KAP-1 Corepressor Protein Interacts and Colocalizes with Heterochromatic and Euchromatic HP1 Proteins: a Potential Role for Krüppel-Associated Box–Zinc Finger Proteins in Heterochromatin-Mediated Gene Silencing (original) (raw)
Abstract
Krüppel-associated box (KRAB) domains are present in approximately one-third of all human zinc finger proteins (ZFPs) and are potent transcriptional repression modules. We have previously cloned a corepressor for the KRAB domain, KAP-1, which is required for KRAB-mediated repression in vivo. To characterize the repression mechanism utilized by KAP-1, we have analyzed the ability of KAP-1 to interact with murine (M31 and M32) and human (HP1α and HP1γ) homologues of the HP1 protein family, a class of nonhistone heterochromatin-associated proteins with a well-established epigenetic gene silencing function in Drosophila. In vitro studies confirmed that KAP-1 is capable of directly interacting with M31 and hHP1α, which are normally found in centromeric heterochromatin, as well as M32 and hHP1γ, both of which are found in euchromatin. Mapping of the region in KAP-1 required for HP1 interaction showed that amino acid substitutions which abolish HP1 binding in vitro reduce KAP-1 mediated repression in vivo. We observed colocalization of KAP-1 with M31 and M32 in interphase nuclei, lending support to the biochemical evidence that M31 and M32 directly interact with KAP-1. The colocalization of KAP-1 with M31 is sometimes found in subnuclear territories of potential pericentromeric heterochromatin, whereas colocalization of KAP-1 and M32 occurs in punctate euchromatic domains throughout the nucleus. This work suggests a mechanism for the recruitment of HP1-like gene products by the KRAB-ZFP–KAP-1 complex to specific loci within the genome through formation of heterochromatin-like complexes that silence gene activity. We speculate that gene-specific repression may be a consequence of the formation of such complexes, ultimately leading to silenced genes in newly formed heterochromatic chromosomal environments.
Regulation of gene expression at the level of transcription initiation by sequence-specific DNA binding proteins has emerged as one of the most important modes of metazoan development and homeostasis (6). The population of transcription factors that are active in the cell nucleus largely dictates the transcriptional output from the nucleus and hence the proliferative or differentiated phenotype of the cell. The dominant theme that has emerged from the study of eukaryotic transcriptional regulatory proteins is that they are highly modular in architecture, with independent, functionally separable domains mediating nuclear localization, sequence-specific DNA binding, hetero- or homo-oligomerization, activation, and repression of transcription. Recently, much effort has been expended to understand how activation and repression domains transmit the signal for modulation of transcription from a DNA-bound protein to the RNA synthesis machinery.
Studies aimed at understanding the mechanisms of transcription repression have been greatly aided by the realization that the domains which mediate repression are often highly conserved amino acid sequence motifs which occur in one or more families of proteins with common DNA binding domains. Examples of these domains include BTB/POZ, WRPW, SNAG, and Krüppel-associated box (KRAB) (2, 5, 11, 20). We have focused on the KRAB domain as a model system for analysis of conserved repression modules (17, 34). The KRAB repression domain was originally identified in humans as a conserved amino acid sequence motif at the amino termini of proteins which contain multiple TFIIIA/Krüppel class Cys2-His2 (C2H2) zinc fingers in their COOH termini (5) and has been identified in frog, rodent, and human zinc finger proteins (ZFPs) (for a review, see reference 24). It has been estimated that between 300 and 700 human genes encode C2H2 zinc finger proteins (28), one-third of which are predicted to contain KRAB domains (5); accordingly, these genes have been designated the KRAB-ZFP family. The KRAB domain homology consists of approximately 75 amino acids (aa) which can function as a potent transferable DNA binding-dependent repression module. Moreover, more than 10 independently encoded KRAB domains have been demonstrated to be potent repressors, and substitutions of conserved residues within this domain abolish repression activity (34). These observations suggested that transcription repression is a common property of this domain (for a review, see reference 24).
Repression mediated by DNA binding proteins has been shown to proceed via several mechanisms, including histone deacetylation (21, 51, 55), template heterochromatinization (19, 38, 45), and direct interaction with components of the transcription machinery (3, 4, 14, 15, 43, 53). Like activators, many eukaryotic repressor proteins recruit specific corepressors via protein-protein interactions, and these interactions appear to be necessary for template silencing. To identify the mechanism(s) of KRAB-ZFP-mediated repression, we previously identified and cloned the gene encoding a KRAB domain binding protein, KAP-1, which shows all the hallmarks of being a universal corepressor for the KRAB domain (17). KAP-1 was subsequently identified by other investigators by using yeast two-hybrid screens as transcription intermediary factor 1β (TIF1β) and KRIP-1 (27, 30, 36). KAP-1 is a 97-kDa nuclear phosphoprotein whose primary amino acid sequence displays a number of interesting structural motifs. The RING finger, B boxes (β1 and β2), and a coiled-coil region at the amino terminus collectively constitute the KRAB interaction, or RBCC, domain (17, 36, 42). Carboxy terminal to this constellation of motifs appears a relatively novel stretch of amino acids, a plant homeodomain (PHD) finger, and a bromodomain, which likely represent at least two or more independent repression domains (17, 42).
A number of lines of evidence have suggested that KAP-1 plays a key role in mediating KRAB domain repression: (i) KAP-1 binds to multiple KRAB repression domains both in vitro and in vivo, (ii) KRAB domain mutations which abolish repression decrease or eliminate the interaction with KAP-1, (iii) overexpression of KAP-1 enhances KRAB-mediated repression in a manner dependent on the presence of the RBCC domain, and (iv) heterologous fusions between KAP-1 and a DNA binding domain can potentiate repression (17, 36, 42). Finally, the KRAB domain does not exhibit repression activity in cells which lack KAP-1 protein (42). These results support a model in which KRAB-ZFPs bind a gene in a DNA sequence-specific manner and repress transcription of the bound gene by recruiting the KAP-1 corepressor. The next question is: what are the molecules downstream of the KAP-1 corepressor which mediate transcription repression?
Clues to the nature of the downstream players in the repression pathway have come from the analysis of KAP-1 homologues and orthologues. In a functional screen for nuclear hormone receptor coactivators, TIF1α was cloned and shown to be similar in overall architecture to KAP-1 (31, 33). Like KAP-1, TIF1α contains an NH2-terminal RBCC motif and carboxy-terminal PHD and bromodomains and can be considered an orthologue of KAP-1. However, there is little functional cross talk between these proteins: KAP-1 does not bind to nuclear hormone receptors, and TIF1α binds very weakly to the KRAB domain. When TIF1α was used in a two-hybrid screen, two of the interacting components were the murine heterochromatin proteins M31 (mMOD1) and mHP1α. Remarkably, a second two-hybrid screen using mHP1α as the bait yielded the murine homologue of KAP-1 (designated TIF1β [30]). Taken together, these data suggest that the KAP-1-mediated repression pathway may involve the local heterochromatinization of DNA templates via interaction with specific heterochromatin proteins.
A long history of studies have shown that heterochromatin is a repressive chromosomal environment (9). For example, when a euchromatic region is juxtaposed to heterochromatin by chromosomal rearrangement, the genes contained within the region become repressed. This gene-specific repression gives rise to phenotypic variegation in tissues where the genes are normally active. This phenomenon, called position effect variegation (PEV) (48), has allowed geneticists to identify second-site mutations that can modify variegation. One of the first modifiers identified at the molecular level, and subsequently the best studied, is Drosophila heterochromatin-associated protein 1 (HP1) (12, 13, 25). The HP1 gene, allelic to Su(var)2-5, is a dosage-dependent modifier of variegation (13), and the protein is diagnostic for heterochromatin (25). The exquisite sensitivity of PEV to changes in the dosage of heterochromatin proteins like HP1 has led to a model whereby heterochromatin may be envisaged as a large macromolecular complex whose constituent components are encoded by modifier genes: it is this complex that is thought to repress gene activity (50). HP1 shares a highly conserved 50- to 60-aa region, termed the chromodomain (39, 47), with another protein, Polycomb (Pc), which is a repressor of the homeotic genes (38, 39). This observation not only suggested that the chromatin-mediated silencing in PEV and repression of the homeotic genes may be mechanistically related (19, 38) but also allowed the identification of chromobox sequences in a variety of animal and plant species and the cloning of genes that are either HP1-like or Pc-like (47).
The HP1 class of chromodomain proteins are characterized by the presence of a chromodomain at the amino terminus preceded by a stretch of glutamic acid residues. Members of this class also share a second conserved domain at the carboxy terminus termed the chromoshadow domain (1). Pc-like proteins are larger and have instead of a chromoshadow domain another carboxy-terminal homology called a Pc domain (40). Three HP1-like genes have now been identified in humans and rodents. In humans, the genes have been termed hHP1α, hHP1β, and hHP1γ (18, 37, 44, 47, 57); in mice, the homologous genes have been termed mHP1α, M31 (mMOD1), and M32 (mMOD2) (22, 23, 32, 40), respectively. The characterization of M31 and M32 has been revealing. M31, which is identical to hHP1β (37), is the closest sequence homologue of Drosophila HP1 and is a component of constitutive heterochromatin in mice and humans (18, 37, 56). M32, the homologue of hHP1γ (57), is also a member of the HP1 class of chromobox genes (23) but is excluded from constitutive heterochromatin and is distributed in a fine-grain, or speckled, pattern of many hundreds of spots throughout the nucleoplasm. This distribution suggests that the M32 gene product is a component of a macromolecular complex that represses gene activity in euchromatic DNA through regional compaction of chromatin into a heterochromatin-like complex (23).
Our present study builds on the convergent findings from studies on transcriptional corepressors (17, 30) and from the work on mammalian HP1-like genes (23, 45). We now describe a detailed structure-function analysis in vitro and in vivo of the KAP-1 interaction with HP1 family proteins. We demonstrate that KRAB-ZFPs and KAP-1 form a stable quaternary complex with DNA and HP1 protein and that the KAP-1 interaction with HP1-like proteins occurs through a specific protein domain called the HP1 binding domain (HP1BD). We demonstrate that the M31 and M32 proteins colocalize with KAP-1 within interphase nuclei and that the location patterns of these proteins indicate that their subnuclear distribution within the nucleus is dynamic and may lead to the formation of discernible regions that may represent locally silenced chromosomal domains.
MATERIALS AND METHODS
Expression plasmids.
Glutathione _S_-transferase (GST) fusions of the entire murine M31 and M32 cDNAs (22) were created by subcloning into the _Eco_RI sites of pGEX-2T and pGEX-3x (Pharmacia), respectively. The hHP1α and hHP1γ cDNAs were subcloned into pGEX-2T and were kindly provided by H. J. Worman (57). GST-KRAB(B) and GST-KRAB(DV) have been described previously (17). The plasmid expressing aa 1 to 90 of KRAB fused to a His6-tagged GAL4 DNA binding domain [6HisGAL4-KRAB (1-90) protein] was constructed via PCR using plasmid pM1-KOX, 1-90 (34) as a template. Briefly, a 5′ oligonucleotide incorporated a _Bam_HI site immediately 5′ to the GAL4 initiator methionine and a 3′ oligonucleotide incorporated a stop codon after aa 90 of KOX-1 followed by a _Hin_dIII site. The resulting, appropriately digested PCR product was cloned into the pQE30 vector (Qiagen Inc.) at the corresponding restriction sites. The protein was purified under denaturing conditions (6 M guanidine-HCl) and then subjected to exhaustive step dialysis. The GAL4–KAP-1 expression construct was described previously (17). The Mut1 and Mut2 GAL4–KAP-1 plasmids were created by standard PCR-mediated mutagenesis. The mutagenic primers contained the following codons: Mut1, GCTGCT (AlaAla) at amino acid positions 519 to 520; and Mut2, GAAGAG (GluGlu) at amino acid positions 487 to 488. To generate the corresponding Escherichia coli expression plasmids for these mutants, each was digested with _Bam_HI and _Xma_I (internal sites in human KAP-1), and the DNA fragments (encoding aa 381 to 618) were subcloned into the pQE31 (Qiagen) expression plasmid at the corresponding restriction sites. These proteins were produced, purified, and eluted from the Ni2+-agarose (Qiagen) with imidazole under native conditions as recommended by the manufacturer. The FLAG epitope-tagged mammalian expression plasmids containing aa 1 to 191 of hHP1α and aa 17 to 173 of hHP1γ were generated by subcloning _Bam_HI/_Xho_I fragments from the pBTF4 plasmids into the corresponding sites of pcDNA3 (Invitrogen), kindly provided by H. J. Worman (57). The cytomegalovirus (CMV)-based mammalian expression plasmids used in COS-1 cells, 6HisKAP-1 delRB (aa 239 to 835), 6HisRBCC (aa 20 to 419), and 6HisPHD/Bromo (aa 619 to 835), have been described elsewhere (17, 42). The 6HisKAP-1 aa 423 to 584 mammalian expression plasmid was generated by subcloning an _Eco_RI/_Hin_dIII fragment from pQE30 (17) into the corresponding sites of pcDNA3.1 (Invitrogen). The GAL4–nuclear hormone receptor corepressor (N-CoR) plasmid expressing aa 1 to 312 of N-CoR repression domain 1 was kindly provided by M. Lazar. All PCR-derived plasmids were subjected to automated DNA sequencing of both strands to confirm sequence integrity.
Cell extract preparation.
COS-1 cells were grown in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and grown in 5% CO2 at 37°C. Whole-cell extracts were prepared by first washing cells with phosphate-buffered saline (PBS) four times and then lysing them in the dish in ELB buffer (50 mM HEPES [pH 7.5], 250 mM NaCl, 0.1% Nonidet P-40 [NP-40], 1 mM EDTA) including protease inhibitors (0.1 mM phenylmethylsulfonyl fluoride [PMSF], 100 μg of aprotinin, 10 μg of leupeptin, and 10 μg of pepstatin per ml, and 1 mM benzamidine). The lysates were aspirated from the dish, particulate matter was clarified from the extract by centrifugation at 100,000 × g for 30 min, at 4°C, and supernatants were collected. COS-1 nuclear extracts (CNE) were prepared from COS-1 cells via a slight modification of the method of Lassar et al. (29). After four washes with PBS, cellular lysis was carried out by first treating cells at 4°C with nonnuclear lysis buffer (10 mM HEPES [pH 7.6], 10 mM NaCl, 1.5 mM MgCl2, 20% glycerol, 0.2 mM EDTA, 0.1% Triton X-100) including protease inhibitors (0.1 mM PMSF, 100 μg of aprotinin, 10 μg of leupeptin, and 10 μg of pepstatin per ml, and 1 mM benzamidine). The nuclei were then collected by centrifugation at 1,250 × g for 5 min at 4°C. The pelleted nuclei were lysed in nuclear extraction buffer (NEB; 10 mM HEPES [pH 7.6], 500 mM NaCl, 1.5 mM MgCl2, 20% glycerol, 0.2 mM EDTA, 0.1% Triton X-100) containing protease inhibitors. Preparations consisted of approximately 2 × 107 nuclei/ml. The extraction was carried out by rotation at 4°C for 1 h, and the tubes were then centrifuged at 100,000 × g for 30 min at 4°C. The final protein concentration in each nuclear extract varied from 1 to 5 mg/ml.
Metabolic labeling.
For metabolic labeling of COS-1 cells, fresh cultures were first starved by incubation in DMEM lacking methionine and cysteine (ICN Biochemicals) for 30 min. The cells were then labeled using Tran35S-label (75% [35S]methionine, 15% [35S]cysteine; ICN) for 30 to 120 min in DMEM containing 10% dialyzed FBS (Sigma). The cell cultures were then washed four times in PBS, and lysates were prepared as whole-cell or nuclear extracts as described above.
GST protein preparation.
Following transformation of the expression plasmids into competent E. coli BL21(DE3) bacteria and identification of highly expressing bacterial colonies, 10-ml overnight cultures were started in 2YT medium. The next day, the entire overnight culture was added to 250 ml of fresh 2YT, and the culture was allowed to grow until the optical density at 600 nm reached 0.4 to 0.6. Isopropyl-β-d-thiogalactopyranoside was then added to 0.5 mM, and the cultures were incubated for an additional 3 to 4 h. The cells were pelleted at 8,000 × g for 10 min at 4°C. The bacteria were resuspended in 4 ml of PBS, and 400 μg of lysozyme was added. After a 15-min incubation on ice, dithiothreitol to 5 mM and protease inhibitors to final concentrations of 0.1 mM PMSF, 100 μg of aprotinin per ml, 10 μg of leupeptin per ml, 10 μg of pepstatin per ml, and 1 mM benzamidine were added. Sarcosyl was added to a final concentration of 3.5%, and the bacterial suspension was sonicated for 30 s, left on ice for 1 min, and then sonicated for an additional 30 s. The sample was centrifuged 16,000 × g for 10 min at 4°C. Triton X-100 was then added to the supernatant to a final concentration of 4%, and the protein extract was snap frozen in small aliquots. Large-scale preparations of purified GST fusion proteins were prepared by eluting the proteins from glutathione-Sepharose (Pharmacia) in GST elution buffer (100 mM Tris [pH 8.0], 150 mM NaCl, 0.1% NP-40, 20 mM freshly added reduced glutathione). Elution was with a buffer volume equal to 2.5 times the packed bead volume, and incubation was at room temperature for 1 h. The beads were centrifuged briefly, and the supernatant was collected. The elution was repeated, and the supernatants were combined and concentrated in 5,000-molecular-weight-cutoff microspin concentrators (Millipore). Protein concentrations were determined for each of the proteins by the DC protein assay (Bio-Rad). The GST proteins were diluted with PBS prior to use in all electrophoretic mobility shift assays (EMSAs).
GST pull-down assays.
Five micrograms of freshly prepared GST fusion protein immobilized on glutathione-Sepharose (Pharmacia) was incubated with either 2 μl of in vitro-translated, 35S-labeled KAP-1 (T3 TnT; Promega), 50 μl of 35S-labeled whole-cell lysate from transiently transfected COS-1 cells, 500 μg of HeLa whole-cell lysate (PBS, 0.1% NP-40), or 1 to 2 μg of Ni2+-agarose (Qiagen)-purified recombinant His6-tagged protein in 200 μl of BB100 (20 mM Tris [pH 7.9], 100 mM NaCl, 0.2 mM EDTA, 10% glycerol, 0.1% NP-40, 1 mM PMSF, 500 μg of bovine serum albumin [BSA; fraction V]) for 1 h at room temperature. Protein complexes were washed four times with BB750 (20 mM Tris [pH 7.9], 750 mM NaCl, 0.2 mM EDTA, 10% glycerol, 0.1% NP-40, 1 mM PMSF), and the bound proteins were eluted in 2× Laemmli buffer by boiling for 10 min. Proteins were resolved by standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) procedures. Retention of 35S-labeled KAP-1 was visualized by fluorography in dimethyl sulfoxide–2,5-diphenyloxazole (Fisher Biotech) followed by exposure to MR X-ray film (Kodak). For evaluation of KAP-1 binding to GST fusion protein by Western blotting, proteins were transferred to Immobilon-P (Millipore) in Towbin buffer–0.1% SDS for 20 h at 4°C and 250 mA. Membranes were blocked in 5% Blotto–Tris-buffered saline (TBS; 50 mM Tris [pH 7.5], 150 mM NaCl). KAP-1 was detected with antigen affinity-purified rabbit polyclonal antiserum (17) diluted 1:200 in TBS–1% BSA. Membranes were washed three times in TBS–0.05% Tween 20 and then incubated with a 1:5,000 dilution of a horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G secondary antibody (Sigma) in 1% Blotto-TBS. Proteins were visualized by chemiluminescence (Pierce) and exposure to MR X-ray film (Kodak).
EMSAs.
The ability of KAP-1 to bind a KRAB domain-containing protein was assayed by EMSA essentially as described previously (16). Two partially overlapping GAL4-upstream activation sequence (UAS) oligonucleotides (5′-GATCCCGGAGGACAGTACTC-3′ and 5′-CTAGACGGAGTACTGTCCTC-3′) were annealed and used for EMSAs. EMSA binding reaction mixtures (20 μl) were assembled by adding 50 ng of purified recombinant 6HisGAL4-KRAB to 2× binding buffer (40 mM HEPES [pH 7.6], 100 mM NaCl, 1 mM dithiothreitol, 10 mM MgCl2, 20% glycerol) containing 1 μg of poly(dI-dC) · poly(dI-dC). Nuclear extract (∼5 μg/l to 2 μl) and/or titrating amounts of GST fusion protein were added, and incubation was continued at room temperature for 15 min. Then γ-32P-end-labeled GAL4-UAS probe was added (∼0.5 to 1 ng, 105 cpm/reaction), and the incubation continued for an additional 10 min at room temperature. The samples were chilled on ice, centrifuged 4 min at 4°C, and then loaded onto 4.5% nondenaturing polyacrylamide gels containing 1× TBE (45 mM Tris [pH 8.3], 45 mM boric acid, 1 mM EDTA), which were preelectrophoresed for 45 min at 4°C in 0.5× TBE buffer. DNA-protein complexes were separated from unbound DNA by electrophoresis at 500 V and 24 mA for 2 more h at 4°C. The gels were dried and exposed to MR X-ray film.
FPLC gel filtration fractionation.
The 35S-labeled CNE was subjected to isocratic fast protein liquid chromatography (FPLC) gel filtration on a Superose 6 column (Pharmacia) in NEB running buffer at a flow rate of 0.5 ml/min at room temperature. The KAP-1 protein was detected in the FPLC fractions by immunoprecipitation using protein G-purified KAP-1 polyclonal antibodies (17). To determine if a KAP-1–heterochromatin protein complex is stable to and detectable after FPLC gel filtration, a nonradioactive CNE containing KAP-1 was incubated with the purified GST-heterochromatin fusion protein, GST-M32, in NEB. After 60 min, incubation at room temperature, the mixture was subjected to isocratic gel filtration as described above. Glutathione-Sepharose resin (20 μl, 50% slurry; Pharmacia) was added to individual FPLC fractions, incubated for 1 h at 4°C followed by 1 h at room temperature, and then washed two times with NEB and five times with PBS; 2× Laemmli sample buffer was added to the resin; samples were boiled and separated by SDS-PAGE (10% gel). The proteins were transferred to Immobilon-P membranes, and recovered KAP-1 was visualized by using protein G-purified anti-KAP-1 polyclonal antibodies in a standard Western blot procedure described above.
Indirect immunofluorescence.
NIH 3T3 cells were grown on glass coverslips in DMEM containing 10% calf serum and immunostained as previously described (35). The murine KAP-1 protein was visualized by indirect immunofluorescence with an antigen affinity-purified rabbit polyclonal antibody previously described (42). The M31 protein was visualized by indirect immunofluorescence using a rat monoclonal antibody (MAb) raised to the COOH-terminal 71 amino acids (anti-M31 MAb MAC 353 [56]). The M32 protein was detected with a rat MAb developed by using a GST fusion protein that included the entire coding region of M32 (anti-M32 MAb MAC 385 [23]). The hHP1 proteins were recognized by using a rabbit polyclonal antibody raised against hHP1, kindly provided by W. C. Earnshaw (44). The secondary antibodies were either Texas red-conjugated goat anti-rabbit or biotinylated goat anti-rat, used in conjunction with an avidin-biotin-linked fluorescein isothiocyanate reagent (Vector Laboratories). All immunofluorescence was performed as described previously (35). DNA was counterstained with Hoechst 33258 (Sigma), and coverslips were mounted with Fluoromount G (Fisher Scientific). Cells were visualized with a scanning confocal microscope (Leica Inc.). The images obtained through image capture were processed with Adobe Photoshop 3.0.4 (Adobe Systems Inc.) from files or scanned slide images.
Transient transfection luciferase assays.
DNA for transfection was prepared by CsCl gradient centrifugation. Protein expression from all plasmids was confirmed by transient transfection of COS-1 cells followed by immunoprecipitation of [35S]methionine-labeled cell extracts as described previously (16). All transcription assay transfections were done with NIH 3T3 cells maintained in DMEM–10% calf serum. A total of 2.0 × 105 cells were plated in a 60-mm-diameter tissue culture dish and transfected in OptiMEM for 5 to 6 h with Lipofectamine (Life Technologies Inc.) under conditions recommended by the manufacturer. The cells were harvested 24 h posttransfection, and luciferase assays were performed as previously described (17). Cotransfection with a pcDNA3-β-galactosidase expression plasmid was used to normalize all luciferase values.
RESULTS
Sequence analysis of KRAB binding corepressor and heterochromatin-associated protein families.
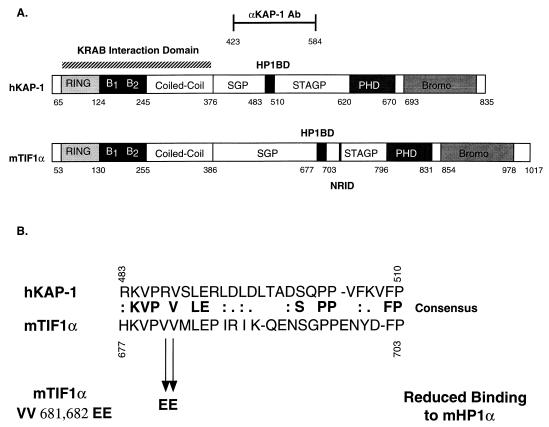
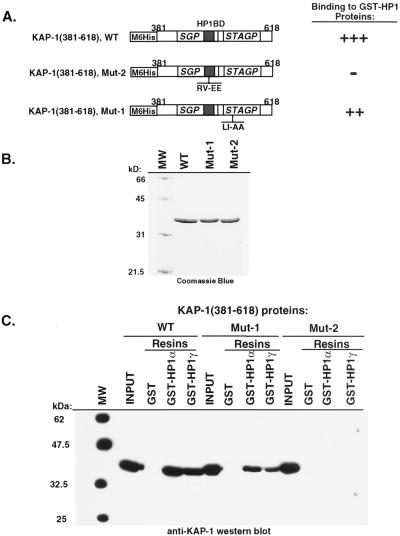
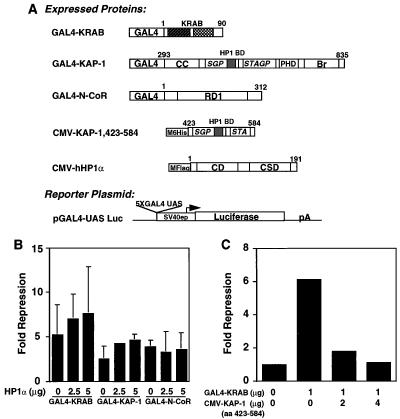
TIF1α and KAP-1 share a number of similar amino acid sequence motifs, including the well-characterized RING finger, B1 and B2 boxes, a coiled-coil, a PHD finger, and an extended bromodomain (17, 30, 36) (Fig. 1A). Overall, KAP-1 is only 33% identical and 45% similar to hTIF1α. Previous studies with mTIF1α delineated a region of the protein (aa 672 to 698) which may be sufficient for binding mHP1α (30) (Fig. 1A). We have completed sequence comparison of these amino acids to KAP-1 and have found that KAP-1 contains a highly homologous domain which we believe is a potential HP1BD (KAP-1 aa 483 to 510). This putative HP1BD in KAP-1 is conserved with 45% identity and 60% similarity to the analogous region in mTIF1α (Fig. 1B). The greatest degree of identity and conservation is shown in the amino-terminal portion of the HP1BD. Two previously identified valines (aa 681 and 682) were mutated to glutamic acid in mTIF1α and shown to substantially reduce binding to mHP1α (30). One of these valines is among the conserved amino acids found in this putative HP1BD (Fig. 1B).
FIG. 1.
Schematic diagram illustrating the architecture of the KAP-1/TIF1α family of transcriptional regulatory proteins. (A) The conserved motifs include the RING finger, B boxes (B1 and B2), coiled-coil, PHD (also known as the LAP domain), and bromodomain (Bromo). Note the overall similar architectures among this family of proteins defined by the layout of the various domains. The putative HP1BD (black boxes) is conserved in each protein; the nuclear receptor interaction domain (NRID) is conserved only in TIF1α. Regions of significantly enriched amino acids, serine-glycine-proline (SGP) and serine-threonine-alanine-glycine-proline (STAGP), are also spatially conserved in this family. The minimal KRAB binding domain comprises the RBCC domain and is marked with a striped bar above the hKAP-1 protein. Also indicated is the region of hKAP-1 expressed as a recombinant protein and used to raise anti-KAP1 polyclonal antisera (αKAP-1 Ab). (B) Amino acid alignment of the putative HP1BD of KAP-1 and mTIF1α. Mutation of valines 681 and 682 of mTIF1α to glutamic acid (VV 681,682 EE) were previously observed to abolish mHP1α binding to mTIF1α (30). The corresponding sequences are from data bank entries 78773 (hKAP-1) and 78219 (mTIF1α).
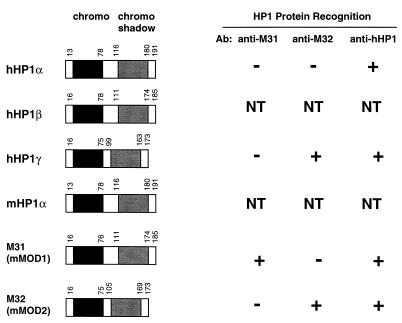
Sequence comparisons among the human and mouse chromodomain-containing HP1 proteins suggest that the murine homologues of hHP1α, -β, and -γ are mHP1α, M31, and M32, respectively. Studies have shown that all of these HP1 proteins contain the highly conserved chromodomains and chromo shadow domains (1) as well as nuclear localization signals, and some may even have potential DNA binding domains (49). To evaluate antibodies raised against the M31, M32, and hHP1 proteins, we measured the binding specificity and cross-reactivity of these antibodies against bacterially expressed and affinity-purified human and murine heterochromatin fusion proteins. The results indicate that the M31 antibody is specific for M31, the M32 antibody recognizes both M32 and hHP1γ, and the hHP1 antibody recognizes all of the heterochromatin proteins (Fig. 2). These observations suggest that the M31 and M32 antibodies are specific reagents that can be used in immunolocalization studies with KAP-1.
FIG. 2.
The HP1 family of human and mouse heterochromatin proteins. The regions of conservation which are diagnostic for the HP1 family of proteins include the NH2-terminal chromodomain (black boxes) and the COOH-terminal chromo shadow domain (grey boxes). On the right is a summary of the immunoreactivities for the various antibodies (Ab) available to recombinant heterochromatin proteins derived from humans and mice. NT, not tested. The corresponding sequences are from the following data bank entries: hHP1α, 60277; hHP1β, 23197; hHP1γ, 26312; mHP1α, 99641; M31, 56690; and M32, 56683.
KAP-1 and HP1 family proteins interact in vitro.
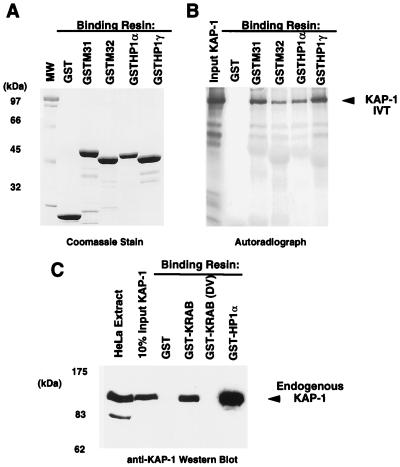
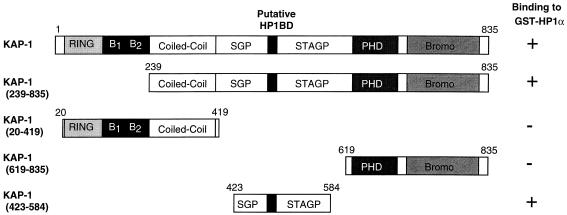
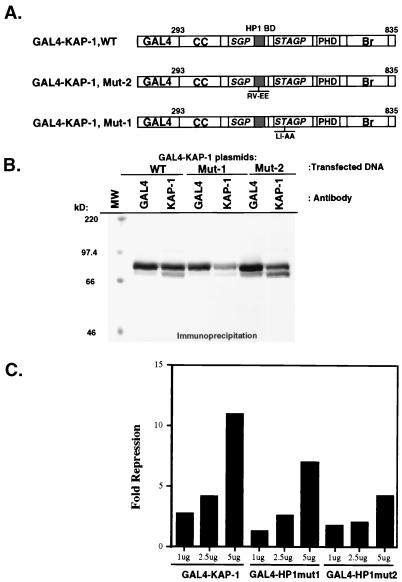
In light of the result that KAP-1 could bind mHP1α and M31 proteins in a yeast two-hybrid assay (30), we initiated a comprehensive analysis of the abilities of other heterochromatin family members to bind KAP-1. After purification of GST-heterochromatin protein fusion proteins (Fig. 3A), the resins were analyzed for the ability to bind KAP-1 protein produced by in vitro transcription and translation. In general, significant binding of KAP-1 was observed for all of the HP1 proteins but was negative for the control GST protein (Fig. 3B). Further analysis revealed that the interaction between in vitro-transcribed-translated KAP-1 and recombinant HP1 proteins in solution is extremely stable, as nearly equal signal intensities were observed between resins washed in either 250 mM NaCl or 1 M NaCl (data not shown). To assay the ability of cell-derived KAP-1 protein to bind HP1 proteins, GST-hHP1α was incubated with HeLa cell extracts. Binding of KAP-1 to the recombinant GST-hHP1α protein was detected by Western blot analysis using polyclonal antiserum raised against KAP-1. As illustrated in Fig. 3C, hHP1α displays significant binding capacity for cell-derived KAP-1; greater than 50% of the input KAP-1 in the extract bound to the GST-hHP1α resin. Identical data were obtained when GST-hHP1γ resin was used in the assays (data not shown). The GST-KRAB and GST-KRAB(DV) resins demonstrate the specificity of KAP-1 binding, since KAP-1 is observed to interact only with GST-KRAB and not the mutant GST-KRAB(DV), which lacks KAP-1 binding activity (Fig. 3C). To grossly localize the HP1 interaction region, we expressed truncated KAP-1 proteins in COS-1 cells and assayed extracts containing these proteins for the ability to bind GST-hHP1α (data not shown). As summarized in Fig. 4, only KAP-1 proteins which contained the central region spanning aa 423 to 584, which includes the putative HP1BD, bound to GST-hHP1α. In addition, two peptides corresponding to aa 435 to 449 and 568 to 581 of KAP-1 when added in molar excess to the binding reaction mixture were unable to block the interaction between KAP-1 and HP1s, suggesting that the putative HP1BD is between aa 450 and 568 of KAP-1 (data not shown). To determine if the KAP-1–HP1 interaction is direct, we used purified, recombinant proteins (Fig. 5). The region of wild-type KAP-1 spanning aa 381 to 618 and two mutant versions thereof (Fig. 5A) were expressed in bacteria and purified to homogeneity (Fig. 5B). These proteins were used in in vitro binding assays utilizing GST-hHP1α and -γ proteins purified to near homogeneity (Fig. 3A). Wild-type KAP-1 efficiently bound to both resins (Fig. 5C). The Mut1 protein, which contains the LI-AA (aa 519 and 520) substitutions carboxy terminal to the putative HP1BD, also bound each resin with moderate efficiency. However, the Mut2 protein, with the RV-EE (aa 487 and 488) substitutions within the putative HP1BD, completely abolished interaction with the GST-hHP1 proteins. Together, these results strongly suggest that the KAP-1–HP1 association is via direct protein-protein interactions and that the region of KAP-1 spanning aa 450 to 568 possesses the interaction domain.
FIG. 3.
The KAP-1 protein binds to both human and murine HP1 family proteins. (A) Analysis of the GST fusion proteins affinity purified as a soluble protein from E. coli extracts and used in the binding assays. Approximately 5 μg of each protein was electrophoresed on an SDS–10% polyacrylamide gel and stained with Coomassie blue. Equivalent quantities of protein (5 μg) were used for each binding reaction as determined by Coomassie blue staining and quantification against BSA. MW, molecular weight markers. (B) hKAP-1 was prepared by in vitro transcription-translation (IVT) of a full-length human cDNA, and the [35S]methionine-labeled protein (arrow) was used in binding reactions with the indicated GST fusion proteins. The input lane represents the total amount of KAP-1 in each reaction mixture. No binding was observed in reactions containing GST alone. (C) A HeLa whole-cell extract containing endogenous KAP-1 was used in binding reactions with the indicated GST fusion protein. GST-KRAB represents a positive control for KAP-1 interaction, and GST-KRAB(DV) is a mutant with reduced affinity for KAP-1, which served as a negative control for KAP-1 interaction. Retention of KAP-1 by the GST fusion protein was detected by Western blot analysis using affinity-purified anti-KAP-1 antibodies. The input lane represents 10% of the KAP-1 in each binding reaction as detected by Western blot analysis of the extract.
FIG. 4.
Localization of the HP1BD in KAP-1. COS-1 cells were transfected with the indicated expression constructs containing different regions of KAP-1, and extracts from these cells were assayed for the ability of KAP-1 to bind GST-hHP1α. All KAP-1 molecules which included the putative HP1BD bound to GST-hHP1α, as indicated by the plus signs. For notation, see the legend to Fig. 1.
FIG. 5.
Amino acid substitutions in the HP1BD abolish HP1 binding in vitro. (A) The indicated wild-type (WT) or mutated segment of KAP-1 (for notation, see the legend to Fig. 1) was expressed in bacteria and purified by nickel chelate chromatography under native conditions. These highly purified proteins (B) were used in GST binding assays using the recombinant proteins shown in Fig. 3A. After the binding reactions and extensive washing, the amount of bound KAP-1 protein was determined by Western blot analysis using affinity-purified anti-KAP-1 sera (C). The input lane represents the total amount of recombinant KAP-1 added to each binding reaction mixture. No binding was detected for GST alone. Note that the recombinant Mut2 protein, which possesses the RV-EE mutation in the putative HP1BD, completely abolishes binding to both hHP1α and hHP1γ. MW, molecular weight markers.
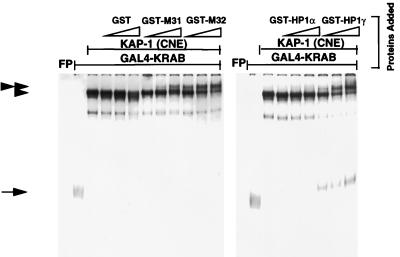
Previously, we have shown that KAP-1 can interact very efficiently with KRAB domain-containing proteins in an EMSA (17, 42). This highly sensitive assay allows for the detection of protein-protein interactions with limiting amounts of proteins at concentrations at or close to their dissociation constants, rather than at extreme excess, as is usually the case in solution-based GST binding assays. In addition, the EMSA requires that the complex once formed be stable to prolonged electrophoresis and has the capability of distinguishing high- and low-affinity interactions. To further assess the ability of the different HP1 proteins to interact with KAP-1, we again used EMSA. The well-characterized GAL4 (aa 1 to 147) DNA binding domain was used to construct a 6HisGAL4-KRAB fusion protein, which was purified to near homogeneity. This protein binds with moderate efficiency to the 32P-labeled GAL4-UAS oligonucleotide (Fig. 6) under EMSA conditions. However, preincubation of the 6HisGAL4-KRAB with a KAP-1-containing CNE results in a dramatic supershift of the DNA binding complex in the EMSA gel. This supershifted complex contains both proteins, as antibodies directed to either GAL4 or KAP-1 efficiently supershifted this complex further (data not shown). Importantly, purified, recombinant HP1 fusion proteins GST-M31, GST-M32, GST-hHP1α, and GST-hHP1γ were all able to bind the 6HisGAL4-KRAB–KAP-1 complex, as evidenced by a new supershifted complex containing the HP1 protein. hHP1α appears to have a lower affinity for the DNA–KRAB–KAP-1 ternary complex (Fig. 6). The addition of purified GST protein alone failed to produce the supershift. We also tested all of the heterochromatin proteins for binding by varying the order of addition of each component (data not shown). In these experiments, the GAL4-UAS–6HisGAL4-KRAB–KAP-1 complex was formed prior to addition of the heterochromatin protein: the binding observed was identical to that seen in Fig. 6, suggesting that interaction of KRAB and KAP-1 does not preclude binding of the heterochromatin protein. These results strongly suggest that a quaternary GAL4-UAS–6HisGAL4-KRAB–KAP-1–HP1 complex is readily formed in vitro and is stable to the EMSA conditions used.
FIG. 6.
Purified recombinant HP1 proteins can bind a DNA–GAL4-KRAB–KAP-1 ternary complex. A gel shift assay was performed by incubating a highly purified recombinant 6HisGAL4-KRAB domain protein with KAP-1 derived from a CNE, the indicated GST fusion protein (increasing concentrations from left to right indicated by open triangles: 50, 100, and 200 ng each of GST, GST-M31, GST-M32, GST-hHP1α, and GST-hHP1γ), and a γ-32P-labeled GAL4-UAS oligonucleotide probe. The complexes were separated by nondenaturing PAGE, and DNA-protein interactions were detected by autoradiography. The symbols on the left denote the following complexes: arrow, DNA–6HisGAL4-KRAB; single arrowhead, DNA–6HisGAL4-KRAB–KAP-1 ternary complex; double arrowhead, DNA–6HisGAL4-KRAB–KAP-1–HP1 quaternary complex. The free probe (FP) was allowed to run off the bottom of the gel to allow for better separation of the complexes at the top of the gel.
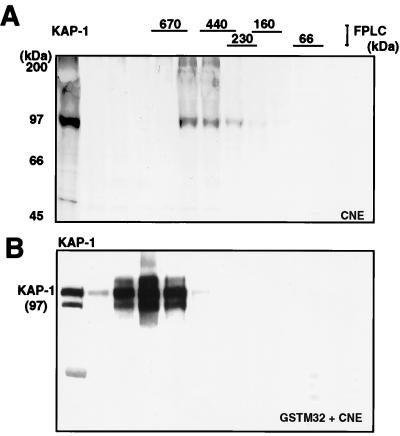
Using gel filtration chromatography, we have shown that the endogenous KAP-1-containing complex found in CNE migrated at approximately 560 kDa (42). We sought to determine if an exogenous source of HP1 protein could influence the migration of the CNE KAP-1 complex in gel filtration (Fig. 7). Purified soluble GST-M32 protein was incubated with CNE, and then the mixture was subjected to gel filtration. The fractions were incubated with glutathione-Sepharose beads to recover the GST-M32 protein and any associated proteins, and the washed beads were analyzed for KAP-1 by Western blotting. Comparison of the native 560-kDa KAP-1 complex from CNE to that produced when GST-M32 was mixed with CNE prior to separation shows that the KAP-1 from the nuclear extract was efficiently bound to and recovered by binding GST-M32 after separation (Fig. 7). More importantly, the resulting KAP-1 complex now migrated at approximately 800 kDa. These results demonstrate that the reconstituted KAP-1–M32 complex is stable to gel filtration and suggest that multiple molecules of M32 may bind to the endogenous KAP-1 complex.
FIG. 7.
Cell-derived KAP-1 complexes increase in apparent size in the presence of the heterochromatin protein M32. (A) A [35S]methionine-labeled CNE was subjected to gel filtration (Superose 6), and the column fractions were analyzed for KAP-1 by immunoprecipitation. (B) A nonradioactive CNE was incubated with purified GST-M32 protein, and the mixture was subjected to gel filtration as for panel A. The column fractions were incubated with glutathione-Sepharose beads and washed, and the retained KAP-1 was detected in a Western blot assay using affinity-purified anti-KAP-1 sera. Lane 1 is a sample of the unfractionated CNE containing endogenous KAP-1 as a positive control and size marker. Indicated above the gels are the positions of the molecular mass standards used to calibrate the Superose 6 column, which were determined by Coomassie blue staining of duplicate gels.
Colocalization of KAP-1 with the M31 and M32 HP1-like proteins.
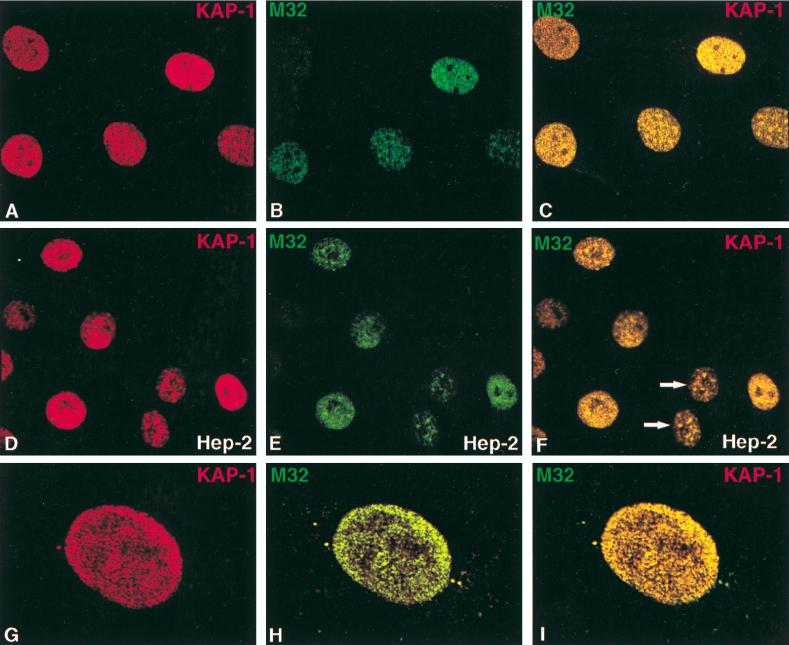
To ascertain a potential in vivo role for the KAP-1–HP1 association, we used antibodies raised to M31 (56) and M32 (23) concurrently with an affinity-purified anti-KAP-1 antibody in dual staining-indirect immunofluorescence studies of asynchronous NIH 3T3 fibroblasts. In a random field of immunostained cells, there appeared to be various levels of KAP-1 staining (Fig. 8 and 9). In general, KAP-1 staining was found exclusively throughout the nucleoplasm as an evenly distributed grainy (speckled) pattern which was almost always excluded from nucleoli. In nearly 50% of the NIH 3T3 nuclei, KAP-1 is significantly concentrated in dot-like structures which could represent regions of pericentromeric heterochromatin commonly observed in murine cells (Fig. 9A). It should be noted that this specific staining pattern for KAP-1 does not overlap with other well-characterized nuclear dot-like structures previously described for PML, SC35, and BRCA1 (34a). KAP-1 staining in human HEp-2 cells yielded very similar results (Fig. 9D to F). An interesting pattern was observed in less than 5% of the nuclei: KAP-1 was concentrated both at the periphery of pericentromeric heterochromatin and around nucleoli (Fig. 8D and G). Since the cell populations illustrated are asynchronously growing, log-phase cells, these data may suggest that the different subnuclear localization patterns of KAP-1 observed in a random field of cells may be due to cell cycle regulation. In support of such a conclusion, apparent daughter cells in many of the fields shown display very similar KAP-1 staining patterns.
FIG. 8.
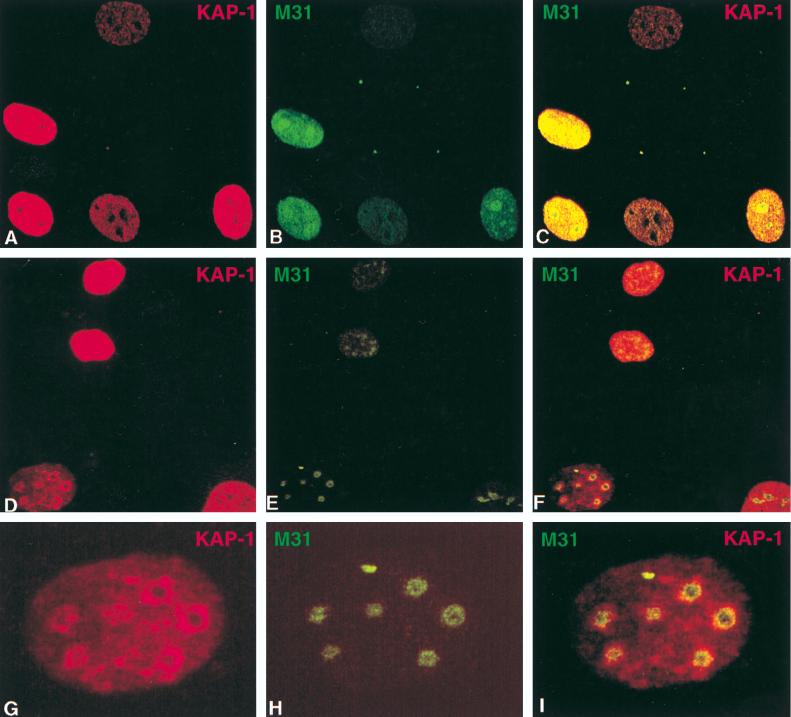
Colocalization of endogenous KAP-1 with the heterochromatin protein M31 in interphase nuclei. All images were derived from the staining of a single population of cells simultaneously for both antigens and viewed by confocal microscopy. (A to F) Each row illustrates an independent random field of asynchronously growing NIH 3T3 fibroblasts to provide a representative view of the various staining patterns observed for both KAP-1 and M31. Staining of individual antigens is identified by the color given in the upper corners of each image. The merged images are shown on the far right for each set. As illustrated, staining for both antigens was exclusively nuclear, with very little background staining in the cytoplasm. KAP-1 staining demonstrated a grainy, speckled pattern throughout the nucleus which was largely excluded from nucleoli. M31 staining displayed a strong association with large heterochromatic granules. (G to I) High-magnification single-cell analysis of KAP-1 and M31 subnuclear distribution. Each panel shows a single nucleus from the field displayed in panels D to F. Note the discrete pericentromeric heterochromatin and nucleoli, as detected by M31 (56), and the concentration of KAP-1 around these structures (I).
FIG. 9.
Colocalization of endogenous KAP-1 with the heterochromatin protein M32 in interphase nuclei. All images were derived from the staining of a single population of cells simultaneously for both antigens and viewed by confocal microscopy. (A to C) Random fields of asynchronously growing NIH 3T3 fibroblasts. (D to F) KAP-1 and hHP1γ staining in human HEp-2 cells, as indicated in the lower right corner of each image (arrows point to two apparent daughter cells). Note the speckled pattern for M32 which is largely excluded from nucleoli and pericentromeric heterochromatin. In several cells, a significant overlap was observed between KAP-1 and M32 staining (C, F, and I). The most dramatic representation of this colocalization is portrayed in panels G to I, representing a high-magnification single-cell analysis of KAP-1 and M32 subnuclear distribution in a NIH 3T3 nucleus, where the green and red speckles show a near complete overlap, yielding a uniform yellow in the merged image (I).
Immunostaining for the M31 protein revealed two distinct patterns in interphase nuclei. The less frequently observed pattern was characterized by diffuse staining throughout the nucleoplasm, with little or no localization to the nucleoli or heterochromatic regions (Fig. 8B). More typically, staining of the M31 protein was restricted to large condensed heterochromatic regions (Fig. 8E and H), similar to results previously shown in interphase mouse C1271 cells (23). When the KAP-1 and M31 staining patterns were directly compared, we found that in some cells, the signals appear to be juxtaposed (Fig. 8C and F). Of particular interest was the significant colocalization of KAP-1 with M31 in the regions bordering the nucleoli and pericentromeric heterochromatin (Fig. 8G to I).
Immunostaining of the M32 protein was generally observed as a fine, speckled pattern throughout the nucleoplasm in regions consistent with euchromatin (Fig. 9B, E, and H). This particular staining pattern is consistent with previous reports for M32 (23). When M32 staining was directly compared to KAP-1 staining, we found that some of these punctate and small speckled M32-reactive regions were also domains where high concentrations of KAP-1 localized (Fig. 9C, F, and I). In some nuclei, M32 and KAP-1 were nearly exclusively colocalized (Fig. 9G to I). Since the M32 MAb also reacts with hHP1γ (Fig. 2), we looked at the localization of HP1γ in human HEp-2 cells. The pattern of hHP1γ staining appeared to be very similar to that observed for M32 in NIH 3T3 cells: punctate nuclear localization in regions that appear to be consistent with euchromatin (Fig. 9D to F). Again significant colocalization of hHP1γ and KAP-1 was observed in the merged images (Fig. 9F). This colocalization may depend on cell cycle stage, which is suggested by the similar staining patterns observed for two daughter cells (arrows) in this field (Fig. 9F). In summary, both the M31 and M32 heterochromatin proteins appear to display substantial colocalization with endogenous KAP-1 in interphase nuclei. These data imply a potential in vivo association between KAP-1 and members of the HP1 family, which may function to epigenetically silence a KRAB-ZFP-regulated gene via heterochromatinization.
Modulation of KRAB–KAP-1 repression activity by hHP1α and the KAP-1 HP1BD.
To determine if there were any functional consequences of the KAP-1–HP1 interaction, we cotransfected NIH 3T3 fibroblasts with a GAL4-KRAB plasmid and the GAL4-UAS luciferase reporter plasmid (Fig. 10A). Under these conditions, the GAL4-KRAB protein was a strong repressor of the luciferase reporter gene activity; however, this repression was further enhanced by cotransfection of the hHP1α expression plasmid in a concentration-dependent manner (Fig. 10B). Cotransfected hHP1α also enhanced GAL4–KAP-1-mediated repression, although to a lesser extent. A GAL4 fusion of the NH2-terminal repression domain of the N-CoR corepressor also displayed significant repression activity, yet this activity was not enhanced by cotransfected hHP1α (Fig. 10B). Thus, N-CoR may not utilize heterochromatin proteins for mediating repression. As a complementary strategy, we attempted to disrupt endogenous KAP-1–HP1 complexes by overexpressing a protein which encodes the segment of KAP-1 (aa 423 to 584) which contains the HP1BD. Ectopic expression of this protein would be predicted to compete with endogenous KAP-1 for binding to endogenous HP1 family proteins, resulting in relief of repression via a squelching mechanism. As predicted, increasing amounts of KAP-1 HP1BD (aa 423 to 584) efficiently relieved GAL4-KRAB-mediated repression but had no effect on the basal level of transcription of the luciferase vector in the absence of GAL4-KRAB (Fig. 10C). We next determined the effect of mutations in KAP-1 which abolish HP1 binding (Fig. 11). The Mut1 and Mut2 mutations (Fig. 5) were introduced into the GAL4-KAP-1,293-835 expression plasmid (Fig. 11A), which has been previously shown to contain the repression functions of KAP-1. Each of these plasmids express equivalent amounts of nuclearly localized (data not shown) proteins which are readily detected by both anti-KAP-1 and anti-GAL4 sera (Fig. 11B). As expected, expression of the heterologous wild-type GAL4–KAP-1 protein demonstrated a strong, dose-dependent repression of the GAL4-UAS-simian virus 40 (SV40) immediate-early promoter luciferase reporter (Fig. 11C). The heterologous Mut2 protein, which is negative for HP1 binding in vitro (Fig. 5C), was strongly reduced in its repression capacity. Similar to its HP1 binding characteristics in vitro, expression of the heterologous Mut1 protein was less strongly affected. The residual repression activity in both Mut1 and Mut2 proteins is not surprising, as the still-intact PHD and bromodomain in each protein are themselves independent repression domains (17, 42). Together, these results suggest that the HP1 family of proteins may serve as one component of the KRAB–KAP-1 repression complex in vivo.
FIG. 10.
Modulation of KRAB/KAP-1 repression activity by exogenous hHP1α and a dominant negative KAP-1. (A) Plasmids used for transfection. The indicated GAL4-DNA binding domain fusion proteins were expressed from the SV40 enhancer/promoter (SV40ep) in mammalian cells. The full-length hHP1α contained a FLAG epitope tag at the amino terminus. The fragment of KAP-1 (aa 423 to 584) which contains the HP1BD was expressed as an NH2-terminal histidine fusion protein from a CMV vector. The SV40 promoter-based luciferase (Luc) reporter plasmid contains five synthetic GAL4-UAS sites and was used in all transfections. CC, coiled coil; Br, bromodomain; RD1, repression domain 1; CD, chromodomain; CSD, chromo shadow domain; pA, poly(A) site. (B) One microgram of each GAL4 expression plasmid was cotransfected into NIH 3T3 cells with increasing amounts of the hHP1α plasmid. Fold repression was calculated by comparing normalized luciferase activities with that of cells transfected in the absence of a GAL4 effector protein. The error bars represent the standard errors observed for three independent transfections, each performed in duplicate. (C) A GAL4-KRAB expression plasmid was cotransfected with increasing amounts of CMV–KAP-1 (aa 423 to 584), which encodes the putative HP1BD. Note that GAL4-KRAB repression activity is almost completely abolished in the presence of 4 μg of the CMV–KAP-1 (aa 423 to 584) plasmid, suggesting that the HP1BD is titrating endogenous HP1 proteins, thus relieving GAL4-KRAB-mediated repression via a squelching mechanism.
FIG. 11.
Mutations in the HP1BD reduce the intrinsic repression activity of the KAP-1 protein. (A) The plasmids used for transfection include GAL4 fusions to the wild-type (WT) KAP-1 or the Mut1 and Mut2 versions depicted in Fig. 5. Each plasmid (for notation, see the legend to Fig. 10) was tested for stable expression via transfection into COS-1 cells followed by immunoprecipitation analyses using both anti-KAP-1 and anti-GAL4 sera (B). These plasmids were then used in transfection assays with the 5XGAL4-UAS-SV40 luciferase reporter plasmid (Fig. 10A). (C) The wild-type (WT) GAL4–KAP-1 protein displayed potent repression of the reporter plasmid. However, the heterologous Mut2 protein, which does not bind HP1 proteins (Fig. 5) is reduced in repression activity, while the heterologous Mut1 protein displayed an intermediate effect on KAP-1-mediated repression. MW, molecular weight markers.
DISCUSSION
The KRAB-ZFP family of zinc finger-containing transcriptional repressors has the potential for becoming the largest single class of human transcription factors which contain a common set of DNA binding (C2H2 zinc fingers) and effector (KRAB domain) motifs. It has been estimated that the human genome contains approximately 700 genes that encode C2H2 zinc finger proteins and that one-third or more of these contain KRAB domains (28). Their prevalence and conservation indicate the need for a molecular understanding of their mechanism of action. A significant advance was the discovery, purification, and cloning of a novel gene encoding a potential universal binding protein which functions as a corepressor for the KRAB domain, KAP-1. The present work represents the beginning of studies aimed at identifying downstream effector components operative in the KRAB–KAP-1 repression pathway.
KAP-1 recruits HP-like proteins which mediate KRAB-ZFP repression.
This report demonstrates that KAP-1 interacts directly with members of the chromodomain- or chromo shadow domain-containing heterochromatin protein family including M31, M32, hHP1α, and hHP1γ. Efficient binding can be detected in solution-based chromatography assays and in EMSAs using recombinant GST-HP1 family proteins. The interaction is a direct protein-protein interaction between KAP-1 and HP1, as shown using highly purified recombinant versions of each. We have localized an HP1BD in KAP-1 and have shown that amino acid substitutions in the HP1BD abolish HP1 binding. The complex formed in vitro is stable to gel filtration chromatography and alters the apparent native molecular mass of a KAP-1-containing complex from 560 kDa to around 800 kDa (Fig. 7). There are several explanations for this apparent increase: (i) multiple molecules of M32 interact with KAP-1 and/or each other, (ii) additional heterochromatin proteins from the nuclear extract bind M32 bound to the KAP-1 complex, and (iii) both heterochromatin proteins and other interacting proteins interact with the new KAP-1–GST-M32 complex to shift the endogenous complex. Given the demonstration that chromodomains of heterochromatin proteins are able to homo and hetero-oligomerize (10, 41, 42), we favor the possibility that between two and four M32 molecules interact with a KAP-1 complex. We have also succeeded in using EMSA to reconstitute a stable, quaternary complex containing DNA, a KRAB domain, KAP-1, and heterochromatin proteins. This finding suggests that chromodomain proteins, exemplified by M32, do not displace KAP-1 from the KRAB domain upon binding. Furthermore, KAP-1–M32 interaction does not abrogate DNA binding by KRAB domain-containing DNA binding proteins. Together, these observations suggest that endogenous KAP-1 has the capability of recruiting HP1 family members to a gene bound by a KRAB-ZFP protein. Consistent with this, we observed that transient expression of hHP1α augmented KRAB–KAP-1-mediated repression. Furthermore, overexpression of a segment of KAP-1 which contains the HP1BD abolished KRAB-mediated repression. Finally, mutations in the HP1BD which abolish HP1 binding significantly reduce the repression potential of a GAL4–KAP-1 protein. Thus, recruitment of heterochromatin proteins may play an important role in KRAB–ZFP–KAP-1-dependent repression (Fig. 12). Consistent with this hypothesis is the observation that HP1 family members themselves are potent, DNA binding-dependent repressors when tethered to a template via fusion to heterologous DNA binding domains (30).
FIG. 12.
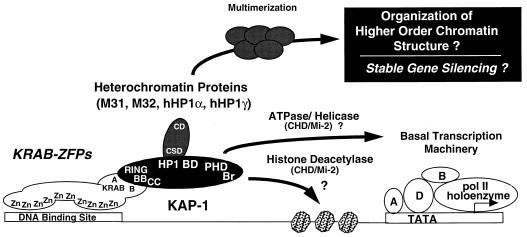
Summary of protein-protein interactions identified in the KRAB-ZFP–KAP-1 repression pathway. We propose that the KRAB-ZFP family of transcriptional repressors function in part as sequence-specific DNA binding proteins which recruit the KAP-1 corepressor to a target gene. This interaction is dependent on the RBCC domain of KAP-1. Together, the HP1BD, PHD, and bromodomain comprise the surfaces which mediate gene silencing via interaction with the indicated potential partners. This work has defined members of the HP1 family of heterochromatin proteins as likely downstream effectors of KAP-1 which may mediate the assembly of stable, higher-order silenced domains in the eukaryotic nucleus. For notation, see the legend to Fig. 10. pol, polymerase.
Heterochromatinization may be only part of KAP-1-mediated repression.
The predicted region of KAP-1 required for binding to HP1 proteins lies between the coiled-coil domain and the PHD (Fig. 1). Consistent with this, analysis of this central domain of KAP-1 revealed a region of amino acid sequence homology termed the HP1BD, shared with the mTIF1α coactivator protein, which was previously shown to bind the mHP1α and M31 heterochromatin proteins (30). This finding suggests that the small-signature amino acid sequence motif may be responsible for protein-protein interactions involving HP1 family members and may be useful in identifying other components which are direct targets bound by HP1 family members. While the HP1BD is sufficient for binding of HP1 family members, we have previously shown that significant repression activity is also exhibited by a GAL4 fusion which contains only the PHD and bromodomains (42). One interpretation of these data is that multiple surfaces of KAP-1 contribute to the protein-protein interactions required for repression and that multiple mechanisms are involved. Paradigms for this scenario have been found for other large nuclear corepressors such as N-CoR, SMRT, Sin3, Mad, and YY1 (see reference 26 and references therein). These corepressor proteins appear to serve as molecular scaffolds for nucleation of multicomponent complexes required for transcriptional repression, the primary specificities of which are provided by the DNA binding components of the complexes. Molecular mechanisms for the actions of these complexes have come from the study of murine Sin3-HDAC complexes containing histone deacetylase activity (26). These complexes apparently mediate localized chromatin changes which occur at the level of the nucleosome. As a consequence of deacetylating core histone, there is a resulting increase in repression of transcription. However, the finding that KAP-1 binds to heterochromatin proteins implies that the KRAB–KAP-1 repression system may function at a different level of chromatin organization than simple histone modification.
Potential for local KRAB–KAP-1/HP1 euchromatin heterochromatic regions.
Using MAbs to M31 and M32, we have shown that these proteins colocalize with KAP-1 within interphase nuclei (Fig. 8 and 9). These data lend support to the suggestion that the KRAB-ZFP–KAP-1 complex and HP1-like proteins are likely to interact in vivo. A thorough inspection of the cells immunostained with specific antibodies against KAP-1 revealed a dynamic staining pattern which may correlate with the cell cycle. Particularly striking is the localization of KAP-1 to distinct territories within the nuclei of some cells. Moreover, some of these territories overlap with the localization patterns of M31 and M32. This colocalization within subnuclear territories between KAP-1 and either M31 or M32 is reminiscent of the observations found with Ikaros, a transcriptional regulator that is essential for lymphoid development. In cells where the lymphoid cell-specific gene CD4 is inactive, the gene is localized, with Ikaros, around centromeric heterochromatin (8). By contrast, the active gene is localized elsewhere in the nucleoplasm. As for Ikaros, KRAB-ZFP binding to a gene may recruit the gene to a heterochromatic territory or form a new, localized heterochromatic domain around the gene, both of which would lead to its stable silencing.
In summary, the KRAB-ZFP family is a large set of proteins, each of which contains multiple contiguous C2H2 zinc fingers and associated KRAB repression domains (Fig. 12). These arrays of zinc fingers may mediate sequence-specific DNA binding, although recent evidence suggests that not all fingers in these long-array ZFPs participate in direct DNA contact. The KRAB domain recruits the KAP-1 corepressor to the DNA template bound by a KRAB-ZFP, binding directly to the RBCC domain of KAP-1. Repression of transcription by a KRAB-ZFP requires recruitment of KAP-1. KAP-1 homo-oligomerizes (unpublished data) upon binding to the KRAB domain and exhibits intrinsic repression activity through the COOH-terminal domains that include the HP1BD, PHD, and bromodomain. The HP1BD of KAP-1 binds to heterochromatic and euchromatic HP1 proteins, resulting in either recruitment of the KRAB-ZFP-bound target gene to heterochromatic chromosomal territories in the nucleus or formation of local heterochromatic chromosomal regions and subsequent silencing of gene expression. Homo- or hetero-oligomerization of HP1 protein family members may play a role in transient and/or stable gene repression, and it is interesting to speculate that this may be catalyzed at a locus by the KRAB-ZFP–KAP-1 system. Additional potential components of the system which still must be considered include the chromodomain-containing CHD/Mi-2 protein, an ATPase/helicase which has been shown to both bind KAP-1 (unpublished data) and be an integral component of a histone deacetylase complex (52, 54, 58).
We suggest that the KAP-1-directed recruitment of HP1-like proteins may provide an example of a mechanism by which euchromatic genes may be locally silenced, through the formation of a heterochromatin-like complex (19, 23). Thus, KAP-1 binding of a KRAB domain-containing protein provides the specificity for the assembly of a mammalian HP1-containing heterochromatin-like complex to a distinct site within the genome, as previously suggested (23), and may be mechanistically related to the mechanisms whereby Hunchback and dYY1 proteins (pleiohomeotic [7]) direct assembly of the Pc-Group complexes in Drosophila (19, 38, 45). Moreover, the formation of a heterochromatin-like complex by the KRAB-ZFP–KAP-1 complex may lead to the dynamic recruitment of silenced genes to a repressive chromosomal environment. The driving energy for such recruitment may come from the complementarity shared between the chromatin components of the repressor complexes, a feature recently described in a model proposed to explain such associations of repressed chromosomal domains (46).
ACKNOWLEDGMENTS
R.F.R., D.C.S., and K.A. contributed equally to the article and should be considered joint first authors.
We thank David E. Jensen for preparation and purification of the 6HisGAL4-KRAB protein, and we thank Qinwu Liu and Gerd Maul, Wistar Institute Microscopy Core Facility, for help in generating and deciphering the immunofluorescence images. We thank M. Lazar for the GAL4–N-CoR plasmid and many helpful discussions. We thank H. Worman for the hHP1γ plasmid.
R.F.R., W.J.F., and D.C.S. were supported by Wistar Basic Cancer Research training grant CA 09171. F.J.R. was supported in part by National Institutes of Health grant CA 52009, Core grant CA 10815, Core grant DK50306, and grants DK 49210, GM 54220, DAMD 17-96-1-6141, and ACS NP-954, the Irving A. Hansen Memorial Foundation, the Mary A. Rumsey Memorial Foundation, and the Pew Scholars Program in the Biomedical Sciences. P.B.S. was supported by the BBSRC.
REFERENCES
- 1.Aasland R, Stewart A F. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 1995;23:3163–3173. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 3.Auble D T, Hansen K E, Mueller C G, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 4.Baniahmad A, Leng X, Burris T P, Tsai S Y, Tsai M J, O’Malley B W. The tau 4 activation domain of the thyroid hormone receptor is required for release of a putative corepressor(s) necessary for transcriptional silencing. Mol Cell Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellefroid E J, Poncelet D A, Lecocq P J, Revelant O, Martial J A. The evolutionarily conserved Kruppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc Natl Acad Sci USA. 1991;88:3608–3612. doi: 10.1073/pnas.88.9.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britten R J, Davidson E H. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 7.Brown J L, Mucci D, Whitely M, Dirksen M L, Kassis J A. The Drosophila polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 8.Brown K E, Guest S S, Smale S T, Hahm K, Merkenschlager M, Fisher A G. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 9.Brown S W. Heterochromatin. Science. 1966;151:417–425. doi: 10.1126/science.151.3709.417. [DOI] [PubMed] [Google Scholar]
- 10.Cowell I G, Austin C A. Self-association of chromo domain peptides. Biochim Biophys Acta. 1997;1337:198–206. doi: 10.1016/s0167-4838(96)00165-3. [DOI] [PubMed] [Google Scholar]
- 11.Dawson S R, Turner D L, Weintraub H, Parkhurst S M. Specificity for the hairy/enhancer of split basic helix-loop-helix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol Cell Biol. 1995;15:6923–6931. doi: 10.1128/mcb.15.12.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eissenberg J C, James T C, Foster-Hartnett D M, Hartnett T, Ngan V, Elgin S C. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eissenberg J C, Morris G D, Reuter G, Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics. 1992;131:345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fondell J D, Brunel F, Hisatake K, Roeder R G. Unliganded thyroid hormone receptor alpha can target TATA-binding protein for transcriptional repression. Mol Cell Biol. 1996;16:281–287. doi: 10.1128/mcb.16.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fondell J D, Roy A L, Roeder R G. Unliganded thyroid hormone receptor inhibits formation of a functional preinitiation complex: implications for active repression. Genes Dev. 1993;7:1400–1410. doi: 10.1101/gad.7.7b.1400. [DOI] [PubMed] [Google Scholar]
- 16.Fredericks W J, Galili N, Mukhopadhyay S, Rovera G, Bennicelli J, Barr F G, Rauscher F J., III The PAX3-FKHR fusion protein created by the t(2;13) translocation in alveolar rhabdomyosarcomas is a more potent transcriptional activator than PAX3. Mol Cell Biol. 1995;15:1522–1535. doi: 10.1128/mcb.15.3.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman J R, Fredericks W J, Jensen D E, Speicher D W, Huang X P, Neilson E G, Rauscher F J., III KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 18.Furuta K, Chan E K L, Kiyosawa K, Reimer G, Luderschmidt C, Tan E M. Heterochromatin protein HP1Hsbeta (p25beta) and its localization with centromeres in mitosis. Chromosoma. 1997;106:11–19. doi: 10.1007/s004120050219. [DOI] [PubMed] [Google Scholar]
- 19.Gaunt S J, Singh P B. Homeogene expression patterns and chromosomal imprinting. Trends Genet. 1990;6:208–212. [PubMed] [Google Scholar]
- 20.Grimes H L, Gilks C B, Chan T O, Porter S, Tsichlis P N. The Gfi-1 protooncoprotein represses Bax expression and inhibits T-cell death. Proc Natl Acad Sci USA. 1996;93:14569–14573. doi: 10.1073/pnas.93.25.14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guarente L. Transcriptional coactivators in yeast and beyond. Trends Biochem Sci. 1995;20:517–521. doi: 10.1016/s0968-0004(00)89120-3. [DOI] [PubMed] [Google Scholar]
- 22.Hamvas R M, Reik W, Gaunt S J, Brown S D, Singh P B. Mapping of a mouse homolog of a heterochromatin protein gene to the X chromosome. Mamm Genome. 1992;2:72–75. doi: 10.1007/BF00570443. [DOI] [PubMed] [Google Scholar]
- 23.Horsley D, Hutchings A, Butcher G W, Singh P B. M32, a murine homologue of Drosophila heterochromatin protein 1 (HP1), localises to euchromatin within interphase nuclei and is largely excluded from constitutive heterochromatin. Cytogenet Cell Genet. 1996;73:308–311. doi: 10.1159/000134363. [DOI] [PubMed] [Google Scholar]
- 24.Huang, X.-P., and F. J. Rauscher III. Unpublished data.
- 25.James T C, Elgin S C. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S S, Chen Y M, O’Leary E, Witzgall R, Vidal M, Bonventre J V. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci USA. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klug A, Schwabe J W. Protein motifs 5. Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- 29.Lassar A B, Davis R L, Wright W E, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 30.Le Douarin B, Nielsen A L, Garnier J M, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 31.Le Douarin B, Nielsen A L, You J, Chambon P, Losson R. TIF1 alpha: a chromatin-specific mediator for the ligand-dependent activation function AF-2 of nuclear receptors? Biochem Soc Trans. 1997;25:605–612. doi: 10.1042/bst0250605. [DOI] [PubMed] [Google Scholar]
- 32.Le Douarin B, vom Baur E, Zechel C, Heery D, Heine M, Vivat V, Gronemeyer H, Losson R, Chambon P. Ligand-dependent interaction of nuclear receptors with potential transcriptional intermediary factors (mediators) Philos Trans R Soc Lond Ser B. 1996;351:569–578. doi: 10.1098/rstb.1996.0056. [DOI] [PubMed] [Google Scholar]
- 33.Le Douarin B, Zechel C, Garnier J M, Lutz Y, Tora L, Pierrat B, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margolin J F, Friedman J R, Meyer W K, Vissing H, Thiesen H J, Rauscher F J., III Kruppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci USA. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Maul, G. G. Unpublished data.
- 35.Maul G G, Jensen D E, Ishov A M, Herlyn M, Rauscher F J., III Nuclear redistribution of BRCA1 during viral infection. Cell Growth Differ. 1998;9:743–755. [PubMed] [Google Scholar]
- 36.Moosmann P, Georgiev O, Le Douarin B, Bourquin J P, Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996;24:4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicol L, Jeppesen P. Human autoimmune sera recognize a conserved 26 kD protein associated with mammalian heterochromatin that is homologous to heterochromatin protein 1 of Drosophila. Chromosome Res. 1994;2:245–253. doi: 10.1007/BF01553325. [DOI] [PubMed] [Google Scholar]
- 38.Paro R. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 1990;6:416–421. doi: 10.1016/0168-9525(90)90303-n. [DOI] [PubMed] [Google Scholar]
- 39.Paro R, Hogness D S. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearce J J, Singh P B, Gaunt S J. The mouse has a Polycomb-like chromobox gene. Development. 1992;114:921–929. doi: 10.1242/dev.114.4.921. [DOI] [PubMed] [Google Scholar]
- 41.Platero J S, Hartnett T, Eissenberg J C. Functional analysis of the chromo domain of HP1. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rauscher, F. J., III, J. R. Friedman, R. F. Ryan, D. C. Schultz, and W. J. Fredericks. Unpublished data.
- 43.Sauer F, Fondell J D, Ohkuma Y, Roeder R G, Jäckle H. Control of transcription by Kruppel through interactions with TFIIB and TFIIE beta. Nature. 1995;375:162–164. doi: 10.1038/375162a0. [DOI] [PubMed] [Google Scholar]
- 44.Saunders W S, Chue C, Goebl M, Craig C, Clark R F, Powers J A, Eissenberg J C, Elgin S C, Rothfield N F, Earnshaw W C. Molecular cloning of a human homologue of Drosophila heterochromatin protein HP1 using anti-centromere autoantibodies with anti-chromo specificity. J Cell Sci. 1993;104:573–582. doi: 10.1242/jcs.104.2.573. [DOI] [PubMed] [Google Scholar]
- 45.Singh P B. Molecular mechanisms of cellular determination: their relation to chromatin structure and parental imprinting. J Cell Sci. 1994;107:2653–2668. doi: 10.1242/jcs.107.10.2653. [DOI] [PubMed] [Google Scholar]
- 46.Singh P B, Huskisson N S. Chromatin complexes as aperiodic microcrystalline arrays that regulate genome organisation and expression. Dev Genet. 1998;22:85–99. doi: 10.1002/(SICI)1520-6408(1998)22:1<85::AID-DVG9>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 47.Singh P B, Miller J R, Pearce J, Kothary R, Burton R D, Paro R, James T C, Gaunt S J. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 1991;19:789–794. doi: 10.1093/nar/19.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spofford J B. Position-effect variegation in Drosophila. In: Ashburner M, Novitski E, editors. The genetics and biology of Drosophila. 1c. New York, N.Y: Academic Press; 1976. pp. 955–1018. [Google Scholar]
- 49.Sugimoto K, Yamada T, Muro Y, Himeno M. Human homolog of Drosophila heterochromatin-associated protein 1 (HP1) is a DNA-binding protein which possesses a DNA-binding motif with weak similarity to that of human centromere protein C (CENP-C) J Biochem. 1996;120:153–159. doi: 10.1093/oxfordjournals.jbchem.a021378. [DOI] [PubMed] [Google Scholar]
- 50.Tartof K D, Bremer M. Mechanisms for the construction and developmental control of heterochromatin formation and imprinted chromosome domains. Dev. Suppl. 1990. pp. 35–45. [PubMed] [Google Scholar]
- 51.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 52.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 53.Um M, Li C, Manley J L. The transcriptional repressor even-skipped interacts directly with TATA-binding protein. Mol Cell Biol. 1995;15:5007–5016. doi: 10.1128/mcb.15.9.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wade P A, Jones P L, Vermaak D, Wolffe A P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 55.Wolffe A P. Histone deacetylase: a regulator of transcription. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 56.Wreggett K A, Hill F, James P S, Hutchings A, Butcher G W, Singh P B. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet Cell Genet. 1994;66:99–103. doi: 10.1159/000133676. [DOI] [PubMed] [Google Scholar]
- 57.Ye Q, Worman H J. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, LeRoy G, Seelig H-P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]