CCR2-64I Polymorphism Is Not Associated with Altered CCR5 Expression or Coreceptor Function (original) (raw)
Abstract
A polymorphism in the gene encoding CCR2 is associated with a delay in progression to AIDS in human immunodeficiency virus (HIV)-infected individuals. The polymorphism, _CCR2_-64I, changes valine 64 of CCR2 to isoleucine. However, it is not clear whether the effect on AIDS progression results from the amino acid change or whether the polymorphism marks a genetically linked, yet unidentified mutation that mediates the effect. Because the gene encoding CCR5, the major coreceptor for HIV type 1 primary isolates, lies 15 kb 3′ to CCR2, linked mutations in the CCR5 promoter or other regulatory sequences could explain the association of _CCR2_-64I with slowed AIDS pathogenesis. Here, we show that _CCR2_-64I is efficiently expressed on the cell surface but does not have dominant negative activity on CCR5 coreceptor function. A panel of peripheral blood mononuclear cells (PBMC) from uninfected donors representing the various CCR5/CCR2 genotypes was assembled. Activated primary CD4+ T cells of CCR2 64I/64I donors expressed cell surface CCR5 at levels comparable to those of CCR2 +/+ donors. A slight reduction in CCR5 expression was noted, although this was not statistically significant. CCR5 and CCR2 mRNA levels were nearly identical for each of the donor PBMC, regardless of genotype. Cell surface CCR5 and CCR2 levels were more variable than mRNA transcript levels, suggesting that an alternative mechanism may influence CCR5 cell surface levels. _CCR2_-64I is linked to the CCR5 promoter polymorphisms 208G, 303A, 627C, and 676A; however, in transfected promoter reporter constructs, these did not affect transcriptional activity. Taken together, these findings suggest that _CCR2_-64I does not act by influencing CCR5 transcription or mRNA levels.
Human immunodeficiency virus (HIV)-infected individuals progress to disease at widely differing rates, with some individuals becoming symptomatic in 2 to 3 years and others remaining asymptomatic for more than 10 to 15 years (14, 44). The factors that influence disease progression rates are not well understood. In some cases, long-term nonprogression can be attributed to infection with a partially attenuated virus (21, 29, 31, 42). However, in many cases, no alterations in the virus are detected (19), suggesting that host factors are likely to contribute to pathogenicity.
Several polymorphisms in the human genome appear to play a role in disease progression rates. A CCR5 allele containing a 32-bp deletion in the CCR5 coding region (CCR5 Δ32) is present in northern European populations at a frequency of about 0.2 (10, 27, 43). The protein encoded by this allele is nonfunctional, both as a chemokine receptor and as an HIV type 1 (HIV-1) coreceptor (27). Homozygotes are strongly although not absolutely (2, 5, 39, 47) protected from infection, while heterozygotes progress to disease with a delay of about 2 years compared to wild types in some (10, 13, 20, 30, 32, 51) but not all (18, 33, 36) cohorts examined. A possible explanation for the association of the mutant allele with decreased pathogenicity derives from the finding that CD4+ T cells from CCR5 +/Δ32 individuals express on average 50% of wild-type levels of CCR5 and as a result support decreased levels of macrophagetropic (M-tropic) HIV-1 replication (40, 50).
A polymorphism in CCR2, the gene encoding the receptor for the C-C chemokines MCP-1 to -4 (4, 15), appears to delay AIDS progression to an extent similar to that of CCR5 Δ32 (22, 36, 45). The polymorphism, a G-to-A transition at position 190, changes CCR2 codon 64 from valine to isoleucine, introducing a conservative change into the first transmembrane domain. The allele is present at a frequency of 0.1 to 0.25 in various populations and is not associated with any clinical abnormality. The effect of _CCR2_-64I was more pronounced in seroconverting than seroprevalent individuals (22, 32) and more pronounced in African Americans than caucasians (36). A polymorphism in the 3′ untranslated region of the gene encoding the ligand for CXCR4, stromal cell-derived factor 1, also appears to influence disease progression rates (36, 49).
The mechanism by which _CCR2_-64I influences AIDS pathogenesis is not clear. It is possible that its effects on pathogenicity are caused by the 64I missense mutation itself. For example, the variant CCR2-64I protein could have diminished coreceptor function. However, this possibility seems unlikely given the minor role of CCR2 as an HIV-1 coreceptor. Alternatively, CCR2-64I could have dominant negative activity that interferes with CCR5 coreceptor function. Another possibility is that the _CCR2_-64I polymorphism does not act directly but is linked to a yet unidentified polymorphism in a gene that influences AIDS pathogenesis. CCR2 is located about 15 kb 5′ to CCR5 in the human genome. CCR5 coding region polymorphisms have not been found associated with _CCR2_-64I; however, linked regulatory mutations could exist that influence CCR5 expression and thereby account for the association of _CCR2_-64I with decreased disease progression.
The CCR5 gene consists of four exons spanning 8 kb: three that constitute the 5′ untranslated region and a fourth that contains the entire coding sequence (37). Transcription appears to initiate at either the first or the third exon and is driven by two different promoters, Pu and Pd, that are separated by approximately 800 bp (16, 28, 35, 37). In activated T cells, the majority of transcripts appear to have initiated from Pd (28).
Several polymorphisms within the CCR5 promoter region have been identified. One of these, 927T, lies in the region 3′ to the Pd transcription start site and is usually found in association with _CCR2_-64I (22, 36). In addition, four other point mutations within the promoter region, at positions 208, 303, 627, and 676, are typically found on _CCR2_-64I alleles (36). Such mutations could conceivably affect CCR5 transcription, altering the ability of T cells to support HIV replication in a manner analogous to that of CCR5 Δ32.
We report here the results of studies aimed at distinguishing between the various mechanisms by which _CCR2_-64I might alter HIV-1 pathogenicity. We tested whether _CCR2_-64I acts directly by influencing CCR5 expression or coreceptor activity in transfected cells or whether it influences HIV-1 replication, CCR5 cell surface levels, or CCR5 mRNA levels in primary CD4+ T cells. These questions were addressed both in cell lines stably expressing retroviral vector-transduced CCR5 and CCR2 or CCR2-64I and in a panel of donor T cells representing each of the CCR5/CCR2 genotype combinations. The results suggest that CCR2-64I does not have dominant negative activity on CCR5 expression or coreceptor activity. In _CCR2_-64I heterozygous and homozygous T cells, the _CCR2_-64I allele is transcribed at wild-type levels and the cells support wild-type levels of M-tropic HIV-1 replication. Furthermore, while there are four single-nucleotide promoter polymorphisms genetically linked to _CCR2_-64I, these did not influence transcriptional activity of the promoter in reporter assays.
MATERIALS AND METHODS
CCR5 and CCR2 genotyping.
Genomic DNA was purified by using a QIAamp kit (Qiagen) from peripheral blood mononuclear cells (PBMC) of 827 uninfected donors randomly selected from the REDS (Retroviral Epidemiology Donor Study) repository (52). DNA was amplified by PCR using primers flanking CCR5 Δ32 or _CCR2_-64I. Primers for CCR5 Δ32 were those previously described by Liu et al. (27) (SP4.760 [5′-CTT CAT TAC ACC TGC AGC TCT] and PM6.942 [5′-CAC AGC CCT GTG CCT CTT CTT C]). CCR2 genotype was detected by using primers described by Michael et al. (32). Amplicons were cleaved with _Bsa_BI to distinguish the wild-type allele from the mutant allele (32) and visualized on ethidium bromide-stained agarose gels.
Expression of chemokine receptors in retroviral vector-transduced cell lines.
Wild-type CCR2 and _CCR2_-64I were cloned into the retroviral vectors pBABE.puro and pBABE.neo (34). Expression vectors for both molecules were constructed by amplifying the coding exons from genomic DNA of individuals with the appropriate CCR2 genotype, using sense and antisense primers containing _Bam_HI and _Xho_I restriction sites (5′-GCT CAG GAT CCT GAG ACA AGC CAC AAG CTG AAC AG and 5′-GTG CCT CTA GAC TGA ATG CGT GAG CCC TTT GCT C, respectively). Amplicons were cleaved and ligated to _Bam_HI- and _Sal_I-cleaved pBABE-neo and pBABE.puro. The complete sequence of each insert was determined. Retroviral stocks were prepared as described previously (24) except that 293 cells instead of COS cells were transfected and vesicular stomatitis virus G-protein (VSV-G) pseudotypes instead of amphotropic murine leukemia virus (MLV) pseudotypes were prepared. HOS.CD4 or CEMx174 cells were infected with 1 ml of CCR5 pBABE.puro retrovirus, 2 days later selected in puromycin (1 μg/ml) for about 2 weeks, then infected with CCR2 or CCR2-64I pBABE.neo virus, and selected in G418 for another 2 weeks. HOS.CD4 cells expressing CCR2 or CCR2-64I without CCR5 were established by using pBABE.puro viruses. Expression levels were verified by fluorescence-activated cell sorting (FACS) analysis with anti-CCR5 monoclonal antibody (MAb) 2D7 and anti-CCR2 MAb 1D9 (25).
HIV replication kinetics.
PBMC were obtained from whole blood from healthy, HIV-1-seronegative individuals. Cells were purified by centrifugation through Ficoll density gradients and frozen in aliquots at −150°C. For infection, cells were thawed at 37°C, washed in phosphate-buffered saline (PBS) and cultured for 2 days in RPMI–10% fetal bovine serum (FBS) containing interleukin-2 (IL-2) (100 U/ml) and phytohemagglutinin (PHA; 5 μg/ml). After 3 days, the cells were centrifuged and resuspended in medium containing IL-2. The next day, the cells (105) were infected in a volume of 300 μl with 1 ng p24_gag_ of each HIV-1 isolate (corresponding to a multiplicity of infection [MOI] of 0.001 to 0.002). Virus isolates MJM and JDC were derived from the lymphocytes of patients recently infected with HIV-1. Both viruses were grown by short-term propagation in PHA-stimulated donor PBMC and were restricted to using CCR5 as a coreceptor. The next day, the cells were washed twice in culture medium and resuspended in 500 μl of culture medium containing IL-2. At indicated days postinfection, half of the medium was collected and replaced with fresh medium containing IL-2. p24_gag_ concentrations were measured by enzyme-linked immunosorbent assay (ELISA).
Luciferase reporter virus assays.
Single-cycle reporter virus assays were used as described previously (9, 11). Briefly, reporter viruses were prepared by cotransfecting 293 cells with NL4-3-based reporter plasmid NL-Luc.R−E− and either M-tropic Env expression vector pSV-JR.FL.Env or VSV-G expression vector. Viruses were harvested and frozen at −80°C in aliquots. Retroviral vector-transduced HOS (5 × 103) or CEMx174 cells (105) were infected in a volume of 100 μl with 10 ng p24_gag_ reporter virus. Cells were lysed 3 days postinfection, and luciferase activity was measured by using commercial reagents (Packard).
RT-PCR.
Total RNA was prepared using Triazol (Gibco/BRL) from fresh or PHA–IL-2-activated PBMC. Contaminating genomic DNA was removed by treating with RNase-free DNase (Boehringer Mannheim). cDNA was generated from 1 μg of RNA by using Moloney murine leukemia virus reverse transcriptase (RT; Gibco/BRL) and oligo(dT). CCR5 and CCR2 cDNAs were amplified as for the genotypic analysis except that 5 μCi [α-32P]dCTP was added to the PCR mixture. In addition, glyceraldehyde phosphate dehydrogenase (GAPDH) cDNAs were amplified as a control, using specific primers. Parallel reactions were prepared in which RT was omitted from each sample to confirm the absence of genomic DNA contamination. PCR products were separated by 6% polyacrylamide gel electrophoresis and quantitated with a STORM PhosphorImager (Molecular Dynamics).
FACS analysis.
Fresh or PHA–IL-2-activated PBMC were stained with anti-CCR5 MAb 2D7-phycoerythrin (PE), anti-CD4 MAb Leu3a-fluorescein isothiocyanate (FITC; Becton Dickinson), and anti-CCR2 MAb 1D9 (25). Cells were incubated with MAb for 15 min at room temperature in 0.1 ml of PBS–1% fetal calf serum (FCS) containing 0.1% sodium azide. For staining with 1D9, cells were washed with PBS and incubated an additional 15 min with goat anti-mouse immunoglobulin G-PE (BioSource). The cells were then washed in PBS and incubated with PBS–10% normal mouse serum. After 15 min, Leu3a-FITC was added. The cells were incubated 15 min, washed, and analyzed. Fluorescence was measured on a FACSCalibur (Becton-Dickinson), and 10,000 events were collected.
Promoter analysis.
A 1-kb genomic DNA fragment containing the CCR5 promoter region from positions 58531 to 59555 (numbered according to GenBank accession no. U95626) was amplified from human genomic DNA of CCR2 +/+ or 64I/64I individuals. PCR was performed with Expand polymerase (Boehringer Mannheim), using a sense primer (5′-GAT CGG TAC CAG CCA AGG TCA CGG AAG C) and an antisense primer (5′-GAT CAA GCT TGG GGA ACG GAT GTC TCA GC) for 25 cycles of 94°C for 45 s, 60°C for 45 s, and 72°C for 1 min. PCR products were cloned into the _Kpn_I and _Hin_dIII sites 5′ to the luciferase reporter gene in pGL3 (Promega), and their nucleotide sequences (nucleotides [nt] 58568 to 59530) were determined on an ABI 370 automated sequencer (Applied Biosystems). A single point mutation was found in 64I(2) at position −133, apparently the result of a PCR. Independent PCR products from the same donor did not contain this mutation.
Promoter activity was quantified as described previously (28). Briefly, Jurkat cells were resuspended in RPMI–10% FCS at 0.5 × 106 cells per ml the day before transfection. The cells (107) were cotransfected by electroporation with 18 μg of reporter plasmid plus 2 μg of plasmid pcβ-gal (Invitrogen) at 200 V and 960 μF in 200 μl of RPMI–10% FBS–37.5 mM NaCl. Two days posttransfection, the cells were lysed in 100 μl of lysis buffer (Promega). Luciferase and β-galactosidase activities in 20 μl of lysate were measured by light emission assays using commercial reagents (Promega and Tropix, respectively). Results are reported as the average relative luciferase activity and are normalized for β-galactosidase activity.
[Ca2+] flux measurement.
[Ca2+] flux was measured essentially as described elsewhere (38). Cells were harvested, washed with SR buffer (136 mM NaCl, 4.8 mM KCl, 5 mM glucose, 1 mM CaCl2, 0.025% [wt/vol] bovine serum albumin, 20 mM HEPES [pH 7.6]), and then incubated in SR buffer containing 10 M Fura-2AM (Sigma) for 60 min at 37°C in the dark. The cells were washed twice with SR buffer, warmed to 37°C, and resuspended in SR buffer to ∼4 × 106 cells/ml. A 2-ml aliquot was then placed in a continuously stirred cuvette at 37°C in a Perkin-Elmer LS50 Spectrometer for 2 min. Fluorescence was recorded every 100 ms at an excitation of 340 nm and an emission of 500 nm. The cell lines were diluted to give identical baseline fluorescence emission. After 40 s of measurement, agonist diluted in PBS was added and fluorescence emission was recorded for an additional 100 s. Lysis with detergent demonstrated equal Fura-2AM loading between the two cell lines.
Chemokine quantitation.
Frozen PBMC from REDS donors were thawed and cultured at 106/ml for 2 days in RPMI–10% FCS with or without IL-2 (10 U/ml) and PHA (5 μg/ml). Culture supernatants were then sampled and chemokines were quantitated by commercial ELISA (R & D Systems).
RESULTS
Assembly of a panel of PBMC from REDS donors representing the various CCR5/CCR2 genotypes.
To establish a panel of uninfected donors representing each of the CCR5 and CCR2 haplotypes, we determined the genotypes of 827 REDS donors (7, 52). Genomic DNA was purified from cryopreserved donor PBMC, and genotypes were determined by PCR using appropriate primers flanking CCR5 Δ32 or _CCR2_-64I as previously described (27, 32). Of the nine possible genotypes, six were represented in the panel (Table 1); 69.8% of the individuals were wild type at both loci, 16.4% were heterozygous for CCR5 Δ32 alone, 11.1% were heterozygous for _CCR2_-64I alone, and 1.3% were heterozygous at both loci. Three individuals (0.36%) were homozygous for CCR5 Δ32, and nine (1.0%) were homozygous for _CCR2_-64I. These frequencies are similar to those reported in previous studies (22, 32, 45). Three genotypes, CCR5 Δ32/Δ32 _CCR2_-64I/64I, CCR5 Δ32/Δ32 CCR2 +/64I, and CCR5 +/Δ32 _CCR2_-64I/64I, were not represented. This absence probably reflects the evolutionary history of the two alleles. Since they are believed to have originated relatively recently and are separated physically by only 15 kb, recombination that would place the two polymorphisms on the same chromosome has not yet occurred at a significant frequency (45). Donors of various haplotypes were randomly chosen for further analysis. We focused primarily on CCR2 64I/64I homozygous rather than heterozygous donors since these were expected to serve as a more sensitive means of detecting subtle effects on CCR5 expression.
TABLE 1.
CCR5 and CCR2 genotype distribution in the San Francisco REDS cohort
| CCR2 | No. (%) of CCR5 individuals | |||
|---|---|---|---|---|
| +/+ | +/Δ32 | Δ32/Δ32 | Total | |
| +/+ | 576 (69.8) | 136 (16.4) | 3 (0.36) | 715 |
| +/64I | 92 (11.1) | 11 (1.3) | 0 | 103 |
| 64I/64I | 9 (1) | 0 | 0 | 9 |
| Total | 677 | 147 | 3 | 827 |
Cell surface expression of CCR5, CCR2, and CXCR4.
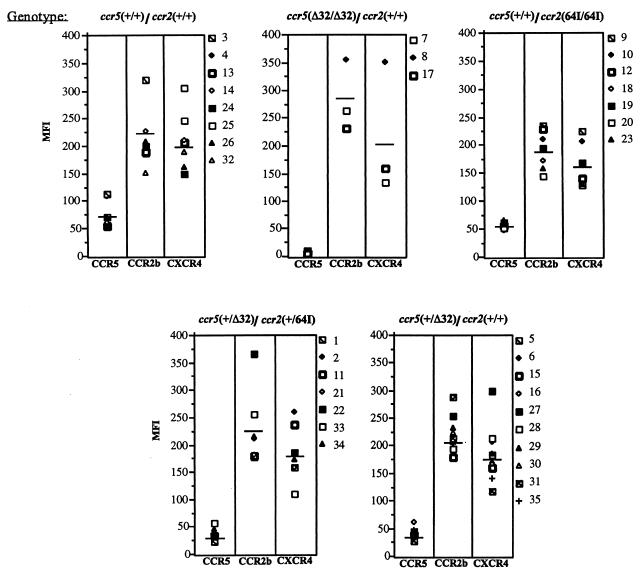
If _CCR2_-64I were associated with linked mutations in CCR5 regulatory sequences, then cells with the allele, especially those homozygous for the mutation, ought to have altered levels of cell surface CCR5. If the linked mutation were to lie in a regulatory sequence controlling expression of a larger region of the chromosome, then it might also affect expression of CCR2 itself. To test these possibilities, we stained PHA–IL-2-activated cells from donors of each genotype with anti-CCR5 MAb 2D7 (50), anti-CCR2 MAb 1D9 (25), and anti-CXCR4 MAb 12G5 (12) (Fig. 1). FACS analysis showed that while CCR5 levels varied up to about threefold from donor to donor, CCR5 was expressed, on average, at similar levels per cell in _CCR2_-64I/64I compared to CCR2 +/+ CD4+ T cells (average mean fluorescence intensities of 54 and 74, respectively). While a slight decrease was noted on the CCR2 64I/64I cells, this difference was not statistically significant (p = 0.121). CCR2 +/64I donor cells (n = 8) had levels of cell surface CCR5 indistinguishable from those of CCR5/CCR2 wild-type cells (n = 7, p = 0.6 [data not shown]). Consistent with earlier reports, CCR5 +/Δ32 CD4+ T cells expressed reduced amounts of cell surface CCR5 (p = 0.002) (40, 50), and CCR5 Δ32/Δ32 cells lacked detectable cell surface CCR5.
FIG. 1.
CCR2, CCR5, and CXCR4 cell surface expression of CD4+ T cells of each CCR2/CCR5 haplotype. Donor PBMC of the indicated genotype were activated for 7 days with PHA–IL-2 and stained with anti-CD4 MAb Leu3a-FITC and anti-CCR5 MAb 2D7 (50), anti-CCR2 MAb 1D9 (25), or anti-CXCR4 MAb 12G5 (12). CD4− cells were gated out, and the mean fluorescence intensity (MFI) of cells expressing the appropriate chemokine receptor is shown.
CCR2 levels on CD4+ T cells of each genotype were similar. CCR5 Δ32/Δ32 CCR2 +/+ cells expressed about 20% more CCR2 than cells of the other genotypes; however, since there were only three donors in this category, the difference was not statistically significant. CXCR4 levels were similar for each haplotype. Taken together, these findings suggest that _CCR2_-64I is not associated with a major alteration in CCR5, CCR2, or CXCR4 expression.
CCR5 and CCR2 mRNA levels in cells of each genotype.
Because _CCR2_-64I is associated with linked polymorphisms in the CCR5 promoter, it is possible that CCR5 mRNA levels are altered in CCR2 64I/64I or +/64I T cells. To test this possibility, RNA was purified from PBMC of members of the REDS repository and used as a template in RT-PCR with primers detecting _CCR5_-Δ32 (27) or _CCR2_-64I (32). In this analysis, compound heterozygotes, CCR5 +/Δ32 CCR2 +/64I, were particularly informative. In such cells, transcripts derived from the two alleles could be distinguished by size, thereby permitting a direct comparison of the transcriptional activity of the _CCR5_-Δ32 allele with that of the _CCR2_-64I-linked CCR5 allele.
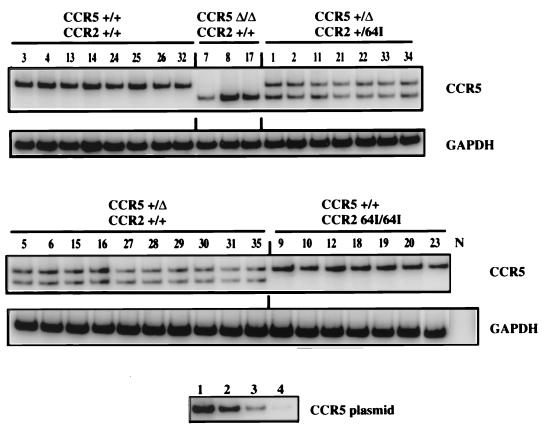
Semiquantitative RT-PCR analysis showed a remarkable similarity in CCR5 transcript abundance in the cells of each donor (Fig. 2). This consistency contrasted with the variability of CCR5 cell surface expression levels found in this and previous studies (50). For example, T cells of donors 3, 18, and 33 contained average amounts of CCR5 mRNA transcript but expressed about twofold more cell surface CCR5 than average (Fig. 1). Quantitation of the band intensities from the cells of each compound heterozygote showed the wild-type CCR5 and _CCR5_-Δ32 transcripts at similar levels (data not shown). The intensity of the band for the CCR5 +/+ donors was close to double that of each of the two bands for the heterozygotes (17.2 × 106 ± 3.5 × 106 versus 6.1 × 106 ± 2.1 × 106, respectively).
FIG. 2.
RT-PCR analysis of chemokine receptor RNA. RNA was isolated from PHA–IL-2-activated donor PBMC of the indicated genotype. CCR5 and GAPDH transcripts were amplified by RT-PCR using specific primers. [32P]dCTP was added to the PCR mixture to allow quantification of the amplified products following polyacrylamide gel electrophoresis. Control reactions in which RT was omitted from the first-strand synthesis were uniformly negative (not shown). Tenfold serial dilutions (lane 1, 100 pg; lane 2, 10 pg; lane 3, 1 pg; lane 4, 0.1 pg) of CCR5 plasmid DNA amplified in parallel are shown below.
_CCR5_-Δ32 alleles are genetically linked to a single nucleotide polymorphism, 29G, in the promoter region (36) that could conceivably affect transcription of the CCR5 Δ32 allele. However, the intensity of the single band in the CCR5 Δ32/Δ32 cells was similar to that of the CCR5 +/+ cells. Thus, 29G does not appear to affect CCR5 promoter function. CCR2 64I/64I cells contained amounts of CCR5 transcript equivalent to those of CCR2 +/+ donors. A similar RT-PCR analysis of CCR2 mRNA transcript levels in each of the donor PBMC showed that this transcript was also present at nearly identical levels in the cells of each haplotype (data not shown).
While the accuracy of RT-PCR is limited, our analysis appears to have been sufficiently accurate to detect small differences mRNA abundance. For example, in CCR5 +/Δ32 cells, the band intensities of the wild-type and CCR5 Δ32 transcript were close to 50% that of the single CCR5 band in CCR5 +/+ cells (Fig. 2). Furthermore, care was taken to ensure that the band intensities detected were within the linear range of the PCR, as demonstrated by amplifying serial dilutions of CCR5 plasmid DNA. Taken together, these data suggest that the _CCR2_-64I polymorphism is not associated with differences in mRNA transcription or stability. The differences found in cell surface CCR5 levels of various donors may thus reflect posttranscriptional control.
HIV growth kinetics on T cells of each genotype.
Because _CCR5_-Δ32 and CCR2_-64I are associated with decreased rates of pathogenesis in infected individuals, we tested whether the mutations are associated with decreased ability of T cells to support virus replication. PBMC of the genotypes CCR5 +/+ CCR2 +/+, CCR5 +/+ CCR2 64I/64I, and CCR5 Δ32/Δ32 CCR2 +/+ were activated with PHA–IL-2 and infected at low MOI with M-tropic (JR.FL, MJM, and JCD) or T-cell-tropic (T-tropic) (SF33) HIV-1 isolates. Supernatant p24_gag concentrations were measured every other day over 3 weeks. The concentration at the day of peak production is shown for each of the donors in Fig. 3. While considerable variability was found in the extent and rate of virus replication in the activated PBMC, this did not correlate with genotype exception in the case of the CCR5 Δ32/Δ32 cells, which, as expected, failed to support replication of the M-tropic isolates. Interestingly, the cells of one donor (donor 24) failed to support efficient replication of either the T- or M-tropic viruses.
FIG. 3.
HIV-1 replication on activated CCR2 +/+, CCR5 Δ32/Δ32, and CCR2_-64I/64I PBMC. PBMC from CCR5 +/+ CCR2 +/+, CCR5 Δ32/Δ32, CCR2 +/+, and CCR5 +/+ CCR2 64I/64I donors were activated for 3 days with PHA–IL-2 and infected at low MOI with T-tropic (SF33) or M-tropic (JR.FL, MJM, and JDC) primary HIV-1 isolates. Supernatants were sampled at 2-day intervals for 3 weeks. Shown are the p24_gag values at the time of peak production (days 7 to 11). Vertical lines indicate average values.
Chemokine production by donor cells of each genotype.
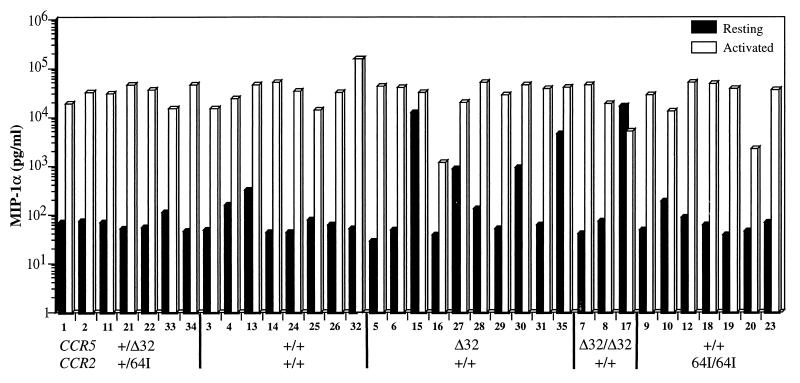
Paxton et al. (41) showed that activated CCR5 Δ32/Δ32 CD4+ T cells produced 5- to 10-fold more of the three CCR5 ligands MIP1-α, MIP1-β, and RANTES than wild-type T cells. CCR5 +/Δ32 T cells expressed slightly elevated levels of the three chemokines (40). Because of the inhibitory properties of these chemokines on HIV replication, we tested whether _CCR2_-64I is associated with increased chemokine production. Chemokine levels in the supernatants of resting and PHA–IL-2-activated T cells from each of the CCR5/CCR2 haplotypes were measured. In the majority of donors cells, activation with PHA and IL-2 induced MIP-1α about 100-fold (0.1 to 35 ng/ml [Fig. 4]). There was no significant difference in chemokine production between the wild-type and CCR2 64I/64I cells (p = 0.92). Interestingly, some of the CCR5 +/Δ32 or Δ32/Δ32 donor cells (donors 15, 27, 30, 35, and 17) produced high levels of chemokine prior to activation, an effect that may be related to the increased chemokine production associated with Δ32 reported previously (40). Similar results were obtained for RANTES and MIP-1β (not shown). Thus, in contrast to CCR5 Δ32, the _CCR2_-64I allele was not associated with increased chemokine secretion.
FIG. 4.
Chemokine production by resting and activated PBMC of each CCR5/CCR2 genotype. PBMC from each donor were cultured with (activated) or without (resting) PHA–IL-2. MIP-1α in the culture supernatant was measured after 48 h by ELISA. Donor genotype and number are indicated at the bottom.
CCR2-64I function in transduced cell lines.
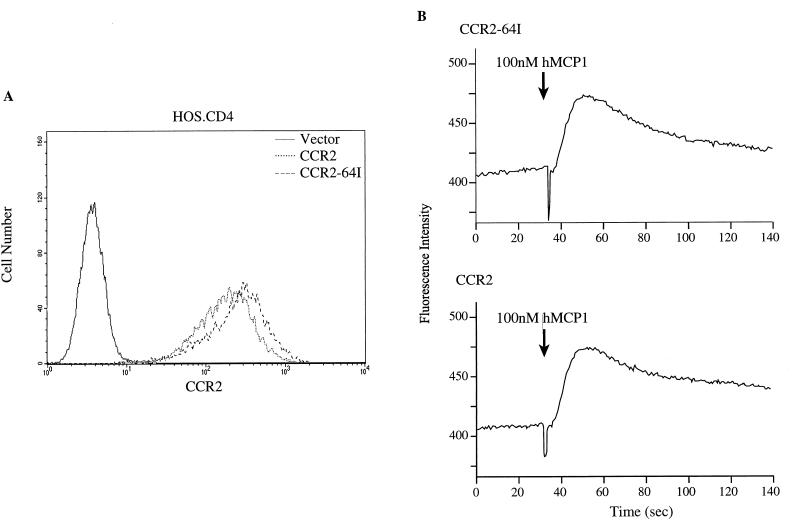
To investigate possible differences in CCR2-64I function, we tested whether CCR2-64I was expressed efficiently at the cell surface and was able to transduce signals in response to ligand binding. HOS.CD4 cell lines stably expressing CCR2 or CCR2-64I were established by infecting them with pBABE retroviral vectors. FACS analysis of the cell lines with anti-CCR2 MAb 1D9 showed that the wild-type and mutant receptors were expressed at comparable levels on the cell surface (Fig. 5A). The cell lines responded to saturating amounts of MCP-1 with similar [Ca2+] fluxes (Fig. 5B). Addition of suboptimal amounts of MCP-1, -2, and -3 showed smaller [Ca2+] fluxes that were indistinguishable in the two different cell lines (not shown).
FIG. 5.
CCR2 and CCR2-64I expression and function in retroviral vector-transduced HOS.CD4 cells. (A) HOS.CD4 cells expressing CCR2 or CCR2-64I transduced by pBABE retroviral vector infection were analyzed for receptor cell surface levels and (B) ability to flux Ca2+ in response to 100 mM human MCP-1 (hMCP1).
Absence of dominant negative effect of CCR2-64I on CCR5.
One mechanism by which the _CCR2_-64I polymorphism could influence AIDS progression rates was if the variant CCR2 molecule interacted with CCR5 to interfere with coreceptor function. Such an interaction was detected previously by Benkirane et al. (3), who reported that the truncated CCR5 protein encoded by the CCR5 Δ32 allele interfered with expression of the wild-type molecule. Oligomerization of CCR5 (3) and other G-protein-coupled receptors (17) has also been reported.
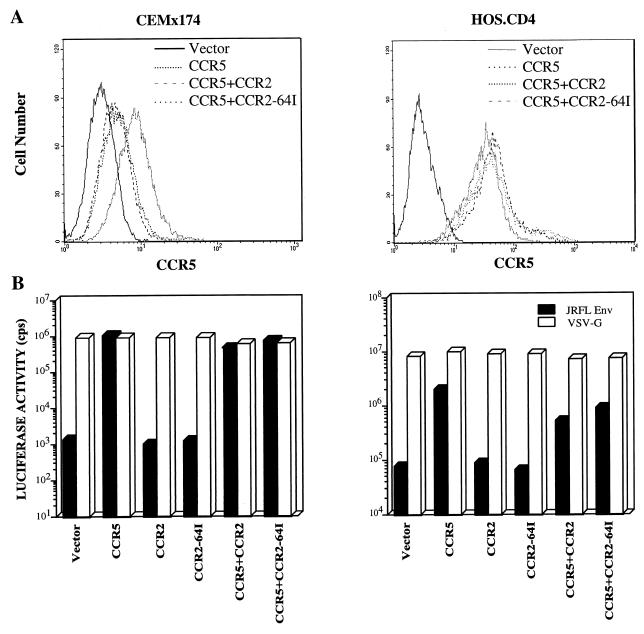
To test whether expression of CCR2-64I could inhibit CCR5 expression, HOS.CD4.CCR5 and CEMx174.CCR5 cell lines stably expressing CCR2 or CCR2-64I were established. FACS analysis with anti-CCR5 MAb 2D7 showed that CCR2-64I or wild-type CCR2 did not affect CCR5 expression levels in HOS.CD4.CCR5 cells (Fig. 6A). A small decrease in CCR5 expression was noted in the CEMx174 cells, but this decrease was similar for both CCR2 and CCR2-64I. This could have resulted either from a masking effect of the 2D7 epitope as a result of CCR2 expression or from a small decrease in expression from the integrated retroviral vector upon long-term culture.
FIG. 6.
HIV entry in CEMx174 and HOS.CD4 cells expressing transduced CCR2 or CCR2-64I with or without CCR5. (A) FACS analysis of HOS.CD4 cells expressing CCR5 with CCR2 or CCR2-64I stained with anti-CCR5 MAb 2D7. (B) Cells expressing the indicated chemokine receptors transduced by pBABE retroviruses were infected with JR.FL- or VSV-G-pseudotyped luciferase reporter virus (5 ng p24_gag_). Luciferase activity is presented as counts per second in lysates of infected cells. Similar results were obtained in two additional repetitions of the experiment.
To test whether CCR2 or CCR2-64I molecules have a dominant negative effect on CCR5 coreceptor function, HOS.CD4.CCR5 and CEMx174.CCR5 cells that stably expressed CCR2 or CCR2-64I were infected with single-cycle luciferase reporter viruses pseudotyped by M-tropic Env glycoprotein derived from HIV-1JR.FL or by VSV-G (11). The JR.FL pseudotypes entered with similar efficiency into HOS.CD4 or CEMx174 cells expressing CCR5 alone or in conjunction with CCR2 or CCR2-64I (Fig. 6B). They did not detectably infect cells expressing CCR2 or CCR2-64I in the absence of CCR5. VSV-G pseudotypes entered with similar efficiency into each of the cell types, confirming that postentry effects did not play a role. Similar analyses were performed on transiently cotransfected 293 cells with similar results (data not shown).
_CCR2_-64I-associated CCR5 promoter activity assay.
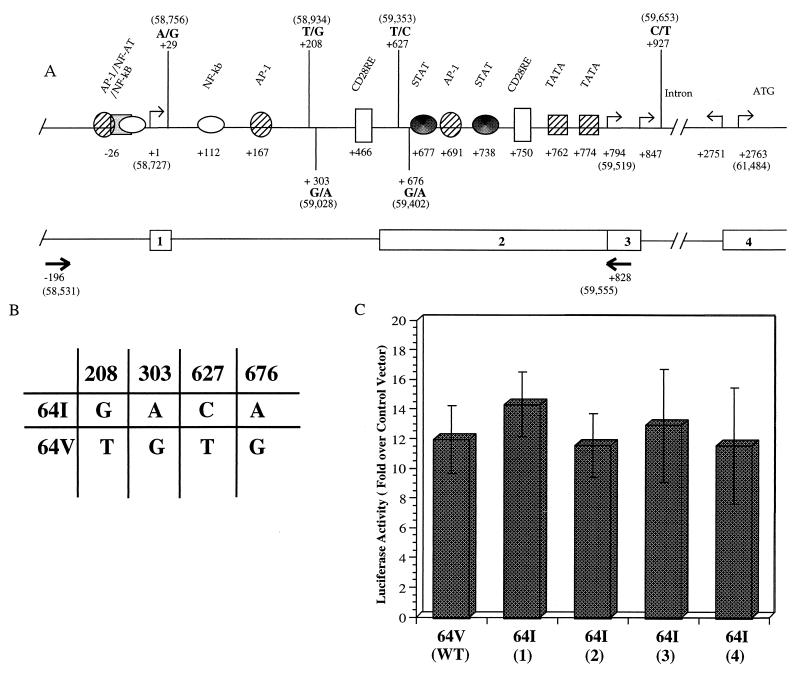
CCR5 transcription initiates at two positions, the first designated nt +1 (37) and a second that has been localized by primer extension (16, 35) or 5′-RACE (rapid amplification of 5′ cDNA ends) (28, 37) to a more proximal region (Fig. 7A). Transcription at the proximal start site is driven by the downstream promoter, Pd, that has been localized to within 1 kb of the start site and that contains consensus binding sites for several transcription factors, including STAT, AP1, TF2D, and CD28RE (16, 28, 35). Introduction of mutations into these sites in reporter plasmids reduces promoter function in transfection analyses (28).
FIG. 7.
Effect of _CCR2_-64I-associated polymorphisms on promoter activity in vitro. (A) Schematic representation of the CCR5 promoter region as reported by Liu et al. (28), with addition of the four polymorphisms frequently found in this region (36). The upstream promoter (Pu) transcription initiation site reported by Mummidi et al. (37) is defined as nt +1. Numbers in parentheses refer to nucleotide positions in GenBank accession no. U95626. Intron/exon organization and positions of the primers (arrows) used for PCR amplification of this region from genomic DNA are shown below. (B) Genotype of the _CCR2_-64I-associated and wild-type CCR2 64V-associated alleles tested for promoter function in panel C. (C) pGL3 reporter plasmids containing 1-kb genomic fragments derived from a CCR2 +/+ donor (wild type [WT]) or from four CCR2 64I/64I donors were tested for promoter activity in transfected Jurkat cells. Activity (mean ± standard deviation) is shown relative to pGL3 without insert, which is defined as 1, and normalized for the efficiency of transfection as measured by cotransfecting with β-galactosidase expression vector. Typical activity of vector without insert was 961 relative light units. Results shown represent averages of two independent experiments.
Sequencing of promoter regions of _CCR2_-64I-linked alleles derived from CCR2 64I/64I genomic DNA revealed four nucleotide substitutions compared to the wild-type _CCR2_-linked allele defined by the sequence previously deposited in GenBank (accession no. U95626). These included 208G, 303A, 627C, and 676A, all of which were present on each of eight CCR2 64I/64I-associated genomic fragments sequenced but not on those from four wild-type donors and are identical to those previously described by others (22, 36). Because these substitutions fall within the Pd promoter, they could conceivably affect CCR5 promoter function.
To determine whether the _CCR2_-64I-associated CCR5 promoter polymorphisms influenced promoter activity, we constructed plasmids in which the 1-kb genomic promoter region (Fig. 7A) derived from CCR2 +/+ or CCR2 64I/64I individuals was linked to a luciferase reporter gene, using the approach that we previously found effective for studying CCR5 promoter activity (28). The plasmids were used to transfect Jurkat cells, and luciferase activity in the cells was measured 3 days later. This analysis showed that each of the four CCR2 64I-associated CCR5 promoters was as active as the wild-type promoter in this assay (Fig. 7C).
DISCUSSION
The findings reported here do not support an association of _CCR2_-64I with decreased CCR5 mRNA or protein expression. The amounts of CCR5 mRNA transcripts derived from wild-type and _CCR2_-64I-associated alleles were nearly identical in resting and activated donor PBMC of each of the CCR5/CCR2 genotypes. Furthermore, there appeared to be no correlation between the _CCR2_-64I genotype and levels of cell surface CCR5 on activated CD4+ T cells. A small reduction in CCR5 levels on the CCR2 64I/64I cells was noted; however, this difference did not reach statistical significance. Analysis of larger numbers of _CCR2_-64I/64I donors will be required to determine whether the mutation has a small effect on CCR5 expression. However, clearly, the 64I polymorphism in its heterozygous state has an effect on AIDS progression similar to that of heterozygosity for Δ32 yet does not cause a similar 50% reduction in CCR5 cell surface levels.
While _CCR2_-64I is linked to at least four mutations in the genomic region containing the two CCR5 promoters, direct measurement of CCR5 promoter transcriptional activity in a reporter assay failed to show any difference in function between the wild-type and _CCR2_-64I-linked CCR5 promoter containing the four nucleotide substitutions. However, with this in vitro assay, it is difficult to completely rule out an association of _CCR2_-64I with altered promoter function. T-cell activation in vivo may differ from PHA–IL-2 activation as used in this study, and thus subtle differences in CCR5 regulation could have been missed. In vivo, CCR5 expression on T cells is influenced by signaling through CD28 (8) and by cytokines such as IL-2 (6) and IL-4 (48). In addition, it is possible that the promoter polymorphisms have an effect on CCR5 expression in monocytes or dendritic cells, which were not tested here.
Our findings also argue against a role for the _CCR2_-64I valine-to-isoleucine missense mutation in HIV-1 replication. Expression of wild-type CCR2 or CCR2-64I in cell lines affected neither CCR5 expression nor infectability by M-tropic virus. Moreover, donor T cells from CCR2 +/64I and _CCR2_-64I/64I individuals were as infectable as CCR2 +/+ cells. The valine-to-isoleucine mutation did not influence ligand binding as measured by ability to flux Ca2+ or by 125I-MCP-1 binding (28a).
While levels of CCR5 and CCR2 mRNA were very similar from donor to donor, cell surface expression levels of the two chemokine receptors varied considerably in our study and that of Wu et al. (50). This could be due to posttranscriptional regulatory mechanisms that influence CCR5 expression. Seven transmembrane G-protein-coupled receptors are endocytosed in response to ligand binding, C-terminal phosphorylation, and association with arrestins (reviewed in reference 23). Following ligand binding, receptors such as CCR1, CCR5, and CXCR4 are rapidly phosphorylated by G-protein receptor kinases and endocytosed (1, 46). Donor differences in any of these processes could conceivably influence CCR5 cell surface levels. Whether any of these mechanisms accounts for the association of _CCR2_-64I with decreased pathogenicity is not clear since none of the genes whose products are involved in these pathways are known to be genetically linked to CCR2.
During the preparation of this report, Lee et al. (26) reported similar findings on lack of influence of the _CCR2_-64I polymorphism on CCR5 expression or coreceptor function. Consistent with the findings reported here, they found that CCR2 +/64I donor T cells supported wild-type levels of virus replication and expressed wild-type levels of cell surface CCR5. In addition, they found that CCR2-64I did not have dominant negative activity on CCR5 function in transiently transfected 293 cells, a finding consistent with those reported here. In that study, _CCR2_-64I was associated with decreased CXCR4 cell surface expression. This difference was not apparent in our study and could be related to differences in the particular donors used in each analysis.
If _CCR2_-64I is not associated with decreased CCR5 transcription and the _CCR2_-64I molecule does not have dominant negative activity, how does it influence HIV-1 pathogenesis? It seems likely that other, as yet unidentified polymorphisms are involved. Such polymorphism could lie in linked genes that are unrelated to CCR5 or other chemokine coreceptors. One possibility is that there is a yet unidentified gene linked to _CCR2_-64I that encodes a polymorphic gene that plays a role in the immune response to HIV. Such a gene remain to be identified.
ACKNOWLEDGMENTS
We thank Anne Guiltinen, Jiamin Chen, Sarah Roberts, Elizabeth Fenamore, Timothy Willingham, and Theresa Gurney for technical assistance; Rong Liu for advice; Simon Monard and Jeremy Segal for FACS analysis; Christina Chiaffarelli for administrative assistance; and Vineet KewelRamani and Derya Unutmaz for critical reading of the manuscript.
This work was supported by grants from the NIH (R01 CA43252, R01 CA72149, R29 AI36057, R01 AI41384, AI42397, and R21 AI42630), AmFAR, Elizabeth Glaser Pediatric AIDS Foundation (77328 to R.M.) and National Heart Lung and Blood Institute in support of the REDS (N01-HB-47114 and N01-HB-97082). N.R.L. and R.A.K. are Elizabeth Glaser Scientists of the Pediatric AIDS Foundation.
REFERENCES
- 1.Amara A, Gall S L, Schwartz O, Salamero J, Montes M, Loetscher P, Baggiolini M, Virelizier J L, Arenzana-Seisdedos F. HIV coreceptor downregulation as antiviral principle: SDF-1alpha-dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. J Exp Med. 1997;186:139–146. doi: 10.1084/jem.186.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balotta C, Bagnarelli P, Violin M, Ridolfo A L, Zhou D, Berlusconi A, Corvasce S, Corbellino M, Clementi M, Clerici M, Moroni M, Galli M. Homozygous delta 32 deletion of the CCR-5 chemokine receptor gene in an HIV-1-infected patient. AIDS. 1997;11:F67–F71. doi: 10.1097/00002030-199710000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Benkirane M, Jin D Y, Chun R F, Koup R A, Jeang K T. Mechanism of transdominant inhibition of CCR5-mediated HIV-1 infection by ccr5delta32. J Biol Chem. 1997;272:30603–30606. doi: 10.1074/jbc.272.49.30603. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout T A, Sarau H M, Moores K, White J R, Elshourbagy N, Appelbaum E, Reape R J, Brawner M, Makwana J, Foley J J, Schmidt D B, Imburgia C, McNulty D, Matthews J, O’Donnell K, O’Shannessy D, Scott M, Groot P H E, Macphee C. Cloning, in vitro expression, and functional characterization of a novel human CC chemokine of the monocyte chemotactic protein (MCP) family (MCP-4) that binds and signals through the CC chemokine receptor 2B. J Biol Chem. 1997;272:16404–16413. doi: 10.1074/jbc.272.26.16404. [DOI] [PubMed] [Google Scholar]
- 5.Biti R, Ffrench R, Young J, Bennetts B, Stewart G, Liang T. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat Med. 1997;3:252–253. doi: 10.1038/nm0397-252. [DOI] [PubMed] [Google Scholar]
- 6.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busch M P. Residual risks of viral transmission by transfusions and projected yields of additional screening tests. Retrovirus Epidemiology Donors Study (REDS) Transfus Clin Biol. 1996;3:7–11. doi: 10.1016/s1246-7820(96)80007-3. [DOI] [PubMed] [Google Scholar]
- 8.Carroll R G, Riley J L, Levine B L, Feng Y, Kaushal S, Ritchey D W, Bernstein W, Weislow O S, Brown C R, Berger E A, June C H, St. Louis D C. Differential regulation of HIV-1 fusion cofactor expression by CD28 costimulation of CD4+ T cells. Science. 1997;276:273–276. doi: 10.1126/science.276.5310.273. [DOI] [PubMed] [Google Scholar]
- 9.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:936–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 10.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, O’Brien S. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 11.Deng D, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 12.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;96:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 13.Eugen-Olsen J, Iversen A K, Garred P, Koppelhus U, Pedersen C, Benfield T L, Sorensen A M, Katzenstein T, Dickmeiss E, Gerstoft J, Skinhoj P, Svejgaard A, Nielsen J O, Hofmann B. Heterozygosity for a deletion in the CKR-5 gene leads to prolonged AIDS-free survival and slower CD4 T-cell decline in a cohort of HIV-seropositive individuals. AIDS. 1997;11:305–310. doi: 10.1097/00002030-199703110-00007. [DOI] [PubMed] [Google Scholar]
- 14.Fauci A S. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Zepeda E A, Combadiere C, Rothenberg M E, Sarafi M N, Lavigne F, Hamid Q, Murphy P M, Luster A D. Human monocyte chemoattractant protein (MCP)-4 is a novel CC chemokine with activities on monocytes, eosinophils, and basophils induced in allergic and nonallergic inflammation that signals through the CC chemokine receptors (CCR)-2 and -3. J Immunol. 1996;157:5613–5626. [PubMed] [Google Scholar]
- 16.Guignard F, Combadiere C, Lee Tiffany H, Murphy P M. Gene organization and promoter function for CC chemokine receptor 5 (CCR5) J Immunol. 1998;160:985–992. [PubMed] [Google Scholar]
- 17.Hebert T E, Bouvier M. Structural and functional aspects of G protein-coupled receptor oligomerization. Biochem Cell Biol. 1998;76:1–11. doi: 10.1139/bcb-76-1-1. [DOI] [PubMed] [Google Scholar]
- 18.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 19.Huang Y, Zhang L, Ho D D. Biological characterization of nef in long-term survivors of human immunodeficiency virus type 1 infection. J Virol. 1995;69:8142–8146. doi: 10.1128/jvi.69.12.8142-8146.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katzenstein T L, Eugen-Olsen J, Hofmann B, Benfield T, Pedersen C, Iversen A K, Sorensen A M, Garred P, Koppelhus U, Svejgaard A, Gerstoft J. HIV-infected individuals with the CCR delta32/CCR5 genotype have lower HIV RNA levels and higher CD4 cell counts in the early years of the infection than do patients with the wild type. Copenhagen AIDS Cohort Study Group. J Acquired Immune Defic Syndr Hum Retrovirol. 1997;16:10–14. doi: 10.1097/00042560-199709010-00002. [DOI] [PubMed] [Google Scholar]
- 21.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 22.Kostrikis L G, Huang Y, Moore J P, Wolinsky S M, Zhang L, Guo Y, Deutsch L, Phair J, U. N A, Ho D D. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat Med. 1998;4:350–353. doi: 10.1038/nm0398-350. [DOI] [PubMed] [Google Scholar]
- 23.Krupnick J G, Benovic J L. The role of receptor kinases and arrestins in G protein-coupled receptor regulation. Annu Rev Pharmacol Toxicol. 1998;38:289–319. doi: 10.1146/annurev.pharmtox.38.1.289. [DOI] [PubMed] [Google Scholar]
- 24.Landau N R, Littman D R. A packaging system for producing murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaRosa, G. J., A. L. Reinhart, L. L. Yang, and N. Kassam. Unpublished data.
- 26.Lee B, Doranz B J, Rana S, Yi Y, Mellado M, Frade J M R, Martinez A C, O’Brien S J, Dean M, Collman R G, Doms R W. Influence of the CCR2-V64I polymorphism on human immunodeficiency virus type 1 coreceptor activity and on chemokine receptor function of CCR2b, CCR3, CCR5, and CXCR4. J Virol. 1998;72:7450–7458. doi: 10.1128/jvi.72.9.7450-7458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiple-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 28.Liu R, Zhao X, Gurney T, Landau N R. Functional analysis of transcriptional regulatory elements in the CCR5 promoter. AIDS Res Hum Retroviruses. 1998;14:1509–1519. doi: 10.1089/aid.1998.14.1509. [DOI] [PubMed] [Google Scholar]
- 28a.Mariani, R. Unpublished data.
- 29.Mariani R, Kirchhoff F, Greenough T C, Sullivan J L, Desrosiers R C, Skowronski J. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J Virol. 1996;70:7752–7764. doi: 10.1128/jvi.70.11.7752-7764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer L, Magierowska M, Hubert J B, Rouzioux C, Deveau C, Sanson F, Debre P, Delfraissy J F, Theodorou I. Early protective effect of CCR-5 delta 32 heterozygosity on HIV-1 disease progression: relationship with viral load. The SEROCO Study Group. AIDS. 1997;11:F73–F78. doi: 10.1097/00002030-199711000-00001. [DOI] [PubMed] [Google Scholar]
- 31.Michael N L, Chang G, d’Arcy L A, Ehrenberg P K, Mariani R, Busch M P, Birx D L, Schwartz D H. Defective accessory genes in a human immunodeficiency virus type 1-infected long-term survivor lacking recoverable virus. J Virol. 1995;69:4228–4236. doi: 10.1128/jvi.69.7.4228-4236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michael N L, Louie L G, Rohrbaugh A L, Schultz K A, Dayhoff D E, Wang C E, Sheppard H W. The role of CCR5 and CCR2 polymorphisms in HIV-1 transmission and disease progression. Nat Med. 1997;3:1160–1162. doi: 10.1038/nm1097-1160. [DOI] [PubMed] [Google Scholar]
- 33.Morawetz R A, Rizzardi G P, Glauser D, Rutschmann O, Hirschel B, Perrin L, Opravil M, Flepp M, von Overbeck J, Glauser M P, Ghezzi S, Vicenzi E, Poli G, Lazzarin A, Pantaleo G. Genetic polymorphism of CCR5 gene and HIV disease: the heterozygous (CCR5/delta ccr5) genotype is neither essential nor sufficient for protection against disease progression. Swiss HIV Cohort. Eur J Immunol. 1997;27:3223–3227. doi: 10.1002/eji.1830271220. [DOI] [PubMed] [Google Scholar]
- 34.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriuchi H, Moriuchi M, Fauci A S. Cloning and analysis of the promoter region of CCR5, a coreceptor for HIV-1 entry. J Immunol. 1997;159:5441–5449. [PubMed] [Google Scholar]
- 36.Mummidi S, Ahuja S S, Gonzalez E, Anderson S A, Santiago E N, Stephan K T, Craig F E, O’Connell P, Tryon V, Clark R A, Dolan M J, Ahuja S K. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med. 1998;4:786–793. doi: 10.1038/nm0798-786. [DOI] [PubMed] [Google Scholar]
- 37.Mummidi S, Ahuja S S, McDaniel B L, Ahuja S K. The human CC chemokine receptor 5 (CCR5) gene. Multiple transcripts with 5′-end heterogeneity, dual promoter usage, and evidence for polymorphisms within the regulatory regions and noncoding exons. J Biol Chem. 1997;272:30662–30671. doi: 10.1074/jbc.272.49.30662. [DOI] [PubMed] [Google Scholar]
- 38.Nibbs R J B, Wylie S M, Pragnell I B, Graham G J. Cloning and characterization of a novel murine beta chemokine receptor, D6. Comparison to three other related macrophage inflammatory protein-1alpha receptors, CCR-1, CCR-3, and CCR-5. J Biol Chem. 1997;272:12495–12504. doi: 10.1074/jbc.272.19.12495. [DOI] [PubMed] [Google Scholar]
- 39.O’Brien T R, Winkler C, Dean M, Nelson J A, Carrington M, Michael N L, White G C N. HIV-1 infection in a man homozygous for CCR5 delta 32. Lancet. 1997;349:1219. doi: 10.1016/s0140-6736(97)24017-1. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 40.Paxton W A, Liu R, Kang S, Wu L, Gingeras T R, Landau N R, Mackay C R, Koup R A. Reduced HIV-1 infectability of CD4+ lymphocytes from exposed-uninfected individuals: association with low expression of CCR5 and high production of β-chemokines. Virology. 1998;244:66–73. doi: 10.1006/viro.1998.9082. [DOI] [PubMed] [Google Scholar]
- 41.Paxton W A, Martin S R, Tse D, O’Brien T R, Skurnick J, VanDevanter N L, Padian N, Braun J F, Kotler D P, Wolinsky S M, Koup R A. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure. Nat Med. 1996;2:412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- 42.Salvi R, Garbuglia A R, Di Caro A, Pulciani S, Montella F, Benedetto A. Grossly defective nef gene sequences in a human immunodeficiency virus type 1-seropositive long-term nonprogressor. J Virol. 1998;72:3646–3657. doi: 10.1128/jvi.72.5.3646-3657.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 44.Schrager L K, Young J M, Fowler M G, Mathieson B J, Vermund S H. Long-term survivors of HIV-1 infection: definitions and research challenges. AIDS. 1994;8:1123–1128. [Google Scholar]
- 45.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O’Brien T R, Jacobson L P, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner M W, O’Brien S J. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 46.Solari R, Offord R E, Remy S, Aubry J P, Wells T, Whitehorn E, Oung T, Proudfoot A E. Receptor-mediated endocytosis of CC-chemokines. J Biol Chem. 1997;272:9617–9620. doi: 10.1074/jbc.272.15.9617. [DOI] [PubMed] [Google Scholar]
- 47.Theodorou I, Meyer L, Magierowska M, Katlama C, Rouzioux C, Group S S. HIV-1 infection in an individual homozygous for CCR5delta32. Lancet. 1997;349:1210–1220. [PubMed] [Google Scholar]
- 48.Valentin A, Lu W, Rosati M, Schneider R, Albert J, Karlsson A, Pavlakis G N. Dual effect of interleukin 4 on HIV-1 expression: implications for viral phenotypic switch and disease progression. Proc Natl Acad Sci USA. 1998;95:8886–8891. doi: 10.1073/pnas.95.15.8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, Buchbinder S, Vittinghoff E, Goedert J J, O’Brien T R, Jacobson L P, Detels R, Donfield S, Willoughby A, Gomperts E, Vlahov D, Phair J, O’Brien S J. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort. Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 50.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zimmerman P A, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, Weissman D, Cohen O, Rubbert A, Lam G, Vaccarezza M, Kennedy P E, Kumaraswami V, Giorgi J V, Detels R, Hunter J, Chopek M, Berger E A, Fauci A S, Nutman T B, Murphy P M. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]
- 52.Zuck T F, Thomson R A, Schreiber G B, Gilcher R O, Kleinman S H, Murphy E L, Ownby H E, Williams A E, Busch M P, Smith J W, Nass C C, Hollingsworth C G, Nemo G J. The Retrovirus Epidemiology Donor Study (REDS): rationale and methods. Transfusion. 1995;35:944–951. doi: 10.1046/j.1537-2995.1995.351196110900.x. [DOI] [PubMed] [Google Scholar]