Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling (original) (raw)
Abstract
Using the OP9-DL1 system to deliver temporally controlled Notch/Delta signaling, we show that pluripotent hematolymphoid progenitors undergo T-lineage specification and B-lineage inhibition in response to Notch signaling in a delayed and asynchronous way. Highly enriched progenitors from fetal liver require ≥3 d to begin B- or T-lineage differentiation. Clonal switch-culture analysis shows that progeny of some single cells can still generate both B- and T-lineage cells, after 1 wk of continuous delivery or deprivation of Notch/Delta signaling. Notch signaling induces T-cell genes and represses B-cell genes, but kinetics of activation of lineage-specific transcription factors are significantly delayed after induction of Notch target genes and can be temporally uncoupled from the Notch response. In the cells that initiate T-cell differentiation and gene expression most slowly in response to Notch/Delta signaling, Notch target genes are induced to the same level as in the cells that respond most rapidly. Early lineage-specific gene expression is also rapidly reversible in switch cultures. Thus, while necessary to induce and sustain T-cell development, Notch/Delta signaling is not sufficient for T-lineage specification and commitment, but instead can be permissive for the maintenance and proliferation of uncommitted progenitors that are omitted in binary-choice models.
Keywords: GATA-3, hematopoietic progenitor cells, lineage commitment, lymphocyte development, Pax-5, transcription factors
The effects of key regulatory molecules in hematopoietic lineage specification of multipotent progenitor cells are considered in terms of either instructive or selective mechanisms. In the first kind of mechanism, molecules such as exogenous cytokines or endogenous transcription factors instructively activate specific transcriptional programs to direct naïve progenitor cells toward a certain lineage (Martin et al. 1993; Nutt et al. 1999; Iwasaki et al. 2003; Xie et al. 2004). In the second kind of mechanism, growth factors support the differential survival or proliferation of particular cell lineages from a mixture of precursors that were specified differentially through some other, possibly stochastic, mechanism. These kinds of mechanistic options are weighted differently in postembryonic hematopoiesis than they would be in many well-characterized embryonic systems of differentiation, where overall timing and coordination of morphogenesis sets strict constraints. Hematopoietic precursors do retain substantial plasticity during differentiation, and selective signals from the microenvironment clearly amplify the effects of transcription-factor combinations to determine cell fate (Graf 2002; Miyamoto et al. 2002). Thus, possible examples of instructive specification in hematopoiesis require close examination to define the direct mechanisms through which cell lineage is chosen.
Within the hematopoietic system, the best-studied candidate for an instructive agent of lineage choice is the role of Notch1 signaling in T-lymphocyte lineage commitment. Notch proteins mediate cell-fate decisions in various developmental systems (Greenwald 1998; Artavanis-Tsakonas et al. 1999). They comprise a family of transmembrane receptors that control cellular differentiation in response to binding of ligands of the Delta or Jagged families. Ligand binding releases an activated intracellular Notch domain (Schroeter et al. 1998) that binds to CSL, thereby converting it from a repressor into an activator and resulting in the expression of downstream target genes (Tamura et al. 1995) such as HES1 and HES5 (Jarriault et al. 1995; de la Pompa et al. 1997; Davis and Turner 2001). Notch signaling appears to be important both to induce T-cell development and to block an alternative pathway leading to B-cell development. Loss of Notch1 function in lymphoid progenitors results in B lymphopoiesis within the thymus, at the expense of T-cell development (Wilson et al. 2001), suggesting that Notch1 has an important role in a T- versus B-cell lineage choice. Gain-of-function studies support this further, as overexpression of a constitutively active form of Notch induces ectopic T-cell development in the bone marrow and inhibits B-cell development that would normally take place there (Pui et al. 1999; De Smedt et al. 2002). In vivo, multiple Notch ligands are expressed by the epithelial cells of the thymus (Anderson et al. 2001), the privileged environment in which T-cell development is induced, and entry of the first embryonic hematopoietic precursors into the thymus is accompanied by induction of HES-1 (Harman et al. 2003; Radtke et al. 2004). T-cell development appears to depend on a Notch1-Delta interaction, since only Delta class ligands can efficiently support T-cell development (Schmitt and Zúñiga-Pflücker 2002; Hozumi et al. 2004; Lehar et al. 2004).
T-cell precursors do not undergo lineage commitment in vivo until after they enter the thymus, and only low-level T-lineage gene expression, if any, is seen in prethymic precursors (for review, see Shortman et al. 1998; Rothenberg 2000; Rothenberg and Dionne 2002; Schwarz and Bhandoola 2004; Rothenberg and Taghon 2005). Thus, an attractive model has been that pluripotent precursors only undertake the T-cell program when triggered by contact with Notch ligands upon entry into the thymic epithelium. Nevertheless, it has remained obscure how Notch/Delta signaling positively initiates the T-lineage differentiation program. No direct pathway has been established to show how a mediator of Notch signaling, such as RBPSuh/CSL, actually induces the complex gene expression changes that result in T-lineage specification. Although implicated in regulating some phases of pTα expression once the T-cell program is already under way (Deftos and Bevan 2000; Reizis and Leder 2002), Notch activation has not been shown to be linked to induction of crucial T-lineage transcription factors such as GATA-3 (Ting et al. 1996; Pai et al. 2003) and TCF (Verbeek et al. 1995; Staal and Clevers 2003) in a pluripotent precursor context. Also, while a binary-choice switching model is widely discussed, it has remained very controversial whether the onset of T-cell gene expression and the inhibition of B-cell development and other developmental alternatives occur through separate or linked mechanisms. While Notch signaling is the leading candidate for an instructive regulator of T-cell specification, there has been no evidence yet to show whether it is sufficient.
A recently established monolayer culture system (Schmitt and Zúñiga-Pflücker 2002) provides a new opportunity to expose the earliest stages of T-cell development for detailed tracking of the commitment process. In this system, the bone marrow stromal line OP9, which normally supports B-cell development in the presence of cytokines Flt3-L and IL-7, is converted to a T-cell inductive stroma by transfection with DL1 (OP9-DL1). Here, we have dissected the kinetics and mechanisms of early T- versus B-lineage differentiation through a detailed analysis of the development of pluripotent fetal liver (FL) progenitor cells on OP9-control and OP9-DL1 stromal cells (Schmitt and Zúñiga-Pflücker 2002), testing time course and the reversibility of gene expression and cell-surface changes down to subpopulation and clonal levels. Our data show that lymphoid progenitor cells actually retain a high degree of flexibility in responses to instructive Notch signaling, and that continuous Notch signaling is needed to maintain development along the T-cell pathway.
Results
Kinetics of early T- and B-cell development
Notch signaling has been shown to induce T-cell specification of multipotent hematopoietic progenitors. To define the exact events triggered and to compare them with those of B-cell differentiation, we cultured c-Kit+Lin- FL cells on OP9-control and OP9-DL1 stromal cells (Schmitt and Zúñiga-Pflücker 2002), and performed a kinetic analysis of changes in cell-surface markers diagnostic of specific hematopoietic differentiation pathways.
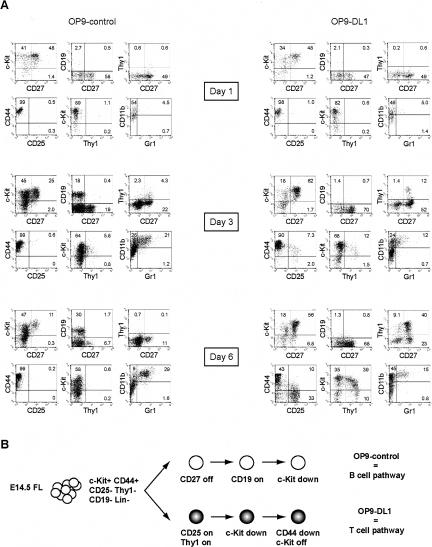
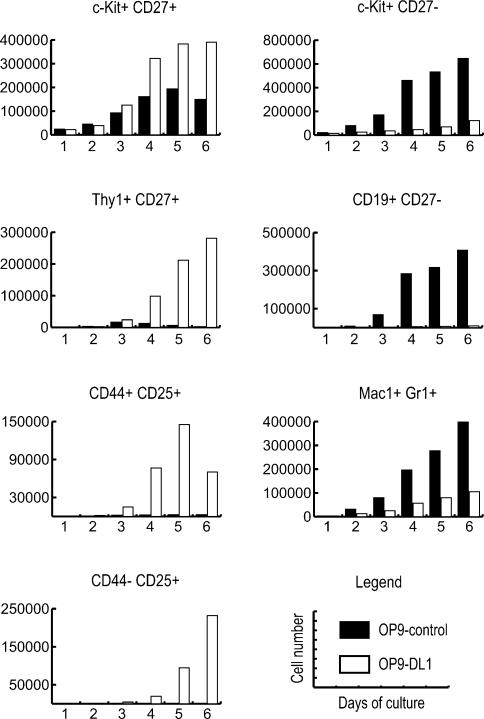
Both OP9-control and OP9-DL1 stromal cultures support the initial proliferative expansion of recognizable lymphoid progenitors. Beside c-Kit, one cell-surface molecule associated with lymphoid precursor cells is CD27 (Igarashi et al. 2002), which is also expressed on early intrathymic progenitors (Allman et al. 2003); CD27 is expressed in ∼30% of the c-Kit+Lin- FL cells initially (data not shown). As expected, in both OP9-control and OP9-DL1 cultures, already after 1 d, an increase in the frequency (Fig. 1A) and number (Fig. 2) of c-Kit+CD27+ cells was observed. In OP9-control cultures, although the absolute cell number continues to increase greatly (Fig. 2, black bars), the frequency of these progenitor cells reproducibly decreases after 3 d of culture, and by day 6, their absolute number also starts to decrease. This is mainly due to a decrease in the percentage of cells that were CD27+ from day 3 onward rather than a reduction in c-Kit+ cells (Fig. 1A). Over the same period, the first CD19+ cells appear, which develop exclusively in OP9-control cultures and do not express CD27, and initially retain c-Kit+. In contrast, in OP9-DL1 cultures, the number of c-Kit+CD27+ cells continues to increase up until day 6 (Fig. 2, white bars). Then, their frequency starts to decrease slightly (Fig. 1A), in this case due to loss of c-Kit expression. From day 3 onward, the first recognizable T-lineage cells are detected based on expression of Thy1 and CD25. Coinciding with the appearance of the first CD44+CD25+ DN2 and CD44-CD25+ DN3 thymocytes, the cells start to express higher levels of Thy1 and first down-regulate, then lose expression of c-Kit, while retaining CD27 (Fig. 1A). Thus, the differential regulation of CD27 relative to c-Kit is correlated with the emergence of cells expressing clear T- or B-lineage markers (Fig. 1B).
Figure 1.
Kinetics of early T- and B-lineage differentiation from multipotent precursors. (A) c-Kit+Lin- fetal liver cells were cocultured on OP9-control and OP9-DL1 stromal cells and analyzed by FACS at various time points as indicated. All dot plots shown are gated on CD45+ hematopoietic cells and representative for three independent experiments. Numbers in quadrants indicate the percentage of each corresponding population. (B) Schematic overview of the initial different phenotypical changes in developing B and T cells, derived from A.
Figure 2.
Absolute cell numbers of early B- and T-lineage developmental intermediates from multipotent precursors. The absolute number of cells for each indicated cell population at each time point were calculated based on the total number of cells in the culture and their percentage in the total cell population. Data shown is derived from cultures shown in Figure 1 and are representative of three independent experiments.
At least some of c-Kit+Lin- cells are capable of myeloid development, producing CD11b+Gr-1+ cells on both OP9-control and OP9-DL1 stroma, with a higher frequency in OP9-control cultures (Fig. 1A), in agreement with previous reports (Schroeder and Just 2000; Kumano et al. 2001; De Smedt et al. 2002). We never observed a significant fraction of NK cells in these cultures (data not shown).
Thus, the first cells with the phenotypes of committed B- and T-lineage cells develop after 3 d of culture, but coexist with undifferentiated precursor-type cells and cells undertaking other differentiation pathways for at least 6 d.
Delayed time course of early lymphoid gene expression
The OP9 culture system provides several stimuli to differentiation, including IL-7, Flt3-L, Jagged-class Notch ligands on both kinds of stroma, and Delta-like 1 only on OP9-DL1 (Schmitt and Zúñiga-Pflücker 2002). To determine how these triggers result in T- versus B-lineage specification of pluripotent precursor cells, we analyzed the kinetics of expression of known important hematopoietic regulatory genes (Rothenberg and Anderson 2002), using quantitative real-time RT-PCR.
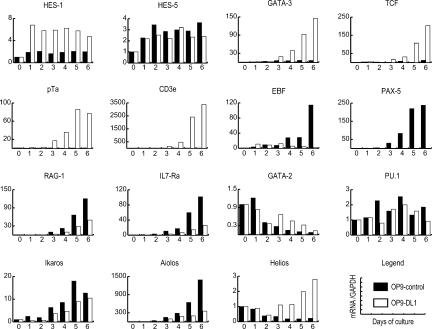
Responses to Notch signaling were evident within the first day. The Notch downstream target HES-1 was specifically up-regulated in FL cells cultured on OP9-DL1, and responded with rapid kinetics, reaching its maximum level at day 1 and remaining constant over 6 d (Fig. 3). Supplementary Figure 1 shows that Deltex1 and Nrarp, as well as HES-1, were rapidly and specifically induced by OP9-DL1 in the c-kit+ CD27+ hematopoietic cells. A different Notch response gene, HES-5, was modestly up-regulated under both conditions at days 1-2, possibly responding to the Jagged-class Notch ligands on both OP9 stromal lines (Fig. 3). pTα and CD3ε, T-lineage-specific differentiation genes, were induced only in the OP9-DL1 cultures. However, the increase was not seen until from day 3 onward (Fig. 3), coincident with CD25 mRNA expression (data not shown) and with the appearance of the first CD44+CD25+ DN2 thymocytes (Fig. 1A). The essential T-cell-specific transcription factors GATA-3 and TCF-1 were also up-regulated dramatically, and this induction was also seen only in OP9-DL1 cultures. Surprisingly, although both factors are required for T-cell development earlier than the expression of CD25, pTα, or CD3ε, the increase in GATA-3 and TCF-1 expression was also delayed until about day 3 of DL1 signaling, in parallel with the T-lineage differentiation genes.
Figure 3.
Time course of gene-expression changes during initial B- and T-lineage specification. Quantitative real-time RT-PCR gene expression analysis of c-Kit+Lin- cells, cocultured for 1-6 d on OP9-control and OP9-DL1 stromal cells as indicated. Samples shown are derived from cultures shown in Figure 1 and are representative of three independent experiments. Units of expression are given relative to levels in the purified starting population.
Conversely, the B-lineage commitment factor Pax-5 was only induced efficiently on OP9-control cultures, and also required ≥3 d to be induced, in accord with the appearance of the first cells expressing CD19 (Fig. 1A), a known Pax-5 target gene. Early B-cell Factor (EBF) was substantially up-regulated in OP9-control cultures and down-regulated in OP9-DL1 cultures from day 4 onward (Fig. 3), in accord with its B-lineage promoting function. Background EBF expression from the OP9 cells themselves masked low-level expression earlier than day 3 (data not shown). RAG-1 and IL7Rα are required for both B and T cells (Mombaerts et al. 1992; Peschon et al. 1994), and were up-regulated in FL cells in both cultures, with similar kinetics to the lineage-specific genes in each case (Fig. 3).
Other lymphoid developmental requirements were met by precursor-cell expression or induction coordinate with the rest of the B- and T-cell programs. The transcription factors E2A or HEB, required for B-cell and T-cell development, were already expressed in the precursors and maintained as differentiation began (data not shown). The Ikaros gene family (Rebollo and Schmitt 2003) showed lineage specificity in expression (Georgopoulos et al. 1994), but did not define any new transitional events between days 1 and 3. Ikaros, expressed in the initial precursors, was up-regulated in FL cells on either OP9-control or OP9-DL1 stromal layers, starting at about day 3 (Fig. 3) and slightly higher in OP9-control, consistent with the early requirement for Ikaros in B-cell development (Allman et al. 2003). Aiolos was also induced from day 3 onward, with higher expression in OP9-control as compared with OP9-DL1 cultures. Helios, thought to be T-lineage specific (Hahm et al. 1998), was expressed in the initial precursors and down-regulated on OP9-control stromal cells, but up-regulated on OP9-DL1 stromal cells in parallel with the T-lineage genes (Fig. 3).
Notch signaling in the OP9 culture system could promote T-cell specification not only by positive regulation of lymphocyte genes, but also by the repression of multipotent stem cell genes. Stem cell-specific regulatory gene expression in these cultures declined slowly, however, while expression of other factors needed for B- and T-cell development was relatively stable. GATA-2, crucial for hematopoietic progenitor cells (Tsai et al. 1994), was gradually down-regulated in both OP9-control and OP9-DL1 cultures, but persisted longer in the presence of extensive Notch/Delta signaling (Fig. 3), in agreement with a recent report (Kumano et al. 2001). Expression of PU.1, another transcription factor expressed in hematopoietic progenitors and required for B- and T-cell development (Scott et al. 1994; McKercher et al. 1996) continued in both B-cell and T-cell culture conditions (Fig. 3). Cell fractionation experiments showed that PU.1 continued to be expressed in c-Kit+ CD27+ lymphoid precursors as well as in myeloid CD11b+Gr-1+ cells (Supplementary Fig. 2). In contrast, induction of GATA-3 and TCF-1 after 4 d in OP9-DL1 cultures was essentially limited to the c-Kit+ CD27+ population, while Pax-5 induction in OP9-control cultures was most enriched in c-Kit+ CD27- CD11b- cells.
Thus, the critical regulatory changes associated with B- and T-lineage differentiation occur coordinately, but only after ≥3 d of culture on OP9-control or OP9-DL1, in parallel with the phenotypic changes shown in Figure 1A. However, these events are significantly delayed from the onset of direct Notch target gene expression.
Delayed commitment relative to initiation of differentiation
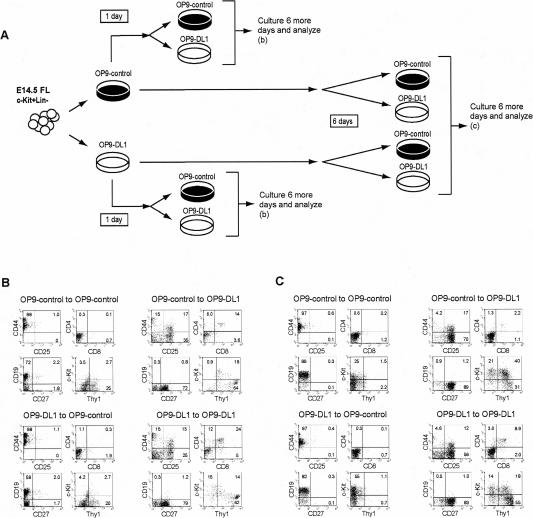
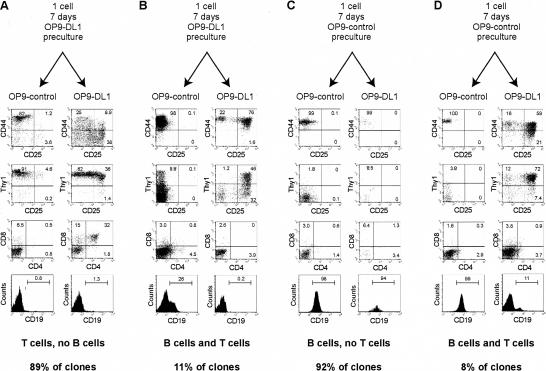
In both stromal cultures, cells resembling uncommitted progenitors were still present at day 6, based on the presence of c-Kit+CD27+ cells in OP9-control cultures and c-Kit+Thy1- cells in OP9-DL1 cultures (Fig. 1A). To investigate whether alternative developmental potentials could persist, we disrupted FL OP9-control and OP9-DL1 cocultures after 1 or 6 d and split each culture between fresh stromal layers of OP9-control and OP9-DL1 cells, thus either maintaining or changing the initial stromal monolayer (Fig. 4A). When transferred to OP9-control monolayers after 1 d of culture on either stromal layer, the progeny of the c-Kit+Lin- FL cells gave rise mainly to B cells as if they had been cultured on OP9-control cells all along (Fig. 4B). Similarly, when plated onto OP9-DL1 after 1 d of preculture on either stromal layer, these cells developed as T-lineage cells of Thy-1+ DN and DP phenotypes, and no B cells could develop (Fig. 4B). Thus a 1-d preculture was not sufficient to constrain the developmental potential of the c-Kit+Lin- FL precursors. Remarkably, though, the results were similar when cells were transferred at any subsequent day up to 6 d of initial culture (Fig. 4; data not shown). Transferring from OP9-control onto OP9-control or from OP9-DL1 to OP9-DL1 stromal cells gave rise to B- and T-lineage cells, respectively (Fig. 4C). However, when the population was switched from OP9-control to OP9-DL1, the B-lineage cells disappeared and cells in the cultures were able to generate T cells, in spite of 6 d in the absence of DL1 (Fig. 4C). Similarly, when cells were switched from OP9-DL1 to OP9-control, T-lineage cells vanished and B-lineage cells were generated (Fig. 4C). Thus, indeed, some of the hematolymphoid progenitors were still capable of giving rise to T- or B-lineage cells, even after 6 d of OP9-control or OP9-DL1 culture, respectively.
Figure 4.
Delayed establishment of T- and B-cell commitment relative to differentiation. (A) Experimental design; c-Kit+Lin- cells were cultured in parallel on OP9-control or OP9-DL1 cells for 1-6 d. At these two time points, both OP9-control and OP9-DL1 cultured were disrupted and replated, half of their progeny onto fresh OP9-control and half onto fresh OP9-DL1 stromal cells. (B) FACS analysis of cultures disrupted and replated after 1 d of preculture, analyzed after an additional 6 d of culture. (C) FACS analysis of cultures disrupted and replated after 6 d of preculture, analyzed after an additional 6 d of culture.
Asynchronous lineage commitment in progeny of single precursors
These results suggest that some precursors remain truly uncommitted after a week of polarizing OP9 culture, but they could alternatively reflect contamination of the starting culture with small numbers of committed cells that survive under adverse conditions of preculture. To determine rigorously whether the same precursor can generate B and T progeny after polarizing preculture, we performed the same switch culture experiments on individual clones. To limit developmental heterogeneity in the starting population, sorted c-Kit+CD27+Lin- cells were used; this fraction also includes all the c-Kit+Lin- cells that could be scored as clonogenic after 7 d of culture (data not shown). Single c-Kit+Lin-CD27+ cells were plated onto either OP9-control or OP9-DL1 stromal cells, where they were allowed to grow for 1 wk. Subsequently, wells in which cellular growth was observed microscopically were resuspended and replated, with half of the progeny transferred onto fresh OP9-control and half onto fresh OP9-DL1 stromal cells, thus maintaining or changing the initial OP9 stromal layer. The wells were then analyzed 1 wk later. On OP9-control stromal cells, 48 of 168 single cells gave rise to microscopically visible progeny, compared with 52 of 168 cells in the case of OP9-DL1 (combined data of two independent experiments with similar results). All of these cultures went on to generate lymphocytes after replating.
Each clone gave rise to T- or B-lineage cells, respectively, when transferred from OP9-DL1 to OP9-DL1 or from OP9-control to OP9-control (Fig. 5). When maintained on OP9-DL1 stromal cells, they differentiated very efficiently into DP thymocytes (Fig. 5A). The majority of the clones did poorly when transferred to the opposite stromal type (Fig. 5A,C). T-cell differentiation was especially inhibited by transfer from OP9-DL1 to OP9-control cultures, and from most clones, only a few arrested Thy1+ DN1 cells, which might be differentiating into the NK-lineage (Schmitt et al. 2004), were recovered (Fig. 5A). These clones were considered committed.
Figure 5.
Asynchronous lineage commitment in progeny of single, differentiating precursor cells. FACS analysis of single c-Kit+CD27+Lin- FL cells that were initially precultured onto OP9-control or OP9-DL1 stromal cells for 7 d. After 7 d, the progeny of each of the outgrowing cells was replated, half onto OP-control and half onto OP9-DL1 stromal cells to determine the frequency of lineage-committed clones. One representative example is shown for lineage commitment (A) and no lineage commitment (B) after OP9-DL1 preculture and for lineage commitment (C) and no lineage commitment (D) after OP9-control preculture.
However, this was not the only result seen. When transferred after 7 d of culture on OP9-DL1 stromal cells, 11% of the clones were not committed to the T-cell lineage (6/52, two experiments). These clones gave rise to CD19+ B-lineage cells when transferred onto OP9-control cells (Fig. 5B), while also giving rise to Thy1++ DN2 and DN3 thymocytes when maintained on OP9-DL1 stromal cells (Fig. 5B). Similarly, when initially cultured on OP9-control cells, 92% of the cells analyzed had committed to the B-cell lineage after 7 d of culture. These cells only generated B cells after transfer onto both OP9-control and OP9-DL1 cells (Fig. 5C). However, still 8% (4/48) had the capacity to initiate the T-cell program, generating Thy1++ DN2 and DN3 thymocytes when transferred onto OP9-DL1 cells, although they very efficiently gave rise to B cells when maintained on OP9-control stromal cells (Fig. 5D).
Thus, the commitment step for FL progenitors to the B- and T-cell lineage can occur at various time points, and the progeny of a single cell can show highly asynchronous differentiation responses, even in this simple monolayer stromal culture environment. Cells do not necessarily lose the potential to initiate T-cell differentiation, even after being cultured without Notch/DL1 interaction for a week. Continuous activation of the Notch-signaling pathway is required to sustain T-cell development, once initiated. However, 7 d of culture on DL1-expressing stroma does not exclusively induce T-cell lineage commitment nor irrevocably block B-cell potential.
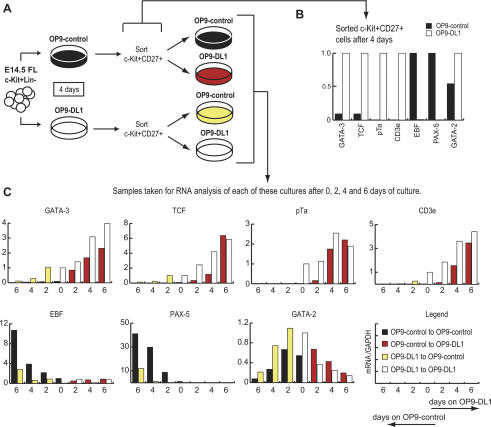
Reversibility of initial lineage-specific gene expression changes
A molecular basis for this developmental plasticity could be shown directly by the reversibility of T- and B-cell gene induction in the precursor cells (experimental design, Fig. 6A). Differential expression of T-cell and B-cell genes initiated while the precursors were still c-Kit+CD27+, as shown by the polarized RNA expression pattern in these cells when sorted after 4 d of OP9-control or OP9-DL1 culture (Fig. 6B). The OP9-DL1 precultured cells expressed pTα and CD3ε and higher levels of GATA-3 and TCF, whereas the OP9-control precultured cells, instead, expressed Pax-5 and elevated levels of EBF. However, these initial positive gene-expression changes remained reversible, as shown by transferring the c-Kit+CD27+ cells to new OP9 stroma after 4 d of initial culture (Fig. 6C).
Figure 6.
Reversibility of initial lineage-specific gene expression changes in c-Kit+ CD27+ progenitors. (A) Experimental design; c-Kit+Lin- cells were cultured in parallel on OP9-control or OP9-DL1 cells for 4 d. At day 4, c-Kit+CD27+ cells were sorted from both cultures and both OP9-control and OP9-DL1 derived c-Kit+CD27+ cells were replated onto both OP9-control and OP9-DL1 stromal cells as indicated. (B) Quantitative real-time gene expression analysis of sorted c-Kit+CD27+ cells after 4 d of culture on OP9-control or OP9-DL1. (C) Quantitative real-time gene expression analysis of sorted 4-d precultured c-Kit+CD27+ cells that were replated onto fresh OP9-control and OP9-DL1 stromal cells with samples taken after 0, 2, 4, and 6 d of additional culture. Units of expression for each gene are relative to their levels in the c-Kit+CD27+ cells from the appropriate preculture, as shown in B.
Cells initially cultured on OP9-control stroma extinguished their initial Pax-5 expression, up-regulated GATA-3 and TCF-1, and turned on pTα and CD3ε when transferred to OP9-DL1 (Fig. 6C). Even the kinetics were similar to those of freshly isolated precursors on OP9-DL1; GATA-3 induction actually appeared to occur somewhat faster. Thus, for cells retaining the precursor phenotype, initial culture under B-cell conditions did not cause any loss of ability to activate a T-cell program in response to Notch/Delta signaling.
More surprisingly, a reciprocal response could be obtained from cells that were precultured on OP9-DL1 for 4 d, and then transferred to OP9-control. These cells showed an immediate loss of pTα and CD3ε expression, with GATA-3 and TCF-1 undergoing down-regulation after 2 d. In spite of their initial DL1 exposure, these cells could still substantially up-regulate EBF and Pax-5 within 6 d of their transfer to OP9-control culture (Fig. 6C), with kinetics again similar to those in cultures initiated with naïve c-Kit+ precursors (cf. Fig. 3 and Supplementary Fig. 3). Importantly, for at least 4 d, the yields of RNA from cells switched from one set of conditions to the opposite set of conditions remained indistinguishable from those returned to the initial culture conditions, ruling out artifactual changes in RNA recovery due simply to extensive cell death.
Thus, the initial induction of lineage-specific gene expression in the c-Kit+CD27+ population remains reversible and subject to reprogramming in an altered environment.
Fast and slow-responding T-cell precursors: differential activation of T-cell program, but similar levels of Notch response
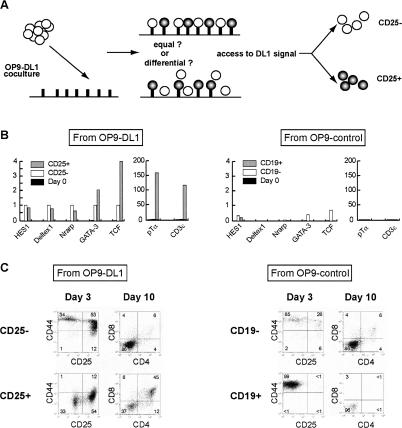
The OP9-DL1 cell system provides precursors with a far more uniform signaling microenvironment than the complex structure of the thymus. The results described here thus suggest that the varied and asynchronous responses of individual precursor cells to Notch signaling are either intrinsically stochastic or else due to developmental differences among the states of the precursors. In either case, variation in response might reflect the levels of discrete, additional regulatory functions needed to cooperate with Notch signals to elicit the T-cell program and/or to block the B-cell program. However, lacking a direct viable-cell indicator for Notch signaling, we also considered the possibility that some precursors escape commitment simply because they escape Notch/DL1 interaction, due to cell-density effects or to possible differences in Notch expression (Fig. 7A).
Figure 7.
Different T-cell developmental kinetics of hematolymphoid precursors are not a result of differences in Notch signaling. (A) Rationale of the experiment: Are the varied and asynchronous responses of individual precursor cells occurring in the presence of equal or differential Notch signaling? (B) Quantitative real-time gene expression analysis of CD27+ c-kit+ CD25- and CD27+ CD25+ sorted cells after 4 d of OP9-DL1 culture (left two graphs) and of CD27+ c-kit+ CD19- and CD19+ cells after 4 d of OP9-control culture (right two graphs). Units of expression are given relative to levels in the purified starting population. Note the change of scale for pTα and CD3ε. (C) T-cell developmental capacities of cells used for gene expression analysis in B. Cells were analyzed by FACS after OP9-DL1 coculture at various time points as indicated. All dot plots shown are gated on CD45+ hematopoietic cells and representative of two independent experiments.
If Notch signaling is the only rate-limiting event in entry into the T-cell pathway, then the earliest responding cells should show greater Notch-response gene induction than cells that have not turned on T-cell genes at the same time point. If additional rate-limiting factors are required, however, then discontinuities may be seen between the patterns of activation of direct Notch target genes and of T-lineage differentiation genes. Only a minority of c-Kit+CD27+ cells up-regulate CD25 and Thy-1 within the first 4 d of OP9-DL1 culture. When sorted to compare gene expression levels, the c-Kit+CD27+ cells that have become CD25+ after 4 d of culture are the only ones to express pTα and CD3ε, with levels ∼100× higher than in the cells remaining CD25- from the same cultures (Fig. 7B). They also show higher GATA-3 and substantially higher TCF-1 expression than the CD25- cells. However, the CD25- and CD25+ subsets have indistinguishable levels of HES-1 induction, both equally elevated as compared with the original uncultured c-Kit+CD27+ cells and compared with OP9-control-derived progeny (Fig. 7B). Similarly, the expression of Deltex1 and Nrarp, two other Notch-specific signaling targets (Deftos et al. 2000; Lamar et al. 2001), is equally induced in unspecified CD25- and specified CD25+ T-cell progenitors after OP9-DL1 coculture (Fig. 7B, left). Conversely, after 4 d in OP9-control cultures, HES-1 is slightly induced in both committed CD19+ and uncommitted CD19- c-kit+ cells, but not Deltex1 or Nrarp (Fig. 7B, right). Yet, GATA-3 and TCF-1 are both detectable in the CD19- c-kit+ cells, while absent in the CD19+ cells in the same cultures. Thus, at least one other regulatory input beside Notch/DL1 signaling is required to explain the differential induction of T-lineage genes between lymphoid precursor subsets on OP9-DL1 and OP9-control stroma.
Cell transfer experiments confirm that the c-Kit+Lin- starting population includes both rapidly and slowly differentiating precursors. When the CD25+ cells from OP9-DL1 cultures are resorted and transferred to fresh cultures, they differentiate rapidly into CD44- DN3/4 and CD4+CD8+ T-lineage cells (Fig. 7C, “CD25+”). They complete differentiation significantly faster than the c-Kit+CD27+ cells that are still CD25- after 4 d of OP9-DL1 culture (Fig. 7C, “CD25-”). However, these “slower” precursors still have clear T-cell lineage potential, as they also give rise to more differentiated CD4+CD8+ cells (Fig. 7C, day 10). Conversely, in OP9-control cultures, the first c-Kit+ cells that turn on CD19 expression within 4 d have almost completely lost the capability to give rise to T cells when transferred to OP9-DL1 culture (Fig. 7C, “CD19+”). This is consistent with other evidence that CD19 expression, driven by Pax-5, is correlated with B-lineage commitment (Nutt et al. 1999). However, the c-Kit+ cells from OP9-control cultures that are still CD27+ CD19- at the same time point retain considerable T-lineage potential (Fig. 7C, “CD19-”), similar to the T-lineage developmental potential of the slower differentiating cells from the OP9-DL1 cultures.
Taken together, these results show that both the timing of the T-lineage gene-induction response to Notch/DL1 signaling and the requirement for Notch/DL1 signaling to preserve the T-cell option are controlled by cell-intrinsic regulatory inputs in addition to the activity of the Notch pathway itself.
Discussion
Notch/Delta signaling has emerged as the only regulatory input for T-cell development that is both essential and consistently positive over a wide dose range. By the final stages of lymphoid precursor specification, it is known that Notch signaling is continuously required to establish and maintain T-lineage identity, and to block B-lineage specification (Pui et al. 1999; Wilson et al. 2001; De Smedt et al. 2002; Schmitt et al. 2004). However, this may not be the only stage at which Notch/Delta interaction promotes T-cell development. Several lines of evidence have recently indicated that multipotent precursors give rise to divergent types of partially restricted lymphoid progenitors, some biased toward T lineage and some biased toward B lineage (Baba et al. 2004; Schwarz and Bhandoola 2004; Rothenberg and Taghon 2005). These studies raise the question of whether Notch signaling plays a role at the multipotent precursor level as well. Traditional approaches of germline transgenesis and steady-state analysis of population phenotypes have not shed much light on mechanisms that may initiate T-lymphocyte development at the level of multipotent precursors. However, the OP9-based stromal cell culture system for T-cell development (Schmitt and Zúñiga-Pflücker 2002) provides a powerful way to study this process in detail. The absence or presence of DL1 expression is the only difference between cultures that support either B- or T-cell development. Thus, this dual culture system is the ideal setup to compare the kinetics of initiation of B- and T-cell commitment from the same hematopoietic progenitor cells.
Our results show that T-lineage commitment from multipotent FL progenitors has three unexpected features. First is the delayed time course and initial reversibility of T-cell regulatory gene induction in response to Notch/Delta signaling. Second, responsiveness to the Notch/DL1 signal is extremely flexible in developmental time; precursors can proliferate in OP9-control culture for days, before the T-cell differentiation cascade is triggered by transfer to OP9-DL1 cells. Third, uncommitted precursors are found to persist for almost a week, although in diminishing numbers, in the OP9-DL1 as well as the OP9-control cultures. This is most dramatically illustrated in functional assays of single-cell progeny, by OP9-DL1 cultured clones remaining capable of B-lineage differentiation despite Notch/Delta antagonism, as well as by OP9-control cultured clones remaining capable of T-lineage differentiation while delivery of the Notch/Delta signal is delayed. These results imply that Notch/Delta triggering may be necessary, but is not sufficient, to trigger the onset of T-cell development or T-lineage commitment, even within a single clone.
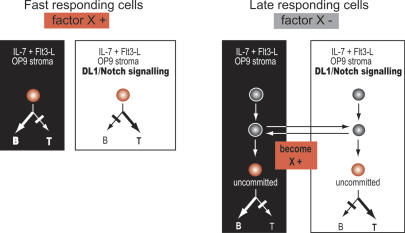
The OP9-DL1 culture system clearly delivers a specific instructive signal for T-cell development. Expression of pTα, GATA-3, TCF-1, and CD3ε is significantly induced in response to the DL1 ligand, contrasting with little if any up-regulation on OP9-control stroma. Yet on OP9-DL1, the T-cell program is induced gradually, in contrast to the direct target HES-1. Differentiation is slow with respect to Notch/Delta signaling, but swift with respect to expression of T-cell-specific transcription factors, since differentiation genes such as pTα and CD3ε are induced with almost the same kinetics as GATA-3 and TCF-1. Furthermore, the program appears to initiate coordinately in the responding cells, with a part of the population beginning a multigene activation of the T-cell program by 3-4 d, while other precursors delay even longer. Subsets that have up-regulated Notch targets HES-1, Deltex1, and Nrarp to similar extents can show an all-or-none difference in their expression of CD25, pTα, and CD3ε. There are only a few hints that GATA-3 itself may be inducible in a broader fraction of Notch-responsive cells than those that are ready to activate the full program. The delayed and asynchronous induction of T-cell differentiation genes shows that some additional rate-limiting regulatory feature, beside Notch/Delta signaling, is required to initiate the expression of these T-lineage-specific genes. We propose that expression or induction of this regulatory input, termed “X” in Figure 8, may also be one of the factors that distinguishes “fast” and “slow” differentiating subsets of precursors, as discussed below.
Figure 8.
Models of the responses of individual hematolymphoid precursors to Notch/Delta signaling. Effect of culture on OP9-DL1 or OP9-control for precursors with ability to delay T/B-lineage commitment. (Left) Fast-responding cells behave as bipotent precursors described by the standard instructive model for Notch/Delta signaling: deterministic model of Notch actions in T-cell development. In this model, the activation of the T-cell alternative automatically excludes the B-cell alternative. (Right) This model does not explain the persistence under polarizing conditions of cells with the capacity for both, as seen in the slow responding cells. In this model, different precursors can respond asynchronously to Notch/Delta signaling or its absence. Cells remaining “multipotent” in this model, as T- or B-cell development is first being induced in other cells, are capable of making the transit to T- or B-cell differentiation at a later time, as permitted by culture conditions, as shown in Figure 5B and D.
The B-cell and T-cell differentiation responses, at a population level, are marked by prolonged plasticity. There are at least two nonexclusive mechanisms that may be responsible; on the one hand, an intrinsic reversibility of the early specification process itself, and on the other hand, the ability of some precursor cells to delay specification. Even once T- or B-lineage gene expression is induced, this program appears to be fully reversible. The T-lineage genes shut off within 2-4 d if the cells are removed from DL1. On OP9-control stroma, among the genes turned on by c-Kit+CD27+ precursors are two B-lineage factors that powerfully antagonize T-cell development in vivo, EBF (Zhang et al. 2003) and Pax-5 (Souabni et al. 2002). However, if the c-Kit+CD27+ cells are transferred to OP9-DL1 at day 4, these genes are rapidly turned off, and the T-cell program begins unimpaired. There is even some evidence in several of our experiments that OP9-DL1 induces GATA-3 faster in cells that have been precultured on OP9-control stroma. These results are fully in accord with the recent report that Pax-5-deficient pro-B cells can also initiate T-lineage gene expression on OP9-DL1 cells, even faster than the multipotent progenitor-dominated populations used here (Höflinger et al. 2004). Notably lacking is evidence for any kind of “lock-down” circuit behavior in early lymphocyte differentiation, during the first 4-6 d, such as operates in many embryonic gene regulatory networks (Davidson et al. 2003). The reversal phenomenon is clear on the population level, and the fact that there is little or no kinetic penalty means that it is likely to apply to many individual cells in the cultures as well, at least as long as they retain the c-Kit+CD27+ phenotype.
The strongest case for plasticity can be made for the slowly differentiating cells in these cultures. There are also a minority of fast-differentiating cells in both OP9-control and OP9-DL1 cultures. These show a more pronounced lineage-specific gene expression pattern compared with those cells remaining CD27+ c-Kit+ CD25- after 4 d, and have less developmental plasticity, as shown at the single-cell level. Thus, the “fast” cells (Fig. 8, left) do contribute disproportionately to the earliest increases in lineage-specific gene expression in the cultures, around days 3-4. However, over time, these are not the only contributors to lymphocyte differentiation in the cultures. Our results emphasize that slowly differentiating cells (Fig. 8, right) are also highly efficient T-cell precursors, but correspond more closely to expectations for cells that retain a multipotent progenitor phenotype in culture, since they also retain B-cell potential. The kinetics of gene expression in transfer experiments (Figs. 6C, 7B) as compared with those of the initial population (Fig. 3) imply that slow-differentiating cells can be recruited continuously into differentiation, in what may be a stochastic process. The differentiation path they follow can lead either to the T- or the B-lineage, irrespective of the stroma from which they came.
Switching cells from one culture condition to another proves that plasticity at the level of distinct responses is retained within a single clone. Remarkably, after 7 d of continuous Notch/DL1 stimulation, uncommitted progenitors are still present in ∼10% of clones undergoing T-lineage differentiation, as these clones are able to give rise to B cells when transferred onto OP9-control cells. This suggests that the initial single cells in these conditions give rise to heterogeneous daughter cells, some of which are clearly not yet committed to the T-cell lineage. Preculture in B-cell conditions for a week also permits survival of cells within the clone with the capacity to undergo T-lineage differentiation.
In at least some cases, the same clone can generate committed and uncommitted cells. Individual clones can start B-lineage development and still give rise to T-lineage cells, since CD19+ cells are still recognizable after transfer to OP9-DL1 culture (Fig. 5B). Committed B cells can survive in the presence of continuous Notch signaling as shown previously (Schmitt and Zúñiga-Pflücker 2002). However, T-lineage-committed cells cannot be detected after transfer to OP9-control culture. Although 7 d of OP9-DL1 culture is sufficient for differentiation to the DN3 stage, if the cells are removed from DL1 at this time, neither DN2, DN3, DN4, or DP cells with high levels of Thy1 expression survive 7 d later. This confirms reports that continuous Notch signaling is required in T-lineage cells for survival and/or for maintenance of their specification, even once T-cell differentiation is under way (Davidson et al. 2003).
One powerful application of this culture system should be to help identify additional molecular requirements for entry into the T-cell pathway. This has remained a problem for the field, because Notch signaling does not have uniquely T-lineage-specific effects (Schroeder et al. 2003), whereas the one known T-cell-specific transcription factor, GATA-3, blocks T-cell development when overexpressed (Chen and Zhang 2001; Taghon et al. 2001; Anderson et al. 2002). Identification of additional regulators that cooperate with Notch signaling to give it specificity would be an important advance. The delay between delivery of the first Notch/Delta signals and the onset of GATA-3 and TCF-1 expression in the OP9-DL1 system precisely defines the temporal window within which any additional positive T-lineage regulators must begin to act, thus providing a kinetic assay for their identification. Fast- and slow-differentiating subsets of precursors, if they can be isolated prospectively, should also differ in their expression or inducibility of such regulators.
In conclusion, we have demonstrated that hematopoietic progenitor cells exhibit a high level of plasticity in their responses to instructive signals provided by Notch/Delta interactions in the context of IL-7, Flt3-L, and OP9 stromal cells. This conclusion contrasts with a common image of the role of Notch signaling in T/B lineage choice, in which the Notch/Delta signal acts as a simple binary switch that blocks other fates as a direct effect of its action at initiating the T-cell program. Instead, these precursors appear to be choosing among at least three possible states, i.e., T-specified, B-specified, and remaining unspecified or multipotent. The first of these is dependent on DL1 and the second is antagonized by it, but the last option is not directly controlled by Notch/DL1, and can remain open in either case. The transit between multipotent and specified states is asynchronous among different precursor cells and initially reversible, as shown at the gene-expression level. These features imply that Notch/Delta signaling cooperates with an additional, time-delayed function(s) in an unstable regulatory network in the T-lineage specification of multipotent precursors.
Materials and methods
Monoclonal antibodies and flow cytometry
All monoclonal antibodies used were from EBioscience: CD4 (clone GK1.5, CY and APC), CD8α (clone 53-6.7, FITC), CD11b (clone M1/70, biotin), CD19 (clone MB19.1, PE and biotin), CD25 (clone PC61.5, FITC and CY), CD27 (clone LG.7F9, FITC, PE and APC), CD44 (clone IM7, PE and APC), CD45 (clone 30F11, PE and APC), c-kit (clone ACK2, PE and CY), F4/80 (clone BM8, biotin), Gr-1 (clone RB6-8C5, FITC and biotin), Ter-119 (clone TER-119, biotin), and Thy1 (CD90, clone 30-H12, FITC and APC). For detection of biotinylated antibodies, streptavidin-CY was used. Before each antibody staining, cells were incubated for 20 min in supernatant from clone 2.4G2 anti-CD32/CD16 (FcγRIII/II) to block Fc receptors. Data was acquired and analyzed using CellQuest software (Becton Dickinson Immunocytometry Systems [BDIS]) on a FACS Calibur (BDIS).
Purification of fetal liver cells and OP9 cocultures
Fetal liver (FL) cells obtained from embryonic day 14-14.5 (E14-E14.5) BDF1 mouse embryos were depleted of Gr-1+, F4/80+, Ter119+, and CD19+ cells through labeling with the corresponding biotinylated antibodies, followed by incubation with streptavidin-coated magnetic beads (Miltenyi Biotec). Labeled cells were subsequently removed by magnetic column separation (Miltenyi Biotec). Finally, lineage-depleted FL cells were stained with c-Kit-PE and streptavidin-CY and c-Kit+Lin- cells were sorted. Cell sorting was performed on a FACS Vantage (BDIS) using CellQuest software (BDIS).
OP9-MIG and OP9-DL1 stromal cells (Schmitt and Zúñiga-Pflücker 2002) were cultured in Minimum Essential Medium Alpha (Invitrogen), supplemented with 20% fetal bovine serum (Hyclone), 100 U/mL penicillin, 100 μg/mL streptomycin, and 1× L-glutamine (all from Invitrogen), and plated 1 or 2 d before use in 24-well plates (Corning) to achieve a confluent monolayer of cells. Cocultures were initiated with 10 × 105 c-Kit+Lin- FL cells in the presence of 5 ng/mL Flt3-L and 5 ng/mL IL-7 (both cytokines from Peprotech). Cocultures were harvested by forceful pipetting at the indicated time points. For transfer experiments, 1/5 of the harvested cells were replated and cultured for 1 wk. For RNA extraction, cells were resuspended in RNAlater (Ambion) and stored at -20°C until used. When transfer experiments were performed to investigate the reversibility in gene expression, OP9-control and OP9-DL1 cocultures with c-Kit+Lin- FL cells were harvested after 4 d, and c-Kit+CD27+ cells were sorted. New cocultures were initiated with these cells on fresh layers of OP9-control and OP9-DL1 stroma and harvested at the indicated time points.
Single-cell cultures
For single-cell experiments, lineage-depleted FL cells were stained with c-Kit-PE, streptavidin-CY, and CD27-APC. Single c-Kit+CD27+Lin- cells were sorted using the Clonecyte software (BDIS) on the FACSVantage (BDIS) into one well of a 96-well plate (Corning) containing a confluent monolayer of OP9-MIG or OP9-DL1 stromal cells in the same conditions as mentioned above. After 1 wk, wells that showed cellular growth (scored microscopically) were resuspended and cells were replated, half of the progeny onto fresh OP9-MIG stromal cells and the other half onto fresh OP9-DL1 stromal cells. After an additional week of culture, cells were harvested and analyzed by flow cytometry. Two stainings were performed on each sample.
RNA extraction and quantitative real-time RT-PCR
RNA was extracted using the RNeasy miniprep kit (Qiagen) and converted into cDNA using Superscript RT II (Invitrogen) following the guidelines of the manufacturer. Real-time PCRs were performed using the SYBRGreen Universal Master Mix (Applied Biosystems) and analyzed on the GeneAmp 7700 sequence detection system (Applied Biosystems). For data analysis, each primer set was used to generate a standard curve with 10-fold dilutions of a positive control sample, and the CT values for experimental samples were converted to relative cDNA levels based on the standard curve for that primer set. In most cases, the standard curve was linear over more than three decades, with a slope close to the theoretical value of -3.3. To correct for differences in inputs among samples, results were then normalized to equivalent levels of GAPDH, calculated similarly from the standard curve for those primers. These normalized values are presented (Figs. 3, 6) in units, such that the expression of each gene in the starting population is taken as “1.” Primer sequences (forward = fw, reverse = rev) were taken from previous reports (Anderson et al. 2002, 2004; Yun and Bevan 2003) or are as follows (5′ to 3′): CD3ε fw, CGTCCGCCATCTTGGTAGAG, and CD3ε rev, ATTCAATGTTCTCGGCATCGT; GATA-2 fw, GGAGACGATTGTGCTGAGTCAA, and GATA-2 rev, AAAATGCTGCCGATTCTTCTCT; HES-5 fw, TCGGGACCGCATCAACA, and HES-5 rev, CAGGTAGCTGACGGCCATCT; TCF-1 fw, TGCTGTCTATATCCGCAGGAAG, and TCF-1 rev, CGATCTCTCTGGATTTTATTCTCT.
Acknowledgments
We thank Michele K. Anderson for primer sequences; Shirley Pease, Bruce Kennedy, and Armando Mayo of the Caltech Transgenic Animal Facility for timed matings; and Rochelle Diamond, Stephanie Adams, and Pat Koen of the Caltech Flow Cytometry/Cell Sorting Facility for fluorescence-activated cell sorting. This work was supported by grants from the NIH (R01 CA98925 and R01 CA90233).
Supplemental material is available at www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1298305.
References
- Allman D., Sambandam, A., Kim, S., Miller, J.P., Pagan, A., Well, D., Meraz, A., and Bhandoola, A. 2003. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 4**:** 168-174. [DOI] [PubMed] [Google Scholar]
- Anderson G., Pongracz, J., Parnell, S., and Jenkinson, E.J. 2001. Notch ligand-bearing thymic epithelial cells initiate and sustain Notch signaling in thymocytes independently of T cell receptor signaling. Eur. J. Immunol. 31**:** 3349-3354. [DOI] [PubMed] [Google Scholar]
- Anderson M.K., Hernandez-Hoyos, G., Dionne, C.J., Arias, A.M., Chen, D., and Rothenberg, E.V. 2002. Definition of regulatory network elements for T cell development by perturbation analysis with PU.1 and GATA-3. Dev. Biol. 246**:** 103-121. [DOI] [PubMed] [Google Scholar]
- Anderson M.K., Pant, R., Miracle, A.L., Sun, X., Luer, C.A., Walsh, C.J., Telfer, J.C., Litman, G.W., and Rothenberg, E.V. 2004. Evolutionary origins of lymphocytes: Ensembles of T cell and B cell transcriptional regulators in a cartilaginous fish. J. Immunol. 172**:** 5851-5860. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand, M.D., and Lake, R.J. 1999. Notch signaling: Cell fate control and signal integration in development. Science 284**:** 770-776. [DOI] [PubMed] [Google Scholar]
- Baba Y., Pelayo, R., and Kincade, P.W. 2004. Relationships between hematopoietic stem cells and lymphocyte progenitors. Trends Immunol. 25**:** 645-649. [DOI] [PubMed] [Google Scholar]
- Chen D. and Zhang, G. 2001. Enforced expression of the GATA-3 transcription factor affects cell fate decisions in hematopoiesis. Exp. Hematol. 29**:** 971-980. [DOI] [PubMed] [Google Scholar]
- Davidson E.H., McClay, D.R., and Hood, L. 2003. Regulatory gene networks and the properties of the developmental process. Proc. Natl. Acad. Sci. 100**:** 1475-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.L. and Turner, D.L. 2001. Vertebrate hairy and Enhancer of split related proteins: Transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene 20**:** 8342-8357. [DOI] [PubMed] [Google Scholar]
- de la Pompa J.L., Wakeham, A., Correia, K.M., Samper, E., Brown, S., Aguilera, R.J., Nakano, T., Honjo, T., Mak, T.W., Rossant, J., et al. 1997. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development 124**:** 1139-1148. [DOI] [PubMed] [Google Scholar]
- De Smedt M., Reynvoet, K., Kerre, T., Taghon, T., Verhasselt, B., Vandekerckhove, B., Leclercq, G., and Plum, J. 2002. Active form of Notch imposes T cell fate in human progenitor cells. J. Immunol. 169**:** 3021-3029. [DOI] [PubMed] [Google Scholar]
- Deftos M.L. and Bevan, M.J. 2000. Notch signaling in T cell development. Curr. Opin. Immunol. 12**:** 166-172. [DOI] [PubMed] [Google Scholar]
- Deftos M.L., Huang, E., Ojala, E.W., Forbush, K.A., and Bevan, M.J. 2000. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity 13**:** 73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos K., Bigby, M., Wang, J.H., Molnar, A., Wu, P., Winandy, S., and Sharpe, A. 1994. The Ikaros gene is required for the development of all lymphoid lineages. Cell 79**:** 143-156. [DOI] [PubMed] [Google Scholar]
- Graf T. 2002. Differentiation plasticity of hematopoietic cells. Blood 99**:** 3089-3101. [DOI] [PubMed] [Google Scholar]
- Greenwald I. 1998. LIN-12/Notch signaling: Lessons from worms and flies. Genes & Dev. 12**:** 1751-1762. [DOI] [PubMed] [Google Scholar]
- Hahm K., Cobb, B.S., McCarty, A.S., Brown, K.E., Klug, C.A., Lee, R., Akashi, K., Weissman, I.L., Fisher, A.G., and Smale, S.T. 1998. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes & Dev. 12**:** 782-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman B.C., Jenkinson, E.J., and Anderson, G. 2003. Entry into the thymic microenvironment triggers Notch activation in the earliest migrant T cell progenitors. J. Immunol. 170**:** 1299-1303. [DOI] [PubMed] [Google Scholar]
- Höflinger S., Kesavan, K., Fuxa, M., Hutter, C., Heavey, B., Radtke, F., and Busslinger, M. 2004. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J. Immunol. 173**:** 3935-3944. [DOI] [PubMed] [Google Scholar]
- Hozumi K., Negishi, N., Suzuki, D., Abe, N., Sotomaru, Y., Tamaoki, N., Mailhos, C., Ish-Horowicz, D., Habu, S., and Owen, M.J. 2004. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat. Immunol. 5**:** 638-644. [DOI] [PubMed] [Google Scholar]
- Igarashi H., Gregory, S.C., Yokota, T., Sakaguchi, N., and Kincade, P.W. 2002. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity 17**:** 117-130. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Mizuno, S., Wells, R.A., Cantor, A.B., Watanabe, S., and Akashi, K. 2003. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity 19**:** 451-462. [DOI] [PubMed] [Google Scholar]
- Jarriault S., Brou, C., Logeat, F., Schroeter, E.H., Kopan, R., and Israel, A. 1995. Signalling downstream of activated mammalian Notch. Nature 377**:** 355-358. [DOI] [PubMed] [Google Scholar]
- Kumano K., Chiba, S., Shimizu, K., Yamagata, T., Hosoya, N., Saito, T., Takahashi, T., Hamada, Y., and Hirai, H. 2001. Notch1 inhibits differentiation of hematopoietic cells by sustaining GATA-2 expression. Blood 98**:** 3283-3289. [DOI] [PubMed] [Google Scholar]
- Lamar E., Deblandre, G., Wettstein, D., Gawantka, V., Pollet, N., Niehrs, C., and Kintner, C. 2001. Nrarp is a novel intracellular component of the Notch signaling pathway. Genes & Dev. 15**:** 1885-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehar S.M., Dooley, J., Farr, A.G., and Bevan, M.J. 2004. Notch ligands Delta1 and Jagged1 transmit distinct signals to T cell precursors. Blood 105**:** 1440-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Strasser, A., Baumgarth, N., Cicuttini, F.M., Welch, K., Salvaris, E., and Boyd, A.W. 1993. A novel cellular model (SPGM 1) of switching between the pre-B cell and myelomonocytic lineages. J. Immunol. 150**:** 4395-4406. [PubMed] [Google Scholar]
- McKercher S.R., Torbett, B.E., Anderson, K.L., Henkel, G.W., Vestal, D.J., Baribault, H., Klemsz, M., Feeney, A.J., Wu, G.E., Paige, C.J., et al. 1996. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 15**:** 5647-5658. [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T., Iwasaki, H., Reizis, B., Ye, M., Graf, T., Weissman, I.L., and Akashi, K. 2002. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell 3**:** 137-147. [DOI] [PubMed] [Google Scholar]
- Mombaerts P., Iacomini, J., Johnson, R.S., Herrup, K., Tonegawa, S., and Papaioannou, V.E. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68**:** 869-877. [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Heavey, B., Rolink, A.G., and Busslinger, M. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401**:** 556-562. [DOI] [PubMed] [Google Scholar]
- Pai S.Y., Truitt, M.L., Ting, C.N., Leiden, J.M., Glimcher, L.H., and Ho, I.C. 2003. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity 19**:** 863-875. [DOI] [PubMed] [Google Scholar]
- Peschon J.J., Morrissey, P.J., Grabstein, K.H., Ramsdell, F.J., Maraskovsky, E., Gliniak, B.C., Park, L.S., Ziegler, S.F., Williams, D.E., Ware, C.B., et al. 1994. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J. Exp. Med. 180**:** 1955-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui J.C., Allman, D., Xu, L., DeRocco, S., Karnell, F.G., Bakkour, S., Lee, J.Y., Kadesch, T., Hardy, R.R., Aster, J.C., et al. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11**:** 299-308. [DOI] [PubMed] [Google Scholar]
- Radtke F., Wilson, A., Mancini, S.J., and MacDonald, H.R. 2004. Notch regulation of lymphocyte development and function. Nat. Immunol. 5**:** 247-253. [DOI] [PubMed] [Google Scholar]
- Rebollo A. and Schmitt, C. 2003. Ikaros, Aiolos and Helios: Transcription regulators and lymphoid malignancies. Immunol. Cell Biol. 81**:** 171-175. [DOI] [PubMed] [Google Scholar]
- Reizis B. and Leder, P. 2002. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes & Dev. 16**:** 295-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E.V. 2000. Stepwise specification of lymphocyte developmental lineages. Curr. Opin. Genet. Dev. 10**:** 370-379. [DOI] [PubMed] [Google Scholar]
- Rothenberg E.V. and Anderson, M.K. 2002. Elements of transcription factor network design for T-lineage specification. Dev. Biol. 246**:** 29-44. [DOI] [PubMed] [Google Scholar]
- Rothenberg E.V. and Dionne, C.J. 2002. Lineage plasticity and commitment in T-cell development. Immunol. Rev. 187**:** 96-115. [DOI] [PubMed] [Google Scholar]
- Rothenberg E.V. and Taghon, T. 2005. Molecular genetics of T cell development. Annu. Rev. Immunol. 23**:** 601-649. [DOI] [PubMed] [Google Scholar]
- Schmitt T.M. and Zúñiga-Pflücker, J.C. 2002. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity 17**:** 749-756. [DOI] [PubMed] [Google Scholar]
- Schmitt T.M., Ciofani, M., Petrie, H.T., and Zúñiga-Pflücker, J.C. 2004. Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions. J. Exp. Med. 200**:** 469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder T. and Just, U. 2000. mNotch1 signaling reduces proliferation of myeloid progenitor cells by altering cell-cycle kinetics. Exp. Hematol. 28**:** 1206-1213. [DOI] [PubMed] [Google Scholar]
- Schroeder T., Kohlhof, H., Rieber, N., and Just, U. 2003. Notch signaling induces multilineage myeloid differentiation and up-regulates PU.1 expression. J. Immunol. 170**:** 5538-5548. [DOI] [PubMed] [Google Scholar]
- Schroeter E.H., Kisslinger, J.A., and Kopan, R. 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393**:** 382-386. [DOI] [PubMed] [Google Scholar]
- Schwarz B.A. and Bhandoola, A. 2004. Circulating hematopoietic progenitors with T lineage potential. Nat. Immunol. 5**:** 953-960. [DOI] [PubMed] [Google Scholar]
- Scott E.W., Simon, M.C., Anastasi, J., and Singh, H. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265**:** 1573-1577. [DOI] [PubMed] [Google Scholar]
- Shortman K., Vremec, D., Corcoran, L.M., Georgopoulos, K., Lucas, K., and Wu, L. 1998. The linkage between T-cell and dendritic cell development in the mouse thymus. Immunol. Rev. 165**:** 39-46. [DOI] [PubMed] [Google Scholar]
- Souabni A., Cobaleda, C., Schebesta, M., and Busslinger, M. 2002. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1. Immunity 17**:** 781-793. [DOI] [PubMed] [Google Scholar]
- Staal F.J. and Clevers, H.C. 2003. Wnt signaling in the thymus. Curr. Opin. Immunol. 15**:** 204-208. [DOI] [PubMed] [Google Scholar]
- Taghon T., De Smedt, M., Stolz, F., Cnockaert, M., Plum, J., and Leclercq, G. 2001. Enforced expression of GATA-3 severely reduces human thymic cellularity. J. Immunol. 167**:** 4468-4475. [DOI] [PubMed] [Google Scholar]
- Tamura K., Taniguchi, Y., Minoguchi, S., Sakai, T., Tun, T., Furukawa, T., and Honjo, T. 1995. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J κ/Su(H). Curr. Biol. 5**:** 1416-1423. [DOI] [PubMed] [Google Scholar]
- Ting C.N., Olson, M.C., Barton, K.P., and Leiden, J.M. 1996. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature 384**:** 474-478. [DOI] [PubMed] [Google Scholar]
- Tsai F.Y., Keller, G., Kuo, F.C., Weiss, M., Chen, J., Rosenblatt, M., Alt, F.W., and Orkin, S.H. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371**:** 221-226. [DOI] [PubMed] [Google Scholar]
- Verbeek S., Izon, D., Hofhuis, F., Robanus-Maandag, E., te Riele, H., van de Wettering, M., Oosterwegel, M., Wilson, A., MacDonald, H.R., and Clevers, H. 1995. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature 374**:** 70-74. [DOI] [PubMed] [Google Scholar]
- Wilson A., MacDonald, H.R., and Radtke, F. 2001. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 194**:** 1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Ye, M., Feng, R., and Graf, T. 2004. Stepwise reprogramming of B cells into macrophages. Cell 117**:** 663-676. [DOI] [PubMed] [Google Scholar]
- Yun T.J. and Bevan, M.J. 2003. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: Multiple Notch1 signaling pathways involved in T cell development. J. Immunol. 170**:** 5834-5841. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Cotta, C.V., Stephan, R.P., deGuzman, C.G., and Klug, C.A. 2003. Enforced expression of EBF in hematopoietic stem cells restricts lymphopoiesis to the B cell lineage. EMBO J. 22**:** 4759-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]