, Mass: Birkhauser Verlag; 1996. pp. 337–354. [Google Scholar]
- 42.Schek N, Bartenschlager R, Kuhn C, Schaller H. Phosphorylation and rapid turnover of hepatitis B virus X-protein expressed in HepG2 cells from a recombinant vaccinia virus. Oncogene. 1991;6:1735–1744. [PubMed] [Google Scholar]
- 43.Schwartz O, Marechal V, Friguet B, Arenzana-Seisdedos F, Heard J-M. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J Virol. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seeger M, Ferrell K, Frank R, Dubiel W. HIV-1 tat inhibits the 20S proteasome and its 11S regulator-mediated activation. J Biol Chem. 1997;272:8145–8148. doi: 10.1074/jbc.272.13.8145. [DOI] [PubMed] [Google Scholar]
- 45.Sitterlin D, Lee T-H, Prigent S, Tiollais P, Butel J S, Transy C. Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J Virol. 1997;71:6194–6199. doi: 10.1128/jvi.71.8.6194-6199.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su F, Schneider R J. Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by tumor necrosis factor alpha. Proc Natl Acad Sci USA. 1997;94:8744–8749. doi: 10.1073/pnas.94.16.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun B S, Zhu X, Clayton M M, Pan J, Feitelson M A. Identification of a protein isolated from senescent human cells that binds to hepatitis B virus X antigen. Hepatology. 1998;27:228–239. doi: 10.1002/hep.510270135. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka K, Yoshimura T, Kumatori A, Ichihara A, Ikai A, Nishigai M, Kameyama K, Takagi T. Proteasomes as 20S ring-shaped particles in a variety of eukaryotic cells. J Biol Chem. 1988;263:16209–16217. [PubMed] [Google Scholar]
- 49.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Treier M, Staszewski L M, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the D domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 51.Truant R, Antunovic J, Greenblatt J, Prives C, Gromlish J A. Direct interaction of the hepatitis B virus HBX protein with p53 leads to inhibition by HBX of p53 response element-directed transactivation. J Virol. 1995;69:1851–1859. doi: 10.1128/jvi.69.3.1851-1859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ueda H, Ullrich S J, Gangemi J D, Kappel C A, Ngo L, Feitelson M A, Jay G. Functional inactivation but not structural mutation of p53 causes liver cancer. Nat Genet. 1995;9:41–47. doi: 10.1038/ng0195-41. [DOI] [PubMed] [Google Scholar]
- 53.Wang X W, Forrester K, Yeh H, Feitelson M A, Gu J-R, Harris C C. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y, Fruh K, Ahn K, Peterson P A. In vivoassembly of the proteasomal complexes, implications for antigen processing. J Biol Chem. 1995;270:27687–27694. doi: 10.1074/jbc.270.46.27687. [DOI] [PubMed] [Google Scholar]
- 55.Yen T S B. Hepadnaviral X protein: review of recent progress. J Biomed Sci. 1996;3:20–30. doi: 10.1007/BF02253575. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, Z., A. Furusaka, N. Malayaman, Z. Hu, and T. J. Liang. 1998. Unpublished data.
- 57.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]