Jab1 antagonizes TGF-β signaling by inducing Smad4 degradation (original) (raw)
Abstract
Tumor suppressor Smad4 is the common signaling effector in the transforming growth factor β (TGF-β) superfamily. Phosphorylated regulatory Smads (R-Smads) interact with Smad4, and the complex translocates into the nucleus to regulate gene transcription. Proper TGF-β signaling requires precise control of Smad functions. Smurfs have been shown to mediate the degradation of R-Smads but not the common-partner Smad4. We report a novel mechanism of Smad4 degradation. Jab1 interacts directly with Smad4 and induces its ubiquitylation for degradation. Jab1 was initially identified as a co-activator of c-Jun, and it also induces degradation of cell cycle inhibitor p27 and tumor suppressor p53. Ectopic expression of Jab1 decreased endogenous Smad4 steady-state levels. The 26S proteasome inhibitors lactacystin and MG132 reduced the degradation rate of Smad4 protein. Examination of the effects of JAB1-induced Smad4 degradation indicates that Jab1 inhibited TGF-β-induced gene transcription. Our data suggest that Jab1 antagonizes TGF-β function by inducing degradation of Smad4 through a distinct degradation pathway.
INTRODUCTION
The transforming growth factor β (TGF-β) superfamily includes TGF-βs, bone morphogenetic proteins (BMPs) and activin, with more than 40 members that play important roles in regulating a broad range of biological activities including proliferation, migration, bone development, differentiation and apoptosis (Kingsley, 1994; Christian, 2000). Aberrant TGF-β signaling results in developmental disorders, human cancers and other diseases (Wall and Hogan, 1994; Moses and Serra, 1996). The signaling responses to TGF-β-like factors are mediated by two types of transmembrane receptors and their intracellular substrates, the Smad proteins (Massague, 1998; ten Dijke et al., 2000; Wrana, 2000). Following ligand binding, the activated receptors directly phosphorylate regulatory Smads (R-Smads). Phosphorylated R-Smads interact with Smad4, and the complex translocates into the nucleus to regulate gene transcription. Thus, Smad4 is the common signaling molecule shared in the whole TGF-β superfamily. Mutations of Smad4 are frequently found in human cancers, accounting for half of pancreatic cancers (O’Brien, 1996) and >30% of invasive metastatic colorectal cancers (Miyaki et al., 1999). Mutations in Smad4 inactivate TGF signaling by targeting Smad4 to the ubiquitin–proteasome pathway (Xu and Attisano, 2000). Therefore, control of Smad4 degradation is an important mechanism in the regulation of functional activity in the TGF-β superfamily. Smurfs have been shown to mediate the degradation of R-Smads and inhibitory Smads (I-Smads) (Zhu et al., 1999; Kavsak et al., 2000; Lin et al., 2000; Ebisawa et al., 2001; Zhang et al., 2001). However, the mechanisms of common-partner Smad4 degradation are not clear.
We report a novel mechanism of Smad4 degradation. Jab1 interacts directly with Smad4. Ectopic expression of Jab1 decreased endogenous Smad4 steady-state levels and induced its ubiquitylation. 26S proteasome inhibitors reduced the degradation rate of Smad4 protein. In addition, Jab1 inhibited TGF-β-induced gene transcription.
RESULTS AND DISCUSSION
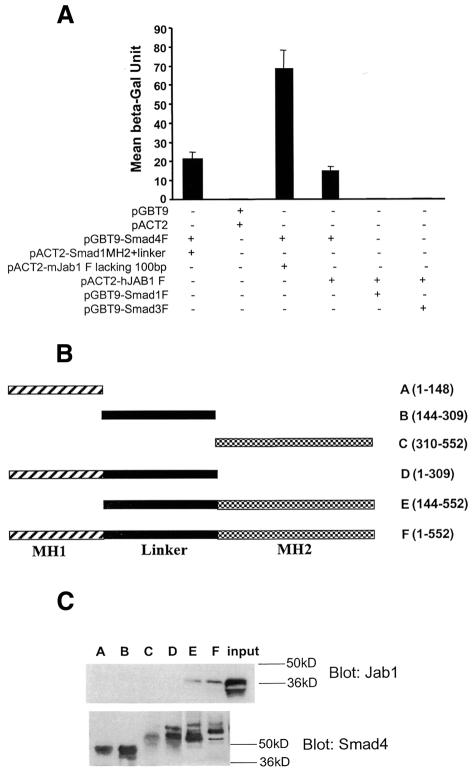
To identify proteins that mediate Smad4 degradation, we screened a human breast cancer cDNA library constructed in pACT2 plasmid with an intact Smad4 cDNA fused with the Gal4 DNA-binding domain as bait. DNA sequence analysis identified two Jab1 clones from 34 positive clones obtained. The interactions were quantified by a liquid β-gal assay (Figure 1A). Smad4, Smad1 and Smad3 were also examined for their interactions with Jab1 in yeast two-hybrid assays. The results of β-gal activity assays indicated that Jab1 interacts with Smad4 but not with Smad1 or Smad3. We then tested the specificity of the interaction and mapped the Smad4 domain(s) that interacts with Jab1 in pull-down assays. In vitro translated Jab1 protein was used in pull-down assays with a series of truncated GST–Smad4 fusion proteins (Figure 1B). The results indicated that full-length Smad4 and MH2 with linker region interacts with Jab1 (Figure 1C). However, neither MH1 nor linker alone interacts with Jab1. It appears that a multi-domain is required for the interaction.
Fig. 1. Interaction of Jab1 with Smad4 in yeast and in vitro. (A) Jab1 interacts with Smad4 in yeast. Intact Jab1 cDNA or Jab1 lacking 100 bp in the N-terminus were fused with the Gal4 DNA-binding domain and transformed in yeast with indicated cDNAs in prey plasmids. The interactions were quantified by a liquid β-gal assay. (B) Smad4 deletion constructs. (C) Smad4 MH2 domain with linker interacts with Jab1. GST–Smad4 (F) or derivatives (A–E) were incubated with in vitro translated Jab1 protein. Bound Jab1 was detected by immunoblotting with antibody against Jab1 (upper panel). The level of GST–Smad4 and derivatives was detected by immunoblotting with antibody against Smad4 (lower panel).
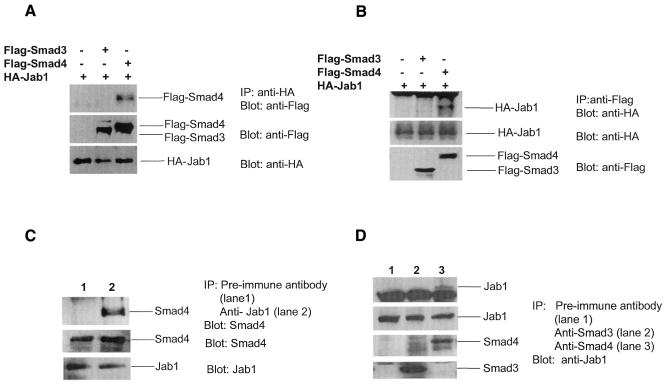
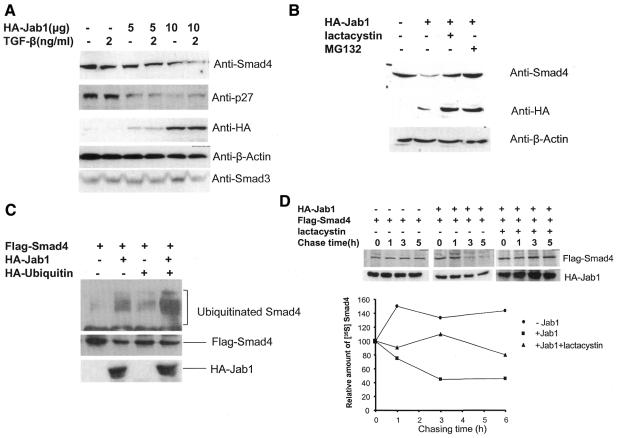
To confirm the interaction in mammalian cells, we performed an immunoprecipitation experiment. HA-Jab1 together with Flag-Smad3 or Flag-Smad4 expression plasmid was co-transfected in 293T cells. The whole-cell lysates were immunoprecipitated by either anti-HA (Figure 2A) or anti-Flag antibody (Figure 2B) and then separated on an SDS–polyacrylamide gel and blotted to nitrocellulose. Immobilized HA-Jab1 was incubated with either anti-Flag or anti-HA antibody, respectively. Thus, Smad4 associated with Jab1 in 293T cells when both proteins were overexpressed. Most importantly, we also demonstrated that endogenous Smad4 interacts with endogenous Jab1 in Mv1Lu cells (Figure 2C and D). Jab1, identified as the Jun activating domain binding protein, enhances AP-1-mediated gene transcription and cell proliferation (Claret et al., 1996). It also induces protein degradation such as cell cycle inhibitor p27 (Tomoda et al., 1999) and tumor suppressor p53 (Bech-Otschir et al., 2001), whose biological functions, like those of TGF-β, inhibit cell proliferation. Thus, it is possible that Jab1 regulates TGF-β activity by inducing degradation of Smad4 in the TGF-β pathway. The interaction between Jab1 and Smad4 suggests a mechanism of Smad4 degradation. We then examined whether the interaction affects endogenous Smad4 steady-state levels in cells. Jab1 expression plasmid was transfected in Mv1Lu cells and the cells treated with or without TGF-β, and the endogenous Smad4 protein levels were measured by western blotting with an antibody against Smad4. Figure 3A demonstrates that the expression of Jab1 reduced Smad4 steady-state levels in a dose-dependent manner. As controls, endogenous p27, Smad3 and β-actin protein levels were also measured. As expected, p27 protein levels were decreased in cells expressing Jab1, whereas Smad3 and β-actin levels remained unchanged. These results suggest that Jab1 induces Smad4 protein degradation and that Jab1-induced reduction of Smad4 protein levels may not be TGF-β dependent. However, TGF-β stimulates Smad4 translocation to the nucleus by phosphorylation of R-Smads, and Jab1 is a component of the COP9 signalosome in the nucleus (Kwok et al., 1998; Deng et al., 2000). This raises the question of whether COP9 signalosome complex is required for Smad4 degradation. To ensure that the decrease in Smad4 steady-state levels is due to protein degradation via the 26S proteasome, the proteasome inhibitors lactacystin and MG132 (Lee and Goldberg, 1998) were added to Mv1Lu cells transfected with Jab1 expression plasmid. Jab1-induced down-regulation of Smad4 protein level was blocked by both lactacystin and MG-132 (Figure 3B). Obviously, the degradation of Smad4 is mediated by the 26S proteasome. Proteins destined for degradation by the 26S proteasome are marked by covalent attachment of ubiquitin chains (DeSalle and Pagano, 2001), which mediate recognition by the 26S proteasome. Thus, we examined whether enhanced Smad4 turnover by Jab1 is through its ability to promote ubiquitylation of Smad4. Flag-Smad4, HA-Jab1 and/or ubiquitin expression plasmids were co-transfected in Mv1Lu cells. Flag-Smad4 protein was double-immunoprecipitated from the cell lysates and blotted with an anti-ubiquitin antibody. As shown in Figure 3C, in the absence of Jab1, little ubiquitylation was observed for Smad4 (lanes 1 and 3) that may be due to endogenous Jab1. A stronger ladder of high-molecular-weight ubiquitin-conjugated Smad4 products was observed when both Jab1 and ubiquitin were ectopically expressed (lane 4). The effects of Jab1 on the turnover of Smad4 were also examined with pulse–chase experiments. The half-life of Smad4 was >10 h in the absence of Jab1. However, ectopic expression of Jab1 shortened the Smad4 half-life significantly. As expected, lactacystin suppressed Jab1-induced Smad4 degradation.
Fig. 2. Interaction of Jab1 with Smad4 in vivo. 293T cells were transfected with 5 µg empty vector, 5 µg Flag-Smad3 or 5 µg Flag-Smad4 per 10-cm dish together with 10 µg HA-Jab1. Immunoprecipitation assays were performed using anti-HA (A) or anti-Flag (B) antibody, and the immunocomplex was detected by western blotting using anti-HA or anti-Flag antibody, respectively. The expression levels of Flag or HA fusion proteins in cells were also detected, as indicated in the lower panels. Mv1Lu cells were incubated overnight in a CO2 incubator in 10-cm dishes. Pre-immune antibody, Jab1 antibody (C), Smad3 antibody or Smad4 antibody (D) were used to immunoprecipitate the endogenous proteins, and the immunocomplex was detected by western blotting using anti-Jab1 or anti-Smad4 antibody, respectively. The expression levels of Jab1, Smad3 or Smad4 in cells were also detected, as indicated in the lower panels.
Fig. 3. Jab1 induces degradation of Smad4. (A) Ectopic expression of Jab1 decreases endogenous Smad4 steady-state levels. Mv1Lu cells were transfected with different doses of HA-Jab1 expression vector or control plasmid and cells were treated with TGF-β (2 ng/ml). After 40 h, total cell extracts were analyzed by immunoblotting using antibodies against Smad4, p27, HA, β-actin or Smad3. (B) Degradation of Smad4 is sensitive to 26S proteasome inhibitors. Mv1Lu cells transfected with HA-Jab1 were incubated with or without the proteasome inhibitors lactacystin (20 µM) and MG132 (20 µM), and expression of endogenous Smad4 was analyzed by immunoblotting. (C) Ubiquitylation of Smad4 mediated by Jab1. COS-1 cells were transfected with Flag-Smad4 together with HA-Jab1 or/and ubiquitin as indicated. Cell lysates were subjected to immunoprecipitation with Flag antiserum, boiled in SDS, and then re-precipitated prior to immunoblotting. Protein expression was confirmed by immunoblotting total cell lysates. (D) Jab1 decreases stability of Smad4 protein. COS-1 cells transfected with Flag-Smad4 plasmid alone (circles) or with HA-Jab1 in the absence (squares) or presence (triangles) of lactacystin were pulse labeled with [35S]methionine for 30 min and chased with excess cold methionine. At the indicated times, cells were collected and 35S-labeled Smad4 in anti-Flag immunoprecipitates was detected by autoradiography of an SDS gel, quantified by phosphorimaging and plotted relative to the amount present at time 0.
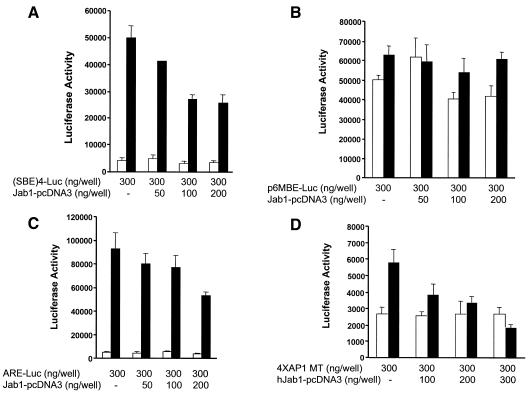
Smad4 is the only common partner for all of the R-Smads and is also known as an activator in R-Smad-induced gene transcription. Therefore, it is likely that degradation of Smad4 affects TGF-β function. To examine the effect of Jab1 on TGF-β-induced gene transcription, Jab1 plasmid was co-transfected with different TGF-β response reporter plasmids, (SBE)4-luc (four repeats of Smad binding element) (Zawel et al., 1998), p6MBE-luc (six repeats of mutant Smad binding element), ARE-luc (activin response element) (Weisberg et al., 1998) and 4×AP1 MT (3TP-luc without AP-1 sites) (Liberati et al., 1999) in Mv1Lu cells. Ectopic expression of Jab1 inhibited TGF-β-induced luciferase activity dose-dependently using (SBE)4-luc, ARE-luc and 4×AP1 MT reporter plasmids (Figure 4A, C and D), whereas expression of Jab1 had no effects on p6MBE-luc luciferase activity since the Smad binding sites were mutated (Figure 4B).
Fig. 4. Jab1 inhibits TGF-β-mediated transcription. Mv1Lu cells were transfected with (SBE)4-luc (A), p6MBE-luc (B), ARE-luc and Fast-1 expression plasmid (C) and 4¥AP1 MT (D) or together with indicated doses of Jab1 expression vector. TGF-β (2 ng/ml) was added in some experiments. Twenty-four hours after transfection, the cells were lysed and the luciferase signal was detected.
We demonstrated that Jab1 interacts specifically with Smad4. The interaction induces ubiquitylation of Smad4, which subsequently undergoes degradation in 26S proteasome. Transfection analysis shows that Jab1 dose-dependently inhibited TGF-β-induced gene transcription in Mv1Lu cells. Since Smad4 is the common partner for all R-Smads, its degradation certainly affects TGF-β transcription activity. Jab1 was initially characterized as a co-activator for AP1-mediated gene transcription, which regulates osteoblast proliferation and bone development (St-Arnaud and Quelo, 1998). Jab1 is also known as the subunit-5 (CSN5) of COP9 signalosome conserved from fission yeast to human (Kwok et al., 1998; Deng et al., 2000; Kapelari et al., 2000). The eight subunit COP9 exhibits homologies to the eight subunits of the 26S proteasome lid complex. Importantly, COP9 signalosome-specific phosphorylation targets human p53 to ubiquitin–26S proteasome-dependent degradation, which is mediated by Jab1/CSN5. The conserved p53 phosphorylation site is found in Smad4 linker region, and part of the Jab1 consensus interaction region of c-Jun, p27 and p53 is also found in the Smad4 MH2 domain. Jab1-mediated phosphorylation of Smad4 could be an important step for its degradation. Recent data indicate that the components of E3 ubiquitin ligase SCF interact with COP9 signalosome and mediate protein degradation (Lyapina et al., 2001; Schwechheimer et al., 2001). This observation provides important information to identify the E3 ligase for Jab1-mediated Smad4 degradation.
METHODS
Two-hybrid library screening. A full-length Smad4 coding sequence was cloned into pGBT9 (Clontech) to generate the pGBT9-Smad4 bait plasmid. The human breast cancer pACT2 cDNA library (Clontech) was screened with the pGBT9-Smad4 bait plasmid according to the manufacturer’s instructions (Clontech).
GST pull-down assay. cDNA fragments corresponding to the different regions of Smad4 were amplified by PCR and sequenced to confirm sequence integrity. cDNA fragments were inserted into pGEX in-frame with GST. GST fusion proteins were expressed in bacteria and purified as described previously (Shi et al., 1999). The pull-down assay was performed as outlined previously (Wan et al., 2001). Briefly, Jab1 protein was synthesized using the TNT-coupled transcription and translation system (Promega) with linearized pcDNA3-Jab1 plasmid according to the manufacturer’s instructions. The production of Jab1 was confirmed by western blotting using antibody against Jab1. An equivalent amount (1 µg) of purified GST–Smad4 or its derivative fusion proteins was preincubated with in vitro translated Jab1 protein (5 µl) for 30 min at 4°C. Following the addition of GST–agarose, the samples were incubated for another 30 min at 4°C. The agarose beads were washed four times in a PBS 0.1% Triton X-100 solution, and bound proteins were eluted by boiling in 2× SDS buffer for 5 min before loading onto a 12% SDS–polyacrylamide gel and blotted to nitrocellulose. Bounded Jab1 was detected by immunoblotting with antibody against Jab1.
Immunoprecipitation and western blotting. Cells were lysed in lysis buffer containing 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholic acid, 50 mM Tris buffer pH 7.5, 1 mM phenylmethylsulfonyl fluoride, 10 µg/ml aprotinin and 10 µg/ml leupeptin. For immunoprecipitation, lysed cells were incubated with different antibodies indicated in the figures. Immunocomplexes were washed and then separated on a 12% SDS–polyacrylamide gel and blotted to nitrocellulose. All blots were developed by the ECL technique (Amersham).
Ubiquitylation and proteasome-dependent degradation assays. COS-1 cells were transfected with Flag-tagged Smad4, HA-tagged Jab1 and HA-tagged HA in the presence or absence of lactacystin. Forty hours after transfection, cell lysates were immunoprecipitated using antibody against Flag (Babco), boiled in SDS and then re-precipitated prior to immunoblotting. To detect ubiquitylation of precipitated Smad4, western blotting was performed using anti-ubiquitin antibody (Santa Cruz). The expression levels of Flag-Smad4 or HA-Jab1 in cell lysates were detected by antibodies against Flag or HA, respectively. Degradation of Smad4 was analyzed by western blotting. Twenty-four hours after transfection with HA-Jab1, Mv1Lu cells were treated for 6 h with or without the proteasome inhibitors lactacystin (20 µM) and MG132 (20 µM). Cell lysates were subjected to SDS–PAGE and western blotting.
Pulse–chase assay. COS1 cells transiently expressing Flag-Smad4 were incubated in methionine-free serum-free medium for 2 h and then pulse-labeled for 30 min with [35S]methionine (125 MBq/ml) (Zhang et al., 2001). After washing with PBS, the cells were then incubated in the medium containing 10% FBS and methionine for 0, 1, 3 and 6 h. The cells were lysed with the lysis buffer. Samples containing equal amounts of protein were immunoprecipitated with an anti-Flag antibody and the proteins were separated on a 12% SDS–polyacrylamide gel. The gel was dried, and proteins were visualized by autophotography.
Transfection and luciferase activity. Mv1Lu cells (5 × 104 cells per 24-well plate) were transfected using Tfx-50 with 0.3 µg luciferase reporter and Jab1 expression plasmid in the absence or presence of TGF-β. Fast-1 expression plasmid was also co-transfected with ARE-luc promoter. Total DNA was kept constant by the addition of pcDNA3 plasmid. Luciferase activities were assayed 24 h after transfection using the Dual-Luciferase™ assay kit (Promega) according to the manufacturer’s instructions. Luciferase values shown in the figures are representative of transfection experiments performed in triplicate in at least three independent experiments.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Mario Ascoli for the HA-Jab1 expression plasmid and Dr Rik Derynck for the Smad4 expression vector. We thank Drs Zafar Nawaz and Bert O’Malley for an expression plasmid for HA-ubiquitin, Dr Malcolm Whitman for the ARE-luc and Fast-1 expression plasmids, Dr Bert Vogelstein for SBE4-luc and p6MBE-luc luciferase reporters and Dr Xiao-Fan Wang for the 4×AP1 MT luciferase reporter gene. We also thank Janice Walker for proofreading the manuscript. This work was supported by National Institutes of Health grants DK53757 and DK57501 (to Xu Cao).
REFERENCES
- Bech-Otschir D., Kraft, R., Huang, X., Henklein, P., Kapelari, B., Pollmann, C. and Dubiel, W. (2001) COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J., 20, 1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian J.L. (2000) BMP, Wnt and Hedgehog signals: how far can they go? Curr. Opin. Cell Biol., 12, 244–249. [DOI] [PubMed] [Google Scholar]
- Claret F.X., Hibi, M., Dhut, S., Toda, T. and Karin, M. (1996) A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature, 383, 453–457. [DOI] [PubMed] [Google Scholar]
- Deng X.W. et al. (2000) Unified nomenclature for the COP9 signalosome and its subunits: an essential regulator of development. Trends Genet., 16, 202–203. [DOI] [PubMed] [Google Scholar]
- DeSalle L.M. and Pagano, M. (2001) Regulation of the G1 to S transition by the ubiquitin pathway. FEBS Lett., 490, 179–189. [DOI] [PubMed] [Google Scholar]
- Ebisawa T., Fukuchi, M., Murakami, G., Chiba, T., Tanaka, K., Imamura, T. and Miyazono, K. (2001) Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem., 276, 12477–12480. [DOI] [PubMed] [Google Scholar]
- Kapelari B., Bech-Otschir, D., Hegerl, R., Schade, R., Dumdey, R. and Dubiel, W. (2000) Electron microscopy and subunit–subunit interaction studies reveal a first architecture of COP9 signalosome. J. Mol. Biol., 300, 1169–1178. [DOI] [PubMed] [Google Scholar]
- Kavsak P., Rasmussen, R.K., Causing, C.G., Bonni, S., Zhu, H., Thomsen, G.H. and Wrana, J.L. (2000) Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF β receptor for degradation. Mol. Cell, 6, 1365–1375. [DOI] [PubMed] [Google Scholar]
- Kingsley D.M. (1994) The TGF-β superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev., 8, 133–146. [DOI] [PubMed] [Google Scholar]
- Kwok S.F., Solano, R., Tsuge, T., Chamovitz, D.A., Ecker, J.R., Matsui, M. and Deng, X.W. (1998) Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell, 10, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H. and Goldberg, A.L. (1998) Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol., 8, 397–403. [DOI] [PubMed] [Google Scholar]
- Liberati N.T., Datto, M.B., Frederick, J.P., Shen, X., Wong, C., Rougier-Chapman, E.M. and Wang, X.F. (1999) Smads bind directly to the Jun family of AP-1 transcription factors. Proc. Natl Acad. Sci. USA, 96, 4844–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Liang, M. and Feng, X.H. (2000) Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-β signaling. J. Biol. Chem., 275, 36818–36822. [DOI] [PubMed] [Google Scholar]
- Lyapina S. et al. (2001) Promotion of NEDD–CUL1 conjugate cleavage by COP9 signalosome. Science, 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- Massague J. (1998) TGF-β signal transduction. Annu. Rev. Biochem., 67, 753–791. [DOI] [PubMed] [Google Scholar]
- Miyaki M. et al. (1999) Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene, 18, 3098–3103. [DOI] [PubMed] [Google Scholar]
- Moses H.L. and Serra, R. (1996) Regulation of differentiation by TGF-β. Curr. Opin. Genet. Dev., 6, 581–586. [DOI] [PubMed] [Google Scholar]
- O’Brien C. (1996) New tumor suppressor found in pancreatic cancer. Science, 271, 294. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M. and Deng, X.W. (2001) Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science, 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Shi X., Yang, X., Chen, D., Chang, Z. and Cao, X. (1999) Smad1 interacts with homeobox DNA-binding proteins in bone morphogenetic protein signaling. J. Biol. Chem., 274, 13711–13717. [DOI] [PubMed] [Google Scholar]
- St-Arnaud R. and Quelo, I. (1998) Transcriptional coactivators potentiating AP-1 function in bone. Front. Biosci., 3, D838–D848. [DOI] [PubMed] [Google Scholar]
- ten Dijke P., Miyazono, K. and Heldin, C.H. (2000) Signaling inputs converge on nuclear effectors in TGF-β signaling. Trends Biochem. Sci., 25, 64–70. [DOI] [PubMed] [Google Scholar]
- Tomoda K., Kubota, Y. and Kato, J. (1999) Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature, 398, 160–165. [DOI] [PubMed] [Google Scholar]
- Wall N.A. and Hogan, B.L. (1994) TGF-β related genes in development. Curr. Opin. Genet. Dev., 4, 517–522. [DOI] [PubMed] [Google Scholar]
- Wan M., Shi, X., Feng, X. and Cao, X. (2001) Transcriptional mechanisms of bone morphogenetic protein-induced osteoprotegrin gene expression. J. Biol. Chem., 276, 10119–10125. [DOI] [PubMed] [Google Scholar]
- Weisberg E., Winnier, G.E., Chen, X., Farnsworth, C.L., Hogan, B.L. and Whitman, M.A. (1998) Mouse homologue of FAST-1 transduces TGF β superfamily signals and is expressed during early embryogenesis. Mech. Dev., 79, 17–27. [DOI] [PubMed] [Google Scholar]
- Wrana J.L. (2000) Regulation of Smad activity. Cell, 100, 189–192. [DOI] [PubMed] [Google Scholar]
- Xu J. and Attisano L. (2000) Mutations in the tumor suppressors Smad2 and Smad4 inactivate transforming growth factor signaling by targeting Smads to the ubiquitin–proteasome pathway. Proc. Natl Acad. Sci. USA, 97, 4820–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L., Dai, J.L., Buckhaults, P., Zhou, S., Kinzler, K.W., Vogelstein, B. and Kern, S.E. (1998) Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell, 1, 611–617. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chang, C., Gehling, D.J., Hemmati-Brivanlou, A. and Derynck, R. (2001) Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc. Natl Acad. Sci. USA, 98, 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Kavsak, P., Abdollah, S., Wrana, J.L. and Thomsen, G.H. (1999) A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature, 400, 687–693. [DOI] [PubMed] [Google Scholar]