Inhibition of Protein Synthesis by Y Box-Binding Protein 1 Blocks Oncogenic Cell Transformation (original) (raw)
Abstract
The multifunctional Y box-binding protein 1 (YB-1) is transcriptionally repressed by the oncogenic phosphoinositide 3-kinase (PI3K) pathway (with P3K as an oncogenic homolog of the catalytic subunit) and, when reexpressed with the retroviral vector RCAS, interferes with P3K- and Akt-induced transformation of chicken embryo fibroblasts. Retrovirally expressed YB-1 binds to the cap of mRNAs and inhibits cap-dependent and cap-independent translation. To determine the requirements for the inhibitory role of YB-1 in P3K-induced transformation, we conducted a mutational analysis, measuring YB-1-induced interference with transformation, subcellular localization, cap binding, mRNA binding, homodimerization, and inhibition of translation. The results show that (i) interference with transformation requires RNA binding and a C-terminal domain that is distinct from the cytoplasmic retention domain, (ii) interference with transformation is tightly correlated with inhibition of translation, and (iii) masking of mRNAs by YB-1 is not sufficient to block transformation or to inhibit translation. We identified a noncanonical nuclear localization signal (NLS) in the C-terminal half of YB-1. A mutant lacking the NLS retains its ability to interfere with transformation, indicating that a nuclear function is not required. These results suggest that YB-1 interferes with P3K-induced transformation by a specific inhibition of translation through its RNA-binding domain and a region in the C-terminal domain. Potential functions of the C-terminal region are discussed.
The phosphoinositide 3-kinase (PI3K) signaling pathway plays a major role in malignant cell transformation. Its molecular participants are frequently deregulated in human cancers (7, 79). A homolog of the catalytic subunit of the lipid kinase PI3K (P3K) and its downstream target, the serine-threonine protein kinase Akt, have been identified as oncogenic elements of transforming retroviruses (11, 14, 71). The PI3K and Akt genes are frequently mutated in cancer (10, 15, 46, 50, 62, 64); constitutively active forms of these proteins are transforming in cell culture and tumorigenic in vivo (1, 14, 47, 70). Negative regulators of PI3K-induced signaling are PTEN (phosphatase and tensin homolog deleted on chromosome 10) and the tuberous sclerosis protein complex, consisting of the proteins TSC1 and TSC2. PTEN is a lipid phosphatase and antagonist of PI3K and shows loss of function in human cancers at a frequency comparable to that of p53 (13, 67). The TSC1/2 complex, located downstream of Akt, functions as a GTPase activating protein and negatively regulates the small GTPase Rheb (33, 34). Germ line loss of function of TSC1 or TSC2 results in the development of benign tumors known as hamartomas (51). Another tumor suppressor, LKB1, controls TSC1/2 through the AMP-dependent protein kinase (16, 65). A loss of LKB1 function causes the Peutz-Jeghers syndrome, a benign tumor histologically similar to hamartomas (17, 30). An important downstream branch of the PI3K signaling pathway is represented by the serine-threonine kinase TOR (target of rapamycin). TOR regulates cell growth in response to PI3K signaling, nutrients, and endogenous energy levels (76). It stimulates translation by activating p70 S6 kinase (S6K) and the eukaryotic translation initiation factor and cap-binding protein 4E (eIF-4E) (29, 56). Phosphorylated and activated S6K phosphorylates ribosomal protein S6, which has been implicated in the recruitment of 5′TOP mRNAs to the ribosome (37). However, recent evidence has amended this hypothesis (74). eIF-4E is regulated by interaction with 4E-binding proteins (4E-BP). 4E-BP are negative regulators of cap-dependent translation. Hypophosphorylated 4E-BP binds to eIF-4E and prevents initiation of translation. TOR-mediated phosphorylation of 4E-BP1 frees eIF-4E from 4E-BP1 and thereby enables eIF-4E to assemble the translational initiation complex eIF-4F.
Increased translational activity is common during malignant transformation (28, 56). Examples documenting the essential role of translation during oncogenic transformation are provided by 4E-BP1, eIF-4E, eIF-4G, and Pdcd4 (programmed cell death 4). A constitutively active form of 4E-BP1 blocks c-Myc-induced transformation (45). eIF-4E and eIF-4G are transforming in cell culture (24, 43); overexpression of eIF-4E cooperates with c-Myc in the development of lymphomas in mice (63). Pdcd4 blocks tetradecanoyl phorbol acetate-induced transformation in susceptible JB6 cells and inhibits the helicase activity of eIF-4A, a component of the 4F translation initiation complex (82). These data are complemented by work with the immunosuppressant rapamycin. Rapamycin, in complex with the cellular protein FKBP12 (FK506-binding protein), binds TOR and inactivates it (76). Rapamycin is able to block P3K- and Akt-mediated transformation and interferes with tumor formation in PTEN-deficient mice (2, 53, 58).
In the present study with the Y box-binding protein 1 (YB-1), we provide more evidence for a fundamental role of translation during oncogenic transformation. YB-1, also known as p50 or DNA-binding protein B (dbpb), regulates transcription and translation. It binds to single-stranded RNA, single-stranded DNA, and double-stranded DNA (35). As a DNA-binding protein, YB-1 binds to promoters containing the Y box element and either activates or represses gene expression (23, 48, 54, 77). As an mRNA-binding protein, YB-1 is the most abundant protein of messenger ribonucleoprotein particles (mRNPs) and controls translation in a dose-dependent fashion (9, 12, 49). Low concentrations stimulate translation, and higher ones are inhibitory. Inactive mRNPs contain about twice as much YB-1 as active mRNPs (49). YB-1 blocks protein synthesis at the initiation stage (52). Masking of mRNA by YB-1 has been suggested as an explanation for YB-1-mediated inhibition of translation (7, 20, 61, 68). In P3K- and Akt-transformed chicken embryo fibroblasts (CEF), YB-1 is transcriptionally repressed (5). Overexpression of YB-1 leads to a specific resistance to oncogenic transformation induced by P3K or Akt. This transformation-resistant cellular phenotype is correlated with a flat cellular morphology, a cytoplasmic localization of YB-1, the ability to bind to the cap of mRNAs, and inhibition of cap-dependent and cap-independent translation. A competition between YB-1 and eIF-4E for cap binding has been suggested (5, 19). A YB-1 mutant, unable to bind to the cap structure of mRNAs, fails to inhibit translation and also does not interfere with transformation (5).
Available data on YB-1-induced resistance to P3K- and Akt-mediated transformation favor a mechanism involving control of protein synthesis. Cycloheximide, a general inhibitor of translation, does not result in an inhibition of P3K- or Akt-induced oncogenesis, suggesting that YB-1 affects the translation of specific mRNAs differentially (unpublished data). In this study, we determined the requirements for interference with transformation in a mutational analysis of the YB-1 protein. We provide evidence that neither mRNA binding nor cap binding of mRNAs is sufficient for inhibition of translation or for interference with transformation. YB-1-induced resistance to transformation by P3K or Akt requires RNA binding and a 90-amino-acid region within the C-terminal domain and is tightly correlated with inhibition of translation. We identified a nonconventional nuclear localization signal (NLS) of YB-1. A mutant lacking the NLS retains its ability to block transformation, indicating that a nuclear function of YB-1 is not required for inhibition of transformation. These results suggest that YB-1 blocks PI3K-induced transformation by a specific inhibition of protein synthesis.
MATERIALS AND METHODS
Cell culture and interference assays.
Fertilized chicken eggs (White Leghorn) were obtained from SPAFAS (Preston, Conn.). Preparation and cultivation of primary CEF have been described previously (80). Stable transfections using 5 μg of RCAS vectors were carried out using 4 × 106 secondary CEF grown in a 100-mm dish overnight in F10 medium (Sigma) supplemented with 5.87% iron-enriched bovine serum (Omega Scientific, Inc., Tarzana, Calif.), 0.93% l-glutamine-penicillin-streptomycin solution (Sigma, St. Louis, Mo.), and 2 ng of Polybrene (Sigma)/ml. Cells were incubated with DNA for 4 h, treated with F10-0.25% dimethyl sulfoxide for 1 min, washed twice with F10, and grown in F10 medium containing 5.87% iron-enriched bovine serum and 0.93% l-glutamine-penicillin-streptomycin solution. Cell culture continued following standard procedures (80). After transfection, CEF were transferred at least twice before they were used for interference assays or immunofluorescence or reporter assays. For interference assays, CEF were transfected with RCAS(B) vectors and seeded onto MP6 wells (5 × 105 cells per 35-mm well). After 10 days, cells were infected with oncogenic RCAS(A)myr-_p3k_Δ72 (3) virus or RCAS(A)myr-AktΔ11-60 virus (1) and overlaid with agar medium on the next day. Fresh agar medium was added every 2 days. Upon formation of transformed cell foci, cells were stained with 2% crystal violet in 20% methanol.
Plasmids.
All RCAS vectors are based on RCAS.Sfi (1), a derivative of RCAS (32) containing SfiI(A) and SfiI(B) sites for unidirectional cloning. RCAS(B)FLAG-YB-1, RCAS(B)FLAG-YB-1*, and pcDNA3.Sfi have been described elsewhere (4, 5). RCAS(B)-IκB-SR was kindly provided by M. Aoki. pBSFI-FLAG-YB-1 was created by moving the FLAG-YB-1 SfiI fragment from pFBnFLAG-YB-1 (5) into the SfiI sites of pBSFI. YB-1 deletion mutants were generated by PCR using pBSFI-FLAG-YB-1 as template and PCR primers as described in Table 1. PCR products were cut with NotI and BamHI and cloned into the NotI and BamHI sites of pBSFI-FLAG. Inserts were recloned as SfiI fragments into RCAS(B) and pcDNA3.Sfi. The chicken YB-1 protein published under accession number AAA02573 was used in this study. Internal deletions were prepared by altering the YB-1 nucleotide sequences in pBSFI-FLAG-YB-1 and pBSFI-FLAG-YB-1* to StuI sites at amino acid positions 183 and 273 (YB-1Δ183-273), 183 and 202 (YB-1Δ183-202 and YB-1*Δ183-202), and 190 and 202 (YB-1Δ190-202 and YB-1*Δ190-202) by using the QuikChange XL site-directed mutagenesis kit from Stratagene (La Jolla, Calif.). Mutated pBSFI vectors were cut with StuI, gel purified, and religated. Inserts were retrieved by SfiI digestion and cloned into the SfiI sites of RCAS(B). YB-1(1-202,4R-4A) was generated by replacing the four arginines at amino acid positions 186 to 189 with alanines by using the QuikChange XL site-directed mutagenesis kit from Stratagene and pBSFI-YB-1(1-202) as template. The particular SfiI cDNA fragments were moved into RCAS(B) or pcDNA3.Sfi as described above. For the generation of pcDNA3-HA-YB-1, a YB-1 cDNA fragment was amplified by PCR using 5′-AACCCATGGTTATGAGCAGCGAGGCCGAGACCCAGCCG-3′ as forward primer and 5′-AACGAATTCTTACTCAGCCCCGCCCTGCTCGGCCTCGGG-3′ as reverse primer and pBSFI-FLAG-YB-1 as template. The DNA fragment was gel purified, cut with NcoI and EcoRI, and cloned into NcoI-EcoRI-cut pBSFI-HA vector (69), creating pBSFI-HA-YB-1. From this vector, the SfiI fragment was cloned into pcDNA3.Sfi. To generate pc-luc, the HindIII-BamHI fragment from pGL3 Basic (Promega) encoding the firefly luciferase gene was cloned into the HindIII and BamHI sites of pcDNA3 (Invitrogen, Carlsbad, Calif.).
TABLE 1.
Forward and reverse primers used for generation of YB-1 deletion mutants
| YB-1 protein | Forward primer | Reverse primer |
|---|---|---|
| AP-CSD | 5′ACGCGGCCGCCCATGAGCAGCGAGGCCGAGACCCAGCCG3′ | 5′ACGGATCCTTATGCGTATTTGCTGCCTTGCACTGGAAC3′ |
| C-term | 5′ACGCGGCCGCCCGCAGACCGTAACCATTACAGACGATAT3′ | 5′ACGGATCCTTACTCAGCCCCGCCCTGCTCGGCCTCGGG3′ |
| YB-1(1-176) | 5′ACGCGGCCGCCCATGAGCAGCGAGGCCGAGACCCAGCCG3′ | 5′TTCGGATCCTTATTCCGGGATGTTCTCTGCTCCTTCGTTC3′ |
| YB-1(1-183) | 5′ACGCGGCCGCCCATGAGCAGCGAGGCCGAGACCCAGCCG3′ | 5′ACGGATCCTTAACGGCGCTGCTGGGCTTGGCCTTCCGG3′ |
| YB-1(1-202) | 5′ACGCGGCCGCCCATGAGCAGCGAGGCCGAGACCCAGCCG3′ | 5′ACGGATCCTTATCGACGCCCGTAGGGCCTACGCATGTA3′ |
| YB-1(1-243) | 5′ACGCGGCCGCCCATGAGCAGCGAGGCCGAGACCCAGCCG3′ | 5′ACGGATCCTTAGCGGAATCGTGGTCTGTAACCTCGATA3′ |
| YB-1(1-264) | 5′ACGCGGCCGCCCATGAGCAGCGAGGCCGAGACCCAGCCG3′ | 5′ACGGATCCTTATTGGTTCTCTTTATCTTCTTCGTTTCC3′ |
| YB-1(1-271) | 5′ACGCGGCCGCCCATGAGCAGCGAGGCCGAGACCCAGCCG3′ | 5′ACGGATCCTTACTGACCTTGGGTCTCATCTCCTTGGTT3′ |
| YB-1(1-290) | 5′ACGCGGCCGCCCATGAGCAGCGAGGCCGAGACCCAGCCG3′ | 5′AACGGATCCTTACTCTGGGCGTCTGCGTCTGTAGTTG3′ |
| YB-1(1-308) | 5′ACGCGGCCGCCCATGAGCAGCGAGGCCGAGACCCAGCCG3′ | 5′TTCGGATCCTTACTCAGCTGGTGGTTCGGCTGTCTTCG3′ |
| YB-1(41-321) | 5′AACGCGGCCGCCCGCGGCGCCTCCCGCCGGCGGGGACAAGAAG3′ | 5′ACGGATCCTTACTCAGCCCCGCCCTGCTCGGCCTCGGG3′ |
| YB-1(41-308) | 5′AACGCGGCCGCCCGCGGCGCCTCCCGCCGGCGGGGACAAGAAG3′ | 5′TTCGGATCCTTACTCAGCTGGTGGTTCGGCTGTCTTCG3′ |
| YB-1(41-290) | 5′AACGCGGCCGCCCGCGGCGCCTCCCGCCGGCGGGGACAAGAAG3′ | 5′AACGGATCCTTACTCTGGGCGTCTGCGTCTGTAGTTG3′ |
Immunofluorescence.
Cells grown overnight on glass coverslips were washed with phosphate-buffered saline (PBS) and fixed with 3.7% formaldehyde for 30 min, rinsed twice with PBS, and incubated with PBS containing 0.1% Triton X-100 (Sigma) for 30 min. After a wash with PBS, the coverslips were incubated with 10% goat serum for 1 h at room temperature in a humidified container followed by incubation with rabbit polyclonal anti-FLAG antibody (Sigma) for another hour. Coverslips were washed with PBS three times and incubated with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (Sigma) for 1 h. During the last 5 min, 4,6-diamidino-2-phenylindole (DAPI) was added. The coverslips were again washed three times with PBS and mounted on glass slides using Slowfade mounting medium (Molecular Probes, Eugene, Oreg.). Primary and secondary antibodies were used at a final dilution of 1:100, and the final concentration of DAPI was 2 ng/μl.
Immunoblotting.
Cells were trypsinized, counted, pelleted for 3 min at 960 × g, and lysed in sample buffer containing 60 mM Tris-HCl (pH 6.8), 10% (vol/vol) glycerol, 3% (wt/vol) sodium dodecyl sulfate (SDS), 5% (vol/vol) β-mercaptoethanol, and 0.005% (wt/vol) bromophenol blue. Lysates from 6.25 × 104 cells were separated on SDS-10% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a nitrocellulose membrane. Membranes were blocked in Tris-buffered saline (TBS)-Tween 20 (0.1%) containing 5% bovine serum albumin for 2 h at room temperature, followed by incubation with horseradish peroxidase-conjugated anti-FLAG M2 antibodies (1:20,000; Sigma) for 40 min at room temperature in TBS-T (0.1%) with 5% bovine serum albumin.
m7GTP pull-down and mRNA-binding assays.
m7GTP pull-down assays were done according to a published protocol (8, 44). Briefly, cells were grown to confluence and suspended in 700 μl of m7GTP lysis buffer containing 20 mM Tris, 100 mM KCl, 20 mM β-glycerophosphate, 1 mM ethylene glycol-bis(2-aminoethylether)-N,N,_N_′,_N_′-tetraacetic acid (EGTA), 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 0.25 mM Na3VO4, 10 mM NaF, and 1× protease inhibitor cocktail from Roche (Indianapolis, Ind.). Native protein extracts were prepared with six cycles of freeze-thaw using a dry ice-methanol bath and thawing in water. Debris was separated by centrifugation, and the protein concentration was determined using the BCA system from Pierce (Rockford, Ill.). For the m7GTP pull down, 300 μg of protein together with 50 μl of 7-methyl GTP-Sepharose 4B beads (Amersham Pharmacia, Piscataway, N.J.) were incubated overnight at 4°C. Beads were washed seven times with m7GTP washing buffer containing 20 mM Tris, 100 mM KCl, 0.2 mM EDTA, and 7 mM β-mercaptoethanol, and bound proteins were applied to an SDS-PAGE (10%). Western blotting was carried out as described above. For the mRNA-binding assay, 150 μg of total protein of lysates that had been used for the m7GTP pull down were immunoprecipitated with anti-FLAG M2 agarose (Sigma) for 30 min at 4°C. Beads were washed four times with m7GTP washing buffer and were incubated with 2 μl of RNase inhibitor (Superase·In; Ambion, Austin, Tex.) and 3.3 × 106 cpm of riboprobe in a total volume of 200 μl of m7GTP washing buffer at room temperature for 2 h. As riboprobe, in vitro-transcribed, [32P]UTP-labeled mRNA was used that had been generated with linearized pBSFI-FLAG-YB-1 vector as template and the MaxiScript kit from Ambion according to the manufacturer's protocol. The approximately 1,000-nucleotide (nt) YB-1 mRNA was purified using MicroSpin G-50 columns from Amersham Pharmacia. After incubation, the beads were washed twice with m7GTP washing buffer and four times with m7GTP washing buffer supplemented with 500 mM NaCl. Beads were suspended in 50 μl of sample buffer (18 mM EDTA, 1× TBE, 3% [wt/vol] SDS, 0.03% [wt/vol] bromophenol blue, 68.5% [vol/vol] formamide) and boiled for 3 min. Results were quantified by measuring Cherenkov radiation of 5 μl. Ten microliters was separated on a 4% polyacrylamide gel with 8 M urea and visualized by autoradiography.
Coimmunoprecipitation.
HEK293 cells were transiently transfected with 3 μg each of pcDNA3-HA-YB-1 and pcDNA3-FLAG encoding various YB-1 proteins. The empty vector pcDNA3.Sfi was cotransfected to reach equal amounts of transfected DNA. Cells were lysed in immunoprecipitation lysis buffer containing 0.5% NP-40, 10% glycerol, 20 mM Tris, 150 mM NaCl, 1 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 50 mM NaF, 1 mM dithiothreitol, 50 mM β-glycerolphosphate, 1 mM Na3VO4, and 1× protease inhibitor cocktail from Roche. Five hundred micrograms of total protein was incubated at 37°C for 2 h with 1 μg of RNase A/μl to ensure complete degradation of mRNAs and applied onto agarose beads conjugated with M2 FLAG-specific antibodies (Sigma) for 30 min at 4°C. Beads were washed five times with immunoprecipitation lysis buffer without protease inhibitors. Bound proteins were separated by SDS-PAGE (10%) and visualized by Western blotting using horseradish peroxidase-conjugated M2 FLAG antibodies (Sigma) or polyclonal HA.11 antibodies (Sigma). Tubulin was detected with monoclonal anti-tubulin from ICN Biomedicals Inc., Aurora, Ohio.
Reporter assays, RNA isolation, RT-PCR, and Northern blotting.
Transient transfections, preparation of cell lysates, and luciferase assays were performed as described previously (5). A total of 1.5 × 105 cells were seeded onto 12-well plates, grown overnight, and transfected with 600 ng of pc-luc; luciferase assays were done using 5 μg of protein. Isolation of total RNA was performed by CsCl gradient centrifugation. Briefly, total protein was lysed in guanidium isothiocyanate buffer containing 4 M guanidine thiocyanate, 25 mM sodium acetate, and 0.835% (vol/vol) β-mercaptoethanol. Lysates were drawn through a 20-gauge needle to shear chromosomal DNA. Lysates were overlaid on a 5.7 M CsCl cushion, and total RNA was pelleted by centrifugation at 107,000 × g for 18 to 24 h, ethanol precipitated, and dissolved in 20 μl of water. Eight microliters of total RNA was used for cDNA synthesis using the ThermoScript reverse transcription-PCR (RT-PCR) system from Invitrogen according to the manufacturer's instructions. PCR was carried out at a 62°C annealing temperature for 25 cycles. The YB-1-specific primers 5′-AACGCGGCCGCCCGCGGCGCCTCCCGCCGGCGGGGACAAGAAG-3′ and 5′-ACGGATCCTTAGCGGAATCGTGGTCTGTAACCTCGATA-3′ were used to amplify a 606-nt cDNA product. For Northern analyses, 4 × 106 CEF were seeded onto 100-mm dishes, grown overnight, and transiently transfected with 3 μg of pc-luc. Cells were serum starved and stimulated with serum growth factors as described elsewhere (5). Isolation of total RNA, RNA gel electrophoresis, Northern blotting, and Northern hybridizations were done as described previously (6). Firefly luciferase and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA fragments were labeled with [α-32P]dCTP using the Random Primed DNA labeling kit from Roche.
RESULTS
The CSD and C-terminal sequences of YB-1 are required to block oncogenic transformation induced by P3K or Akt.
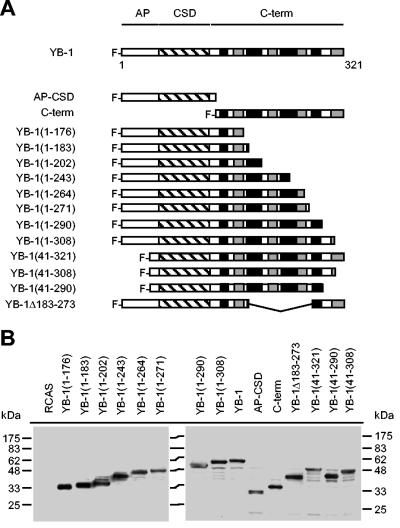
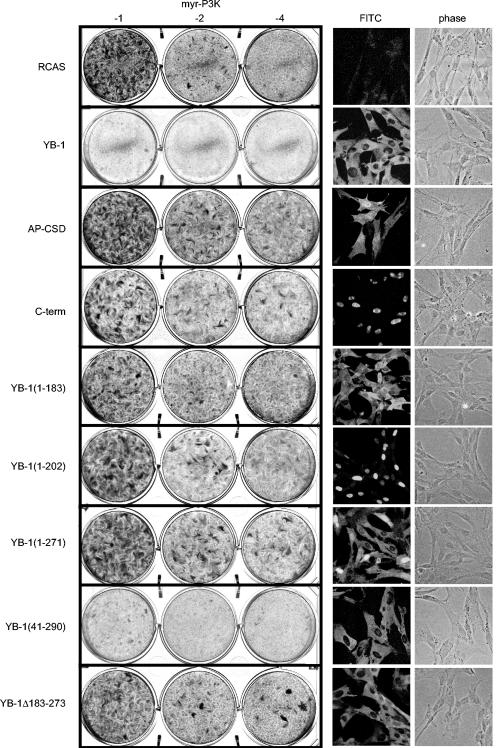
YB-1 consists of three major domains: an N-terminal alanine-proline-rich domain (AP), a central cold shock domain (CSD), and a C-terminal tail domain comprising four alternating clusters of basic and acidic residues (Fig. 1A). The core of the protein is the CSD, which forms a distinct structure and facilitates binding to the nucleic acid; it also contains an RNA-binding motif (20). Mutation of the RNA-binding motif eliminates RNA binding and the ability to interfere with P3K-induced transformation, suggesting that RNA binding is essential for YB-1 to block P3K-mediated transformation (5). In a search for domains of YB-1 necessary for interference with transformation, we generated various deletion mutants (Fig. 1A). All mutants were derived from the chicken YB-1 protein consisting of 321 residues. We cut the protein in half, generating a C-terminal fragment (residues 137 to 321) and an N-terminal fragment consisting of the AP domain and the CSD (AP-CSD, residues 1 to 136). We deleted portions of the C terminus, deleted almost the entire AP domain, and created a YB-1 mutant with an internal deletion within the C-terminal domain. All cDNAs contained an N-terminal FLAG tag and were expressed from the retroviral expression vector RCAS. cDNAs were entirely sequenced, and correct expression of the individual YB-1 proteins was verified by Western analysis (Fig. 1B). To test for interference with Akt- or P3K-induced transformation, CEF transfected with RCAS vectors encoding the YB-1 deletions were superinfected with oncogenic P3K or Akt virus carrying an N-terminal myristylation signal (myr-P3K and myr-Akt). As shown in Fig. 2, neither the AP-CSD nor the C-terminal domains were able to block transformation. Interference with transformation required the CSD and a major portion of the C-terminal domain adjacent to the CSD up to amino acid 290 (Table 2). The mutant lacking amino acids 183 to 273 was also unable to block transformation, highlighting the importance of the C-terminal domain. Protein sequences that are dispensable for interference are the AP domain and C-terminal sequences beyond amino acid 290.
FIG. 1.
YB-1 deletion mutants. (A) Schematic diagram of YB-1 mutants. The three major domains of the YB-1 protein are outlined at the top: AP domain, CSD (hatched box), and C-terminal domain (C-term). The four alternating clusters of acidic and basic residues within the C-terminal domain are indicated by black (basic) and grey (acidic) boxes. All YB-1 proteins contain an N-terminal FLAG tag. The nomenclature of deletion mutants refers to amino acid positions of the chicken YB-1 protein. (B) Protein expression of YB-1 deletion mutants. Total lysates of 6.25 × 104 CEF transfected with RCAS vectors as indicated were subjected to Western analysis with FLAG-specific antibodies.
FIG. 2.
Inhibition of transformation and subcellular localization of YB-1 deletion mutants. (Left) CEF were stably transfected with RCAS vectors encoding YB-1 deletion mutants and superinfected with oncogenic myr-P3K virus. Individual RCAS vectors and the log10 of virus dilutions are shown. Cells were fed with agar medium and stained after 15 days with crystal violet. Data obtained with myr-Akt were equivalent to those of myr-P3K and are not shown. (Right) YB-1 proteins were visualized by immunofluorescence as described in Materials and Methods. Fluorescein (FITC) and phase-contrast (phase) micrographs were taken using a 40× objective lens.
TABLE 2.
Effects of various YB-1 deletion mutants on transformation by myr-P3k and their subcellular localization in CEF
| YB-1 protein | EOTa | Localization |
|---|---|---|
| YB-1 | 0.02 | Cytoplasm |
| AP-CSD | 1.69 | Cytoplasm, nucleus |
| C-term | 0.79 | Nucleus |
| YB-1(1-176) | 0.89 | Cytoplasm |
| YB-1(1-183) | 0.67 | Cytoplasm |
| YB-1(1-202) | 1.19 | Nucleus |
| YB-1(1-243) | 1.27 | Nucleus |
| YB-1(1-264) | 1.29 | Nucleus |
| YB-1(1-271) | 1.33 | Nucleus, cytoplasm |
| YB-1(1-290) | 0.00 | Cytoplasm |
| YB-1(1-308) | 0.02 | Cytoplasm |
| YB-1(41-321) | 0.05 | Cytoplasm |
| YB-1(41-308) | 0.00 | Cytoplasm |
| YB-1(41-290) | 0.02 | Cytoplasm |
| YB-1Δ183-273 | 0.90 | Cytoplasm |
| RCAS | 1.00 | NAb |
Subcellular localization of YB-1 deletion mutants.
The full-length YB-1 protein localizes predominantly in the cytoplasm (5, 41, 59, 73). However, derivatives of YB-1 have also been found in the nucleus (38, 73). The YB-1 mutant YB-1*, which no longer binds to RNA, is retained in the nucleus (5). The ability to interfere with transformation could simply be controlled by the subcellular distribution of YB-1. Therefore, we determined the localization of the YB-1 deletion mutants by immunofluorescence (Fig. 2; Table 2). As expected, wild-type YB-1 was predominantly found in the cytoplasm. Some of the deletion mutants were nuclear and some were cytoplasmic, depending on the protein sequence that had been deleted. The results allowed the following conclusions: (i) the C-terminal half of the protein contains a potential NLS between residues 183 and 202. This is demonstrated by the cytoplasmic YB-1(1-183) and the nuclear YB-1(1-202) proteins. (ii) YB-1 residues 264 to 290 encode a cytoplasmic retention site (CRS) that prevails over the NLS (Table 2). This is shown by YB-1(1-264) and YB-1(1-290). YB-1(1-264) localizes to the nucleus, whereas YB-1(1-290) is cytoplasmic even in the presence of the NLS. (iii) A stabilization of the RNA-binding module in the cytoplasm is required but not sufficient for interference with transformation. YB-1(1-183) and YB-1Δ183-273 are two examples that localize in the cytoplasm and contain an intact CSD but are unable to block transformation. This observation also shows that the CRS and the C-terminal domain, which mediates interference, are two distinct domains.
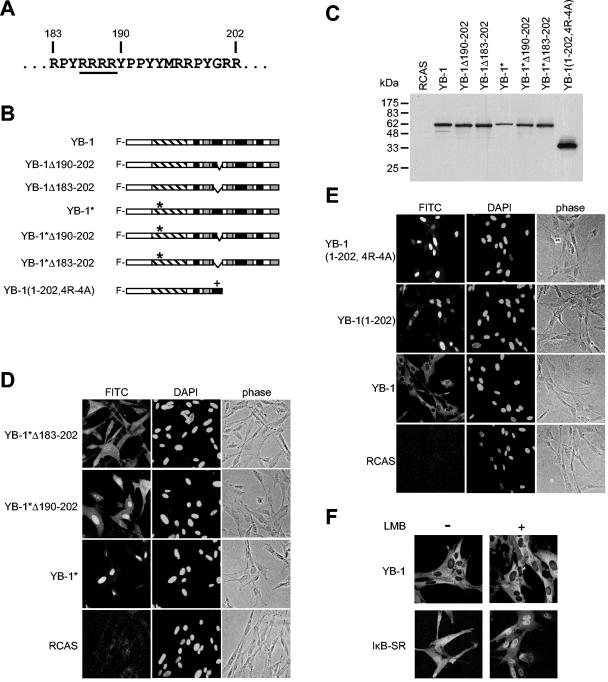
The noncanonical NLS of YB-1.
As shown above, YB-1 residues 183 to 202 encode a potential NLS. To verify that this site is biologically active in the full-length protein, we further analyzed the NLS with the YB-1* mutant that contains a disrupted RNA-binding motif and localizes in the nucleus (5). We generated YB-1* mutants lacking parts of or the entire sequence encompassing residues 183 to 202 and expressed these proteins in CEF using RCAS (Fig. 3A, B, and C). A partial deletion mutant of that site, lacking amino acids 190 to 202, resulted only in an incomplete release of the protein into the cytoplasm (Fig. 3D). Deleting all 20 amino acids (183 to 202) increased cytoplasmic localization and suggested that the residues 183 to 202 play an important role in the control of cellular localization of YB-1 (Fig. 3D). We concede that the cytoplasmic staining is not absolute. However, the staining obtained with YB-1*Δ183-202 strongly resembled that of the AP-CSD fragment (cf. Fig. 2), indicating that the N-terminal half of the protein, consisting of the AP domain and CSD, has residual nuclear localization activity.
FIG. 3.
NLS of YB-1. (A) The chicken YB-1 protein sequence encompassing amino acids 183 to 202 is shown. The four arginines that have been replaced by alanines in the YB-1(1-202,4R-4A) mutant are underlined. (B) Schematics of YB-1 mutants lacking the amino acid sequences 190 to 202 or 183 to 202. The major protein domains are highlighted by the same patterns as in Fig. 1A. The asterisks refer to two point mutations in the RNA-binding motif; the cross indicates the replacement of arginines 186 to 189 with alanines in the mutant YB-1(1-202,4R-4A). All proteins contain an N-terminal FLAG tag. (C) Immunoblot of mutants shown in panel B. Total lysates of 6.25 × 104 cells were probed for YB-1 proteins with anti-FLAG antibodies. (D and E) Subcellular localization of YB-1 proteins outlined in panel B. YB-1 proteins were visualized with anti-FLAG fluorescein-conjugated secondary antibodies. DAPI and phase-contrast images are shown to the right. All micrographs were taken using the 40× objective lens. (F) CEF expressing full-length YB-1 or IκB-SR were incubated in the presence of 5 ng of leptomycin B (Sigma)/ml for 24 h. Controls were treated with solvent (70% methanol) only. Cells were fixed and subjected to immunofluorescence as described in Materials and Methods.
Conventional NLSs are either monopartite, such as the NLS of the SV40 large T-antigen (KKKRK), or bipartite, composed of the structure K/R K/R K/R (X)10-12 K/R K/R (18, 39, 40). Considering the relative position and the high content of arginines within the sequence encompassing residues 183 to 202, this sequence could act as a classic NLS. We therefore determined whether the cluster of four arginines at position 186 to 189 is essential for nuclear localization. We generated the mutant YB-1(1-202,4R-4A), encoding YB-1 residues 1 to 202 with alanines in positions 186 to 189 (Fig. 3A, B, and C). We compared the localization of this mutant with its unaltered counterpart YB-1(1-202). Both proteins were retained in the nucleus, suggesting that the arginines are not essential for nuclear localization (Fig. 3E). Even with the further deletion of the two terminal arginines in positions 201 and 202, the mutant protein remained in the nucleus (data not shown). These data suggest that residues 183 to 202 form a noncanonical NLS that functions independently of basic residues.
In a subsequent experiment we examined whether nuclear export of YB-1 is mediated by CRM1 (chromosome region maintenance 1). CRM1 belongs to the exportin proteins and controls nuclear export of many shuttling proteins, such as MDM2/p53 (murine double minute 2), cyclin B1, and IκB (inhibitor of NF-κB) (22, 31, 83). We treated YB-1-expressing cells with leptomycin B, which prevents CRM1-mediated nuclear export by competing for CRM1 (21, 42, 72). After a 24-h treatment with leptomycin B, the IκB superrepressor accumulates in the nuclei due to inhibition of nuclear export. In contrast, there was no measurable increase in nuclear localization of YB-1, suggesting that nuclear export of YB-1 occurs independently of CRM1 (Fig. 3F). It may be directed through mRNA export instead.
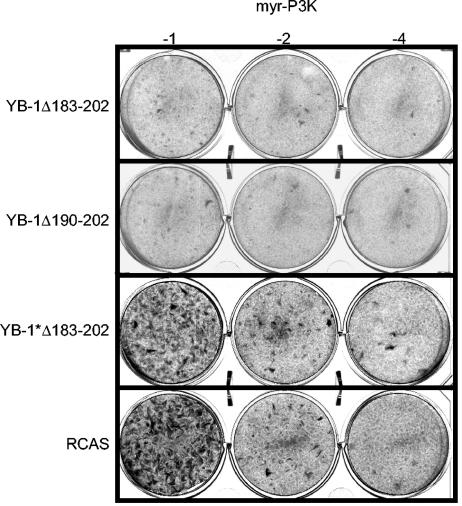
Nuclear localization is not required for interference with transformation by P3K.
Full-length YB-1 localizes to the cytoplasm (5, 41, 59, 73). However, it is still possible that traces of YB-1 travel into the nucleus, mediating interference with P3K and Akt by a mechanism that is distinct from its role as a cytoplasmic translational regulator. Therefore, we deleted the NLS from wild-type YB-1 and tested this mutant in an interference assay. As shown in Fig. 4, interference was neither abolished nor diminished by the lack of an intact NLS, suggesting that the nuclear function of YB-1 is not necessary to prevent transformation. The C-terminal domain that is required for YB-1 function can be further confined to a 90-amino-acid region between residues 202 and 290 that includes the CRS. In vitro, the C-terminal half of the YB-1 protein alone effectively blocks translation (52). To test whether this domain is sufficient to block transformation when localized in the cytoplasm, we used the mutant YB-1*Δ183-202 that lacks RNA-binding activity and the NLS but retains the C-terminal domain 202-290 and localizes to the cytoplasm (Fig. 3D). The data revealed that the C-terminal domain of YB-1 alone is not able to prevent transformation and suggested that the CSD in concert with the C-terminal region is required to block transformation.
FIG. 4.
YB-1 proteins lacking the NLS in an interference assay. CEF stably transfected with RCAS vectors encoding YB-1Δ183-202, YB-1Δ190-202, or YB-1*Δ183-202 were superinfected with transforming myr-P3K virus in a serial dilution as shown. Cells were stained after 15 days.
Cap binding is required but not sufficient for interference with P3K-induced transformation.
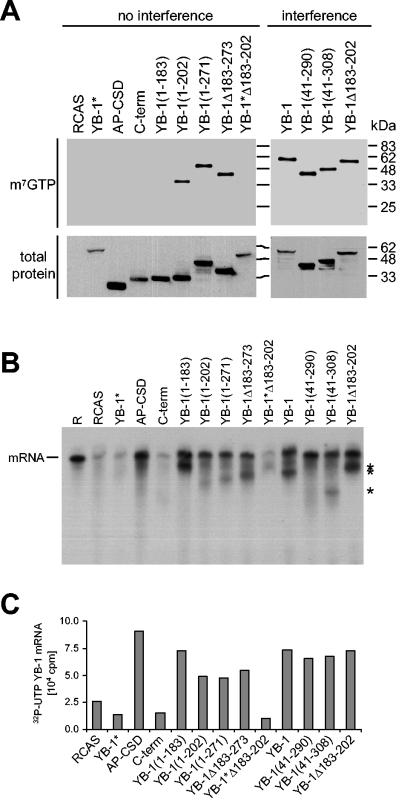
YB-1 binds to the cap region of mRNAs and could function as a competitor of eIF-4E (5). The antioncogenic effect of YB-1 could therefore be mediated by binding to the mRNA cap. In order to explore this possibility, we harvested cells expressing wild-type or mutant YB-1 proteins and applied native cell lysates onto a 7-methyl GTP resin (m7GTP). m7GTP beads mimic the cap region of mRNAs and allow the isolation of cap-binding proteins (44). Bound proteins were identified by Western blotting with FLAG-specific antibodies (Fig. 5A). All YB-1 mutants that were able to block transformation were also bound to the beads. However, some of the mutants that failed to interfere with transformation were also able to bind to the m7GTP resin. Among these were proteins that were abundantly expressed in the cytoplasm, such as YB-1(1-271) and YB-1Δ183-273. This result suggests that mere cap binding is not sufficient for interference. Competition with eIF-4E is therefore not a likely mechanism for the antioncogenic activity of YB-1.
FIG. 5.
mRNA-binding and cap-binding activities of YB-1 mutants. (A) Cap-binding assay. Three hundred micrograms of total protein of native cell lysates was incubated with m7GTP resin at 4°C overnight. Bound YB-1 proteins were detected on a Western analysis using FLAG-specific antibodies (top panel). To show expression of YB-1 proteins, 25 μg of total cell lysates was probed in an immunoblot assay (bottom panel). YB-1 mutants were sorted by the ability to interfere with P3K-induced cell transformation. (B) mRNA binding assay. YB-1 proteins from total lysates as used in panel A were immunoprecipitated with anti-FLAG antibodies and incubated with [32P]UTP-labeled in vitro-transcribed YB-1 mRNA. Beads were washed with 500 mM NaCl to diminish nonspecific protein-mRNA interactions. Bound mRNAs were separated on a 4% PAGE with 8 M urea. As a control, 105 cpm of the mere riboprobe (R) was used. Asterisks indicate mRNA breakdown products. (C) Quantification of the results shown in panel B.
YB-1 nonspecifically binds to mRNAs, preferring sequences with a high G content (84). Masking mRNAs could be another possible way by which YB-1 inhibits translation. Therefore, we investigated mRNA-binding activities of various YB-1 mutants with in vitro-transcribed YB-1 mRNA as an arbitrary mRNA. YB-1 proteins were immunoprecipitated with FLAG-specific antibodies and incubated with [32P]UTP-labeled mRNA. Beads were washed with high salt to break nonspecific protein-RNA interactions. Bound mRNAs were quantified and visualized by PAGE. A majority of YB-1 mutants was able to bind to the mRNA, including mutants that failed to interfere with P3K-induced transformation (Fig. 5B and C). This result is similar to the cap-binding assay, with the exception that cap binding requires additional C-terminal sequences whereas mRNA binding solely depends on a functional CSD.
Oligomerization of YB-1 is required but not sufficient to block P3K-mediated transformation.
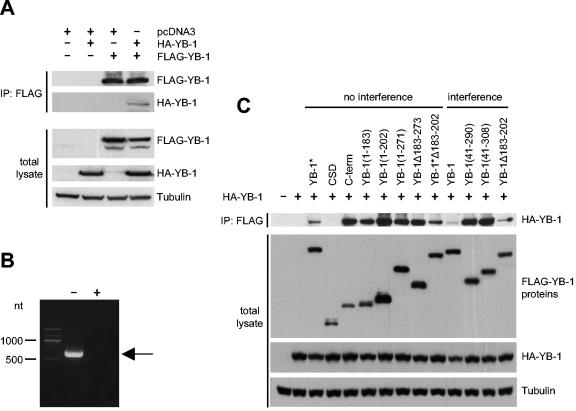
Cap binding or mRNA binding are not sufficient for YB-1 function as shown above. However, it still possible that mRNA binding in concert with homodimerization is required to prevent transformation. YB-1 is able to dimerize, forming homodimers and oligomers (25, 35). The domain necessary for oligomerization has been mapped to the C-terminal half of the protein (35). We confirmed homodimerization by coimmunoprecipitation of full-length YB-1 with full-length YB-1 by using FLAG-tagged and hemagglutinin (HA)-tagged proteins (Fig. 6A). Before precipitation, protein lysates were treated with RNase A to ensure complete degradation of mRNAs (Fig. 6B). To test the possibility that oligomerization of YB-1 bound to RNA causes interference with transformation, we performed coimmunoprecipitation experiments with full-length HA-YB-1 and various YB-1 mutants containing the FLAG tag. The capacity to homodimerize was compared to the ability to interfere with P3K-induced transformation. As shown in Fig. 6C, a great number of YB-1 mutants, including loss-of-function mutants, were able to form homodimers. The domain facilitating homodimerization can be mapped to residues 137 to 183, a domain C-terminally adjacent to the CSD. The data suggest that masking the mRNA as a homodimeric protein complex is not sufficient for the antioncogenic activity of YB-1.
FIG. 6.
Oligomerization of YB-1 mutants. (A) Coimmunoprecipitation of full-length YB-1 with full-length YB-1. FLAG-YB-1 and HA-YB-1 were transiently expressed in HEK293 cells. FLAG-YB-1 and HA-YB-1 proteins were singly expressed to show the specificity of the assay. Empty pcDNA3 vector was added when necessary to reach equal amounts of transfected DNA. A 500-μg aliquot of total protein was preincubated with 1 mg of RNase A/ml at 37°C for 2 h. Lysates were precipitated on a FLAG-conjugated resin and probed for coimmunoprecipitated HA-YB-1. Western analysis using 25 μg of total protein was performed to show expression of YB-1 proteins (bottom panels). Tubulin was used as a loading control. (B) Degradation of mRNA by RNase A treatment. A 500-μg aliquot of total protein of HEK293 cells expressing FLAG-YB-1 was treated with (+) or without (−) RNase A as described above. Total RNA was isolated, reverse transcribed, and amplified using YB-1-specific primers. The arrow indicates a 606-nt PCR product. (C) Coimmunoprecipitation of YB-1 mutants. HA-YB-1 and various YB-1 mutants containing an N-terminal FLAG tag were coexpressed in HEK293 cells and subjected to coimmunoprecipitation as described in the legend for panel A. Five micrograms (FLAG) and 10 μg (HA and tubulin) of total lysates were used in an immunoblot assay to show expression of YB-1 proteins. YB-1 mutants were sorted by the ability to interfere with P3K-induced transformation. CSD indicates the AP-CSD fragment.
Interference with transformation is tightly correlated with inhibition of translation.
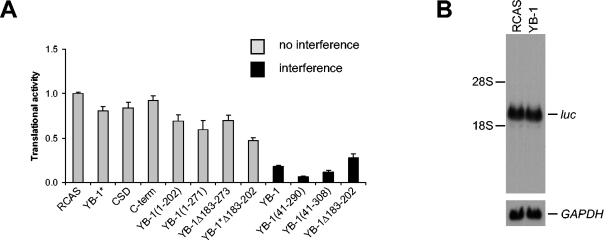
YB-1-expressing cells resistant to transformation by P3K and Akt show a dramatic decrease in translational activity. Cap-dependent and cap-independent translation are affected equally by YB-1 (5). Protein synthesis in cells expressing the YB-1 mutants was therefore measured and compared to the sensitivity to transformation. Cap-dependent translation was measured with the reporter construct pc-luc, encoding the luciferase gene that is driven by a cytomegalovirus promoter and translated in a cap-dependent manner. Transient transfection of pc-luc into CEF that expressed various YB-1 mutants was followed by a luciferase assay as a readout for translational activity (Fig. 7A). The data revealed that inhibition of translation is tightly correlated with the ability to block P3K-induced transformation. All mutants that retained the ability to interfere with transformation also greatly reduced translation, similar to wild-type YB-1. Mutants unable to affect transformation barely reduced translation. The reduction in translational activity was independent of transcriptional activity, as RCAS-YB-1- and RCAS-infected cells showed equal levels of luciferase mRNAs (Fig. 7B). The data suggest that the two regions critical for antioncogenic activity, the RNA-binding domain and the C-terminal domain, function in concert to interfere with protein synthesis and induce resistance to transformation.
FIG. 7.
Inhibition of translation by YB-1 mutants. (A) CEF expressing YB-1 proteins as indicated were transiently transfected with pc-luc encoding the luciferase gene. Five-microgram aliquots of protein were subjected to luciferase assays. The luciferase readout was normalized to that of cells transfected with the empty vector RCAS (1.00). YB-1 proteins able to interfere with P3K-induced transformation are indicated by black bars; data for YB-1 proteins unable to block transformation are shown in grey. The standard deviations of three independent experiments are shown in the graph. CSD indicates the AP-CSD fragment. (B) Analysis of luciferase mRNA levels of CEF stably infected with RCAS-YB-1 or RCAS only. Cells were treated as for panel A and harvested for total RNA isolation. Luciferase mRNAs were detected by Northern blotting. GAPDH was used as a loading control.
DISCUSSION
YB-1 functions as an mRNA-binding protein regulating translation and as a DNA-binding protein controlling gene expression. In addition, YB-1 is a nucleo-cytoplasmic shuttling protein that may be involved in DNA repair and RNA splicing (38, 59, 78). Heterozygous knockout of YB-1 results in major defects of the cell cycle in DT40 cells (75). Overexpression of YB-1 induces a specific resistance to P3K- and Akt-mediated oncogenic cell transformation (5). The mutational analysis of YB-1 presented here seeks to correlate the antioncogenic activity of YB-1 with specific domains and functions of the protein.
In an analysis of the subcellular localization of YB-1 mutants, we found that a cytoplasmic localization of YB-1 is required but not sufficient for interference with transformation. A YB-1 mutant lacking the NLS retained its capacity to block translation and P3K-induced transformation, suggesting that a nuclear function and—by implication—transcriptional regulation are not required. We mapped the domains that control subcellular localization: the NLS encompasses amino acids 183 to 202, and the CRS resides between residues 264 to 290. The sites of these domains are similar to those that have been determined previously (38, 73). We found that the CRS is necessary for cytoplasmic localization but is dispensable for mRNA binding. Loss of either RNA binding or the CRS results in a nuclear localization (5, 73). Under these conditions, the NLS prevails and directs YB-1 into the nucleus. YB-1 has a noncanonical NLS, since it functions independently of its basic residues. Inhibition of the CRM1-mediated nuclear export by leptomycin B fails to block cytoplasmic localization of YB-1, suggesting that nuclear export is facilitated through mRNA export. The data favor a model in which YB-1 interferes with oncogenic transformation by functioning as a cytoplasmic translational regulator and not as a nuclear protein regulating transcription. Nuclear export and cytoplasmic localization are accomplished by association of YB-1 with mRNA, whereas nuclear localization is mediated by the NLS in conjunction with a loss of the CRS. This is demonstrated by site-specific cleavage of YB-1 in response to thrombin, which leads to a loss of the CRS and nuclear localization (73). Nuclear localization can also be achieved by other stimuli, such as UV treatment, phosphorylation, or association with p53 (41, 78, 85). Whether the CRS stabilizes YB-1 in the cytoplasm by interacting with mRNA or another component of mRNPs remains to be determined.
YB-1 is frequently overexpressed in ovarian, breast, and lung cancers (27, 36, 66, 81). In many of these cancers, YB-1 shows increased nuclear localization. YB-1 positively controls the expression of the matrix metalloproteinase 2 and the multidrug resistance gene 1, both of which are important for tumor progression (48, 55). Nuclear localization of YB-1 correlates with tumor progression and a poor prognosis in tumor patients (27, 66). The prooncogenic activity of YB-1 may be explained by elevated levels of YB-1 in the nucleus. In this context, YB-1 may act as a transcriptional regulator, rather than as a cytoplasmic mRNA-binding protein.
As an mRNA-binding protein, YB-1 controls translation in a dose-dependent manner (49). High concentrations are inhibitory and block cap-dependent and cap-independent translation (5). Masking the mRNA and, thereby, making the mRNA inaccessible to the translational machinery has been a plausible explanation for this inhibitory activity (7, 20, 61, 68). YB-1 would bind mRNA as a monomer or as an oligomer, condensing mRNAs intramolecularly and intermolecularly. Our data do not support this model but rather suggest that the inhibition of translation involves a more specific mechanism. mRNA binding and the ability to oligomerize are not sufficient for inhibition of translation. Inhibition requires a C-terminal domain that is distinct from both the CRS and the domain that facilitates dimerization. Nekrasov et al. have shown that YB-1 interferes with translation at the initiation stage of translation. YB-1 binds to the cap region of mRNAs, but there is no apparent competition between YB-1 and eIF-4E in a 7-methyl cap-binding assay (5). Several deletion mutants of YB-1 are able to bind to the cap but fail to inhibit translation and also fail to interfere with P3K-mediated transformation. Cooperative binding of YB-1 and eIF-4E to the cap may occur, since YB-1 binds to the sugar component of mRNAs, leaving nucleotide bases free to interact with other mRNA-binding proteins (57). Initiation of cap-dependent translation is negatively controlled by 4E-BPs and eIF-2α kinases (26). However, 4E-BP1 levels bound to the cap are not increased in the presence of YB-1. Likewise, phosphorylation of eIF-2α at Ser51 remains unaffected by YB-1 (unpublished data). High levels of YB-1 impair the formation of the 48S preinitiation complex (57). Therefore, YB-1 may not prevent the assembly of the translational initiation complex 4F but may interfere at a later stage of initiation. It is conceivable that YB-1 interferes with mRNA scanning, assembly of the 80S ribosome, or recognition of the start codon. We speculate that the CSD facilitates the association of YB-1 and mRNAs, whereas the C-terminal domain has a yet-to-be-defined _trans_-acting function.
Our data establish a fundamental role of translation during P3K-induced oncogenicity. YB-1 inhibits translation and as a consequence prevents P3K- and Akt-induced transformation. Cycloheximide, a general inhibitor of translation, fails to block transformation by P3K or Akt, suggesting that YB-1 differentially affects certain mRNAs that are essential for the transformed phenotype (unpublished data). In this view, the susceptibility to inhibition by YB-1 would be encoded by cis- or _trans_-acting elements of the particular mRNAs. A recent study described mRNAs that are differentially regulated during Ras or Akt activation (60). While total mRNA levels remained largely unchanged, a specific subset of mRNAs associated with polysomes and entered translation. Many of these mRNAs encode proteins that regulate growth, transcription, cell-to-cell interactions, and morphology. The challenge is now to identify the mRNAs that are crucial for P3K-induced oncogenesis and how they are regulated by YB-1.
Acknowledgments
We thank G. Denning, J. Shi, and L. Zhao for helpful discussions and H. Filoteo-Velasco for excellent technical assistance.
This work was supported by National Institutes of Health research grant CA78230. A. Bader is the recipient of Erwin-Schrödinger fellowships J2101 and J2278-B04 from the Austrian Science Foundation (FWF).
Footnotes
†
This is manuscript no. 16934-MEM of The Scripps Research Institute.
REFERENCES
- 1.Aoki, M., O. Batista, A. Bellacosa, P. Tsichlis, and P. K. Vogt. 1998. The akt kinase: molecular determinants of oncogenicity. Proc. Natl. Acad. Sci. USA 95**:**14950-14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, M., E. Blazek, and P. K. Vogt. 2001. A role of the kinase mTOR in cellular transformation induced by the oncoproteins P3k and Akt. Proc. Natl. Acad. Sci. USA 98**:**136-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki, M., C. Schetter, M. Himly, O. Batista, H. W. Chang, and P. K. Vogt. 2000. The catalytic subunit of phosphoinositide 3-kinase: requirements for oncogenicity. J. Biol. Chem. 275**:**6267-6275. [DOI] [PubMed] [Google Scholar]
- 4.Aoki, M., V. Sobek, D. J. Maslyar, A. Hecht, and P. K. Vogt. 2002. Oncogenic transformation by beta-catenin: deletion analysis and characterization of selected target genes. Oncogene 21**:**6983-6991. [DOI] [PubMed] [Google Scholar]
- 5.Bader, A. G., K. A. Felts, N. Jiang, H. W. Chang, and P. K. Vogt. 2003. Y box-binding protein 1 induces resistance to oncogenic transformation by the phosphatidylinositol 3-kinase pathway. Proc. Natl. Acad. Sci. USA 100**:**12384-12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bader, A. G., M. Hartl, and K. Bister. 2000. Conditional cell transformation by doxycycline-controlled expression of the ASV17 v-jun allele. Virology 270**:**98-110. [DOI] [PubMed] [Google Scholar]
- 7.Bader, A. G., and P. K. Vogt. 2004. An essential role for protein synthesis in oncogenic cellular transformation. Oncogene 23**:**3145-3150. [DOI] [PubMed] [Google Scholar]
- 8.Barash, I. 1999. Prolactin and insulin synergize to regulate the translation modulator PHAS-I via mitogen-activated protein kinase-independent but wortmannin- and rapamycin-sensitive pathway. Mol. Cell. Endocrinol. 155**:**37-49. [DOI] [PubMed] [Google Scholar]
- 9.Barrieux, A., H. A. Ingraham, S. Nystul, and M. G. Rosenfeld. 1976. Characterization of the association of specific proteins with messenger ribonucleic acid. Biochemistry 15**:**3523-3528. [DOI] [PubMed] [Google Scholar]
- 10.Bellacosa, A., D. de Feo, A. K. Godwin, D. W. Bell, J. Q. Cheng, D. A. Altomare, M. Wan, L. Dubeau, G. Scambia, V. Masciullo, et al. 1995. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int. J. Cancer 64**:**280-285. [DOI] [PubMed] [Google Scholar]
- 11.Bellacosa, A., J. R. Testa, S. P. Staal, and P. N. Tsichlis. 1991. A retroviral oncogene, akt, encoding a serine-threonine kinase containing an SH2-like region. Science 254**:**274-277. [DOI] [PubMed] [Google Scholar]
- 12.Blobel, G. 1972. Protein tightly bound to globin mRNA. Biochem. Biophys. Res. Commun. 47**:**88-95. [DOI] [PubMed] [Google Scholar]
- 13.Cantley, L. C., and B. G. Neel. 1999. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc. Natl. Acad. Sci. USA 96**:**4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, H. W., M. Aoki, D. Fruman, K. R. Auger, A. Bellacosa, P. N. Tsichlis, L. C. Cantley, T. M. Roberts, and P. K. Vogt. 1997. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science 276**:**1848-1850. [DOI] [PubMed] [Google Scholar]
- 15.Cheng, J. Q., A. K. Godwin, A. Bellacosa, T. Taguchi, T. F. Franke, T. C. Hamilton, P. N. Tsichlis, and J. R. Testa. 1992. AKT2, a putative oncogene encoding a member of a subfamily of protein-serine/threonine kinases, is amplified in human ovarian carcinomas. Proc. Natl. Acad. Sci. USA 89**:**9267-9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corradetti, M. N., K. Inoki, N. Bardeesy, R. A. DePinho, and K. L. Guan. 2004. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 18**:**1533-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devroede, G., B. Lemieux, S. Masse, J. Lamarche, and P. S. Herman. 1988. Colonic hamartomas in tuberous sclerosis. Gastroenterology 94**:**182-188. [DOI] [PubMed] [Google Scholar]
- 18.Dingwall, C., and R. A. Laskey. 1991. Nuclear targeting sequences—a consensus? Trends Biochem. Sci. 16**:**478-481. [DOI] [PubMed] [Google Scholar]
- 19.Evdokimova, V., P. Ruzanov, H. Imataka, B. Raught, Y. Svitkin, L. P. Ovchinnikov, and N. Sonenberg. 2001. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 20**:**5491-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evdokimova, V. M., and L. P. Ovchinnikov. 1999. Translational regulation by Y-box transcription factor: involvement of the major mRNA-associated protein, p50. Int. J. Biochem. Cell Biol. 31**:**139-149. [DOI] [PubMed] [Google Scholar]
- 21.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90**:**1051-1060. [DOI] [PubMed] [Google Scholar]
- 22.Freedman, D. A., and A. J. Levine. 1998. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 18**:**7288-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukada, T., and N. K. Tonks. 2003. Identification of YB-1 as a regulator of PTP1B expression: implications for regulation of insulin and cytokine signaling. EMBO J. 22**:**479-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukuchi-Shimogori, T., I. Ishii, K. Kashiwagi, H. Mashiba, H. Ekimoto, and K. Igarashi. 1997. Malignant transformation by overproduction of translation initiation factor eIF4G. Cancer Res. 57**:**5041-5044. [PubMed] [Google Scholar]
- 25.Gaudreault, I., D. Guay, and M. Lebel. 2004. YB-1 promotes strand separation in vitro of duplex DNA containing either mispaired bases or cisplatin modifications, exhibits endonucleolytic activities and binds several DNA repair proteins. Nucleic Acids Res. 32**:**316-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gebauer, F., and M. W. Hentze. 2004. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 5**:**827-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gessner, C., C. Woischwill, A. Schumacher, U. Liebers, H. Kuhn, P. Stiehl, K. Jurchott, H. D. Royer, C. Witt, and G. Wolff. 2004. Nuclear YB-1 expression as a negative prognostic marker in non-small cell lung cancer. Eur. Respir. J. 23**:**14-19. [DOI] [PubMed] [Google Scholar]
- 28.Graff, J. R., and S. G. Zimmer. 2003. Translational control and metastatic progression: enhanced activity of the mRNA cap-binding protein eIF-4E selectively enhances translation of metastasis-related mRNAs. Clin. Exp. Metastasis 20**:**265-273. [DOI] [PubMed] [Google Scholar]
- 29.Hay, N., and N. Sonenberg. 2004. Upstream and downstream of mTOR. Genes Dev. 18**:**1926-1945. [DOI] [PubMed] [Google Scholar]
- 30.Hemminki, A., D. Markie, I. Tomlinson, E. Avizienyte, S. Roth, A. Loukola, G. Bignell, W. Warren, M. Aminoff, P. Hoglund, H. Jarvinen, P. Kristo, K. Pelin, M. Ridanpaa, R. Salovaara, T. Toro, W. Bodmer, S. Olschwang, A. S. Olsen, M. R. Stratton, A. de la Chapelle, and L. A. Aaltonen. 1998. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature 391**:**184-187. [DOI] [PubMed] [Google Scholar]
- 31.Huang, T. T., N. Kudo, M. Yoshida, and S. Miyamoto. 2000. A nuclear export signal in the N-terminal regulatory domain of IκBα controls cytoplasmic localization of inactive NF-κB/IκBα complexes. Proc. Natl. Acad. Sci. USA 97**:**1014-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes, S. H., J. J. Greenhouse, C. J. Petropoulos, and P. Sutrave. 1987. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 61**:**3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoki, K., Y. Li, T. Xu, and K. L. Guan. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17**:**1829-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoki, K., Y. Li, T. Zhu, J. Wu, and K. L. Guan. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4**:**648-657. [DOI] [PubMed] [Google Scholar]
- 35.Izumi, H., T. Imamura, G. Nagatani, T. Ise, T. Murakami, H. Uramoto, T. Torigoe, H. Ishiguchi, Y. Yoshida, M. Nomoto, T. Okamoto, T. Uchiumi, M. Kuwano, K. Funa, and K. Kohno. 2001. Y box-binding protein-1 binds preferentially to single-stranded nucleic acids and exhibits 3′→5′ exonuclease activity. Nucleic Acids Res. 29**:**1200-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janz, M., N. Harbeck, P. Dettmar, U. Berger, A. Schmidt, K. Jurchott, M. Schmitt, and H. D. Royer. 2002. Y-box factor YB-1 predicts drug resistance and patient outcome in breast cancer independent of clinically relevant tumor biologic factors HER2, uPA and PAI-1. Int. J. Cancer 97**:**278-282. [DOI] [PubMed] [Google Scholar]
- 37.Jefferies, H. B., S. Fumagalli, P. B. Dennis, C. Reinhard, R. B. Pearson, and G. Thomas. 1997. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 16**:**3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jurchott, K., S. Bergmann, U. Stein, W. Walther, M. Janz, I. Manni, G. Piaggio, E. Fietze, M. Dietel, and H. D. Royer. 2003. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J. Biol. Chem. 278**:**27988-27996. [DOI] [PubMed] [Google Scholar]
- 39.Kalderon, D., W. D. Richardson, A. F. Markham, and A. E. Smith. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311**:**33-38. [DOI] [PubMed] [Google Scholar]
- 40.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39**:**499-509. [DOI] [PubMed] [Google Scholar]
- 41.Koike, K., T. Uchiumi, T. Ohga, S. Toh, M. Wada, K. Kohno, and M. Kuwano. 1997. Nuclear translocation of the Y-box binding protein by ultraviolet irradiation. FEBS Lett. 417**:**390-394. [DOI] [PubMed] [Google Scholar]
- 42.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell. Res. 242**:**540-547. [DOI] [PubMed] [Google Scholar]
- 43.Lazaris-Karatzas, A., K. S. Montine, and N. Sonenberg. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345**:**544-547. [DOI] [PubMed] [Google Scholar]
- 44.Lin, T. A., X. Kong, T. A. Haystead, A. Pause, G. Belsham, N. Sonenberg, and J. C. Lawrence, Jr. 1994. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 266**:**653-656. [DOI] [PubMed] [Google Scholar]
- 45.Lynch, M., C. Fitzgerald, K. A. Johnston, S. Wang, and E. V. Schmidt. 2004. Activated eIF4E-binding protein slows G1 progression and blocks transformation by c-myc without inhibiting cell growth. J. Biol. Chem. 279**:**3327-3339. [DOI] [PubMed] [Google Scholar]
- 46.Ma, Y. Y., S. J. Wei, Y. C. Lin, J. C. Lung, T. C. Chang, J. Whang-Peng, J. M. Liu, D. M. Yang, W. K. Yang, and C. Y. Shen. 2000. PIK3CA as an oncogene in cervical cancer. Oncogene 19**:**2739-2744. [DOI] [PubMed] [Google Scholar]
- 47.Mende, I., S. Malstrom, P. N. Tsichlis, P. K. Vogt, and M. Aoki. 2001. Oncogenic transformation induced by membrane-targeted Akt2 and Akt3. Oncogene 20**:**4419-4423. [DOI] [PubMed] [Google Scholar]
- 48.Mertens, P. R., S. Harendza, A. S. Pollock, and D. H. Lovett. 1997. Glomerular mesangial cell-specific transactivation of matrix metalloproteinase 2 transcription is mediated by YB-1. J. Biol. Chem. 272**:**22905-22912. [DOI] [PubMed] [Google Scholar]
- 49.Minich, W. B., and L. P. Ovchinnikov. 1992. Role of cytoplasmic mRNP proteins in translation. Biochimie 74**:**477-483. [DOI] [PubMed] [Google Scholar]
- 50.Miwa, W., J. Yasuda, Y. Murakami, K. Yashima, K. Sugano, T. Sekine, A. Kono, S. Egawa, K. Yamaguchi, Y. Hayashizaki, and T. Sekiya. 1996. Isolation of DNA sequences amplified at chromosome 19q13.1-q13.2 including the AKT2 locus in human pancreatic cancer. Biochem. Biophys. Res. Commun. 225**:**968-974. [DOI] [PubMed] [Google Scholar]
- 51.Montagne, J., T. Radimerski, and G. Thomas. 2001. Insulin signaling: lessons from the Drosophila tuberous sclerosis complex, a tumor suppressor. Sci. STKE 2001**:**PE36. [DOI] [PubMed] [Google Scholar]
- 52.Nekrasov, M. P., M. P. Ivshina, K. G. Chernov, E. A. Kovrigina, V. M. Evdokimova, A. A. Thomas, J. W. Hershey, and L. P. Ovchinnikov. 2003. The mRNA-binding protein YB-1 (p50) prevents association of the eukaryotic initiation factor eIF4G with mRNA and inhibits protein synthesis at the initiation stage. J. Biol. Chem. 278**:**13936-13943. [DOI] [PubMed] [Google Scholar]
- 53.Neshat, M. S., I. K. Mellinghoff, C. Tran, B. Stiles, G. Thomas, R. Petersen, P. Frost, J. J. Gibbons, H. Wu, and C. L. Sawyers. 2001. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc. Natl. Acad. Sci. USA 98**:**10314-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norman, J. T., G. E. Lindahl, K. Shakib, A. En-Nia, E. Yilmaz, and P. R. Mertens. 2001. The Y-box binding protein YB-1 suppresses collagen alpha 1(I) gene transcription via an evolutionarily conserved regulatory element in the proximal promoter. J. Biol. Chem. 276**:**29880-29890. [DOI] [PubMed] [Google Scholar]
- 55.Ohga, T., T. Uchiumi, Y. Makino, K. Koike, M. Wada, M. Kuwano, and K. Kohno. 1998. Direct involvement of the Y-box binding protein YB-1 in genotoxic stress-induced activation of the human multidrug resistance 1 gene. J. Biol. Chem. 273**:**5997-6000. [DOI] [PubMed] [Google Scholar]
- 56.Pandolfi, P. P. E. 2004. Translation and cancer. Oncogene 23**:**3134-3248. [DOI] [PubMed] [Google Scholar]
- 57.Pisarev, A. V., M. A. Skabkin, A. A. Thomas, W. C. Merrick, L. P. Ovchinnikov, and I. N. Shatsky. 2002. Positive and negative effects of the major mammalian messenger ribonucleoprotein p50 on binding of 40S ribosomal subunits to the initiation codon of beta-globin mRNA. J. Biol. Chem. 277**:**15445-15451. [DOI] [PubMed] [Google Scholar]
- 58.Podsypanina, K., R. T. Lee, C. Politis, I. Hennessy, A. Crane, J. Puc, M. Neshat, H. Wang, L. Yang, J. Gibbons, P. Frost, V. Dreisbach, J. Blenis, Z. Gaciong, P. Fisher, C. Sawyers, L. Hedrick-Ellenson, and R. Parsons. 2001. An inhibitor of mTOR reduces neoplasia and normalizes p70/S6 kinase activity in Pten+/− mice. Proc. Natl. Acad. Sci. USA 98**:**10320-10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raffetseder, U., B. Frye, T. Rauen, K. Jurchott, H. D. Royer, P. L. Jansen, and P. R. Mertens. 2003. Splicing factor SRp30c interaction with Y-box protein-1 confers nuclear YB-1 shuttling and alternative splice site selection. J. Biol. Chem. 278**:**18241-19248. [DOI] [PubMed] [Google Scholar]
- 60.Rajasekhar, V. K., A. Viale, N. D. Socci, M. Wiedmann, X. Hu, and E. C. Holland. 2003. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell 12**:**889-901. [DOI] [PubMed] [Google Scholar]
- 61.Ranjan, M., S. R. Tafuri, and A. P. Wolffe. 1993. Masking mRNA from translation in somatic cells. Genes Dev. 7**:**1725-1736. [DOI] [PubMed] [Google Scholar]
- 62.Ruggeri, B. A., L. Huang, M. Wood, J. Q. Cheng, and J. R. Testa. 1998. Amplification and overexpression of the AKT2 oncogene in a subset of human pancreatic ductal adenocarcinomas. Mol. Carcinog. 21**:**81-86. [PubMed] [Google Scholar]
- 63.Ruggero, D., L. Montanaro, L. Ma, W. Xu, P. Londei, C. Cordon-Cardo, and P. P. Pandolfi. 2004. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 10**:**484-486. [DOI] [PubMed] [Google Scholar]
- 64.Samuels, Y., Z. Wang, A. Bardelli, N. Silliman, J. Ptak, S. Szabo, H. Yan, A. Gazdar, S. M. Powell, G. J. Riggins, J. K. Willson, S. Markowitz, K. W. Kinzler, B. Vogelstein, and V. E. Velculescu. 2004. High frequency of mutations of the PIK3CA gene in human cancers. Science 304**:**554. [DOI] [PubMed] [Google Scholar]
- 65.Shaw, R. J., N. Bardeesy, B. D. Manning, L. Lopez, M. Kosmatka, R. A. DePinho, and L. C. Cantley. 2004. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6**:**91-99. [DOI] [PubMed] [Google Scholar]
- 66.Shibahara, K., K. Sugio, T. Osaki, T. Uchiumi, Y. Maehara, K. Kohno, K. Yasumoto, K. Sugimachi, and M. Kuwano. 2001. Nuclear expression of the Y-box binding protein, YB-1, as a novel marker of disease progression in non-small cell lung cancer. Clin. Cancer Res. 7**:**3151-3155. [PubMed] [Google Scholar]
- 67.Simpson, L., and R. Parsons. 2001. PTEN: life as a tumor suppressor. Exp. Cell. Res. 264**:**29-41. [DOI] [PubMed] [Google Scholar]
- 68.Sommerville, J., and M. Ladomery. 1996. Masking of mRNA by Y-box proteins. FASEB J. 10**:**435-443. [DOI] [PubMed] [Google Scholar]
- 69.Sonderegger, C. K., M. Narisawa-Saito, and P. K. Vogt. 2003. The C-terminal region of cellular Qin oligomerizes: correlation with oncogenic transformation and transcriptional repression. Oncogene 22**:**1908-1915. [DOI] [PubMed] [Google Scholar]
- 70.Staal, S. P., and J. W. Hartley. 1988. Thymic lymphoma induction by the AKT8 murine retrovirus. J. Exp. Med. 167**:**1259-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Staal, S. P., J. W. Hartley, and W. P. Rowe. 1977. Isolation of transforming murine leukemia viruses from mice with a high incidence of spontaneous lymphoma. Proc. Natl. Acad. Sci. USA 74**:**3065-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90**:**1041-1050. [DOI] [PubMed] [Google Scholar]
- 73.Stenina, O. I., K. M. Shaneyfelt, and P. E. DiCorleto. 2001. Thrombin induces the release of the Y-box protein dbpB from mRNA: a mechanism of transcriptional activation. Proc. Natl. Acad. Sci. USA 98**:**7277-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stolovich, M., H. Tang, E. Hornstein, G. Levy, R. Cohen, S. S. Bae, M. J. Birnbaum, and O. Meyuhas. 2002. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol. Cell. Biol. 22**:**8101-8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swamynathan, S. K., B. R. Varma, K. T. Weber, and R. V. Guntaka. 2002. Targeted disruption of one allele of the Y-box protein gene, Chk-YB-1b, in DT40 cells results in major defects in cell cycle. Biochem. Biophys. Res. Commun. 296**:**451-457. [DOI] [PubMed] [Google Scholar]
- 76.Thomas, G., D. M. Sabatini, and N. M. Hall (ed.). 2004. TOR—target of rapamycin. Current topics in microbiology and immunology, vol. 279. Springer, Berlin, Germany.
- 77.Ting, J. P., A. Painter, N. J. Zeleznik-Le, G. MacDonald, T. M. Moore, A. Brown, and B. D. Schwartz. 1994. YB-1 DNA-binding protein represses interferon gamma activation of class II major histocompatibility complex genes. J. Exp. Med. 179**:**1605-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Uramoto, H., H. Izumi, T. Ise, M. Tada, T. Uchiumi, M. Kuwano, K. Yasumoto, K. Funa, and K. Kohno. 2002. p73 Interacts with c-Myc to regulate Y-box-binding protein-1 expression. J. Biol. Chem. 277**:**31694-31702. [DOI] [PubMed] [Google Scholar]
- 79.Vivanco, I., and C. L. Sawyers. 2002. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat. Rev. Cancer 2**:**489-501. [DOI] [PubMed] [Google Scholar]
- 80.Vogt, P. K. 1969. Focus assay of Rous sarcoma virus, p. 198-211. In K. Habel and N. P. Salzman (ed.), Fundamental techniques in virology. Academic Press, New York, N.Y.
- 81.Yahata, H., H. Kobayashi, T. Kamura, S. Amada, T. Hirakawa, K. Kohno, M. Kuwano, and H. Nakano. 2002. Increased nuclear localization of transcription factor YB-1 in acquired cisplatin-resistant ovarian cancer. J. Cancer Res. Clin. Oncol. 128**:**621-626. [DOI] [PubMed] [Google Scholar]
- 82.Yang, H. S., A. P. Jansen, A. A. Komar, X. Zheng, W. C. Merrick, S. Costes, S. J. Lockett, N. Sonenberg, and N. H. Colburn. 2003. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol. 23**:**26-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang, J., E. S. Bardes, J. D. Moore, J. Brennan, M. A. Powers, and S. Kornbluth. 1998. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 12**:**2131-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zasedateleva, O. A., A. S. Krylov, D. V. Prokopenko, M. A. Skabkin, L. P. Ovchinnikov, A. Kolchinsky, and A. D. Mirzabekov. 2002. Specificity of mammalian Y-box binding protein p50 in interaction with ss and ds DNA analyzed with generic oligonucleotide microchip. J. Mol. Biol. 324**:**73-87. [DOI] [PubMed] [Google Scholar]
- 85.Zhang, Y. F., C. Homer, S. J. Edwards, L. Hananeia, A. Lasham, J. Royds, P. Sheard, and A. W. Braithwaite. 2003. Nuclear localization of Y-box factor YB1 requires wild-type p53. Oncogene 22**:**2782-2794. [DOI] [PubMed] [Google Scholar]