A Novel Pathway for Mammary Epithelial Cell Invasion Induced by the Helix-Loop-Helix Protein Id-1 (original) (raw)
Abstract
Mammary epithelial cells undergo changes in growth, invasion, and differentiation throughout much of adulthood, and most strikingly during pregnancy, lactation, and involution. Although the pathways of milk protein expression are being elucidated, little is known, at a molecular level, about control of mammary epithelial cell phenotypes during normal tissue morphogenesis and evolution of aggressive breast cancer. We developed a murine mammary epithelial cell line, SCp2, that arrests growth and functionally differentiates in response to a basement membrane and lactogenic hormones. In these cells, expression of Id-1, an inhibitor of basic helix-loop-helix transcription factors, declines prior to differentiation, and constitutive Id-1 expression blocks differentiation. Here, we show that SCp2 cells that constitutively express Id-1 slowly invade the basement membrane but remain anchorage dependent for growth and do not form tumors in nude mice. Cells expressing Id-1 secreted a ∼120-kDa gelatinase. From inhibitor studies, this gelatinase appeared to be a metalloproteinase, and it was the only metalloproteinase detectable in conditioned medium from these cells. A nontoxic inhibitor diminished the activity of this metalloproteinase in vitro and repressed the invasive phenotype of Id-1-expressing cells in culture. The implications of these findings for normal mammary-gland development and human breast cancer were investigated. A gelatinase of ∼120 kDa was expressed by the mammary gland during involution, a time when Id-1 expression is high and there is extensive tissue remodeling. Moreover, high levels of Id-1 expression and the activity of a ∼120-kDa gelatinase correlated with a less-differentiated and more-aggressive phenotype in human breast cancer cells. We suggest that Id-1 controls invasion by normal and neoplastic mammary epithelial cells, primarily through induction of a ∼120-kDa gelatinase. This Id-1-regulated invasive phenotype could contribute to involution of the mammary gland and possibly to the development of invasive breast cancer.
The epithelial cells of the mammary gland undergo coordinate changes in growth, differentiation, and invasion of the surrounding ECM during embryonic development and puberty, and throughout much of adulthood during each menstrual cycle. Particularly striking changes occur during pregnancy, lactation, and involution. The molecular mechanisms that control the growth and functional differentiation of mammary epithelial cells are slowly being elucidated, but far less is known about the transient invasive behavior of normal breast epithelial cells.
Normal breast epithelial cells proliferate and invade the surrounding ECM during the fetal and postnatal development of the gland, and then more vigorously at puberty as the branches of the mammary epithelial tree are formed. After puberty, there are minor waves of mammary epithelial-cell proliferation during each estrous cycle (16, 46). The most striking activity of mammary epithelial-cell proliferation and invasion occurs during pregnancy, as the gland expands in preparation for lactation (45). The proliferation and invasion of breast epithelial cells cease during late pregnancy, whereupon the cells functionally differentiate—that is, they express and secrete milk proteins (44). The epithelial cells remain proliferatively quiescent and functionally differentiated throughout lactation. At the end of lactation, the mammary gland undergoes involution, during which time there is an early and transient reactivation of epithelial-cell proliferation, followed by extensive ECM degradation and epithelial-cell death by apoptosis. The extensive remodeling of the mammary gland that occurs during involution entails the stepwise activation of several MMPs by the stromal and epithelial cells of the gland (29, 41). The involuting gland eventually returns to its prepregnancy structure.
Invasion of the ECM by normal epithelial cells must be tightly regulated and self-limiting. This control is clearly important for the mammary gland to develop and function normally. Control over normal invasive properties is also important in order to prevent neoplastic cells from invading the surrounding ECM. Most cancers develop from epithelial cells, and a hallmark of malignancy is invasion of the ECM by neoplastic epithelial cells (38). In many experimental models of tumorigenesis, an invasive phenotype develops subsequent to neoplasia and often entails expression of ECM-degrading enzymes commonly expressed by mesenchymal or stromal cells. These enzymes include the MMPs stromelysin and the 72- and 92-kDa collagenases (19, 48). It is not clear whether tumor cells express these MMPs because they are normally expressed when epithelial cells transiently invade the ECM during normal tissue morphogenesis or because they frequently acquire mesenchymal characteristics upon transformation. It was recently shown by in situ hybridization that these MMPs are expressed by stromal fibroblasts during certain stages of ductal and alveolar mammary morphogenesis as well as during involution (29, 49).
In order to study normal and abnormal mammary epithelial-cell phenotypes, we developed a murine mammary epithelial-cell line, SCp2, whose growth and differentiation can be controlled in culture (8). SCp2 cells are an immortal line that originated from a heterogeneous cell population derived from a midpregnancy mouse mammary gland (7, 37). SCp2 cells grow well in serum on tissue culture plastic, where they express keratins and exhibit other epithelial characteristics. When serum is removed and they are given lactogenic hormones (insulin, prolactin, and hydrocortisone) and basement membrane components, SCp2 cells first arrest growth, then aggregate and form alveolar structures, and finally express high levels of several milk proteins (8, 36).
We have shown that the differentiation of SCp2 cells requires a sharp decline in the expression of the HLH protein Id-1 (9). Id genes encode a small family of proteins that prevent bHLH transcription factors from binding DNA (4). bHLH transcription factors comprise a large family of sequence-specific DNA binding proteins that activate the transcription of cell- and tissue-specific genes. bHLH proteins act as obligate dimers: they dimerize through the HLH domains and bind DNA through the composite basic domain. Id proteins contain HLH domains and therefore dimerize with bHLH proteins. However, because Id proteins lack basic domains, Id-bHLH heterodimers cannot bind DNA. Thus, Id proteins negatively regulate bHLH transcription factors. The bHLH superfamily contains both ubiquitous and lineage-specific transcription factors that direct many developmental and differentiation processes (20). Two of the four known Id proteins (Id-1 and Id-3) are nearly ubiquitously expressed, whereas the other two Id proteins (Id-2 and Id-4) have a more restricted pattern of expression (35). Thus, lineage-specific differentiation is determined by tissue-specific bHLH genes, which, in turn, are posttranslationally regulated by a small number of Id genes. Whether and how bHLH proteins participate in the differentiation of breast epithelial cells is not yet known.
Id-1 was the first Id protein to be identified (4). Since its initial discovery in myoblasts, it has been shown to be expressed by a variety of cell types and to inhibit the differentiation of myoblasts (18), several hematopoietic cell types (23, 26, 40), trophoblasts (6), and mammary epithelial cells (9). Id-1 was also found to be serum inducible in fibroblasts, where its expression is essential for progression into the S phase of the cell cycle (14). In contrast to the closely related Id-2 protein, Id-1 does not physically associate with the retinoblastoma tumor suppressor protein pRb (15, 17) but can functionally interact with a pRb-regulated pathway for entry into S phase (15).
Id-1 expression declines rapidly when SCp2 cells are induced to differentiate. As long as the cells remain in contact with a basement membrane and lactogenic hormones, Id-1 remains repressed and the cells do not proliferate, but they express milk proteins. By contrast, SCp2 cells that constitutively express Id-1 fail to differentiate, as judged by the expression of milk proteins, but nonetheless transiently arrest growth and form loose alveolar structures. After several days, cells that constitutively express Id-1 dissociate from each other and subsequently resume growth (9).
Here, we show that Id-1 expression confers upon SCp2 cells the ability to migrate and invade the basement membrane. However, cells that constitutively express Id-1 neither grow in soft agar nor form tumors in nude mice. Id-1 expression correlates strongly with expression of an apparently novel gelatinase of approximately 120 kDa, an MMP, which is also expressed during involution. The activity of this MMP was critical for the Id-1-regulated invasive phenotype. We also show that Id-1 expression correlates with the degree of differentiation and invasiveness of human breast cancer cells. The least-differentiated and most highly invasive cells express constitutively high levels of Id-1 and also secrete a 120-kDa gelatinase. Our results suggest that Id-1 is a regulator of the invasive phenotype of normal and neoplastic mammary epithelial cells and that it acts, at least in part, by controlling expression of a 120-kDa gelatinase. The invasive phenotype conferred by Id-1 is not a consequence of tumorigenic transformation, although it may be appropriated in a subset of aggressive breast cancers. Our data provide new insights into the control of breast epithelial-cell invasion and suggest that one or more bHLH transcription factors may repress the invasive phenotype in normal as well as neoplastic breast epithelial cells.
MATERIALS AND METHODS
Abbreviations.
AEBSF, 4-(2-aminoethyl)-benzenesulfonyl fluoride; bHLH, basic helix-loop-helix; BSA, bovine serum albumin; DAPI, 4′,6′-diamidino-2-phenylindole; DMEM, Dulbecco’s modified Eagle’s medium; DMSO, dimethyl sulfoxide; EHS, Englebreth Holm Swarm tumor; ECM, extracellular matrix; F12, Ham’s F-12 medium; FBS, fetal bovine serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HLH, helix-loop-helix; MMP, matrix metalloproteinase; PBS, phosphate-buffered saline; PMSF, phenylmethylsulfonyl fluoride; RT-PCR, reverse transcription-PCR.
Cell culture.
SCp2 cells were grown in a 1:1 mixture of DMEM and F12 (DMEM-F12) containing 5% heat-inactivated FBS, insulin (5 μg/ml), and gentamicin (50 μg/ml) (growth medium) at 37°C in a humidified 5% CO2 atmosphere, as previously described (8). To induce differentiation, cells were plated at 5 × 104/cm2 atop basement membrane components in DMEM-F12 lacking serum but containing lactogenic hormones (insulin, 5 μg/ml; hydrocortisone, 1 μg/ml; prolactin, 3 μg/ml) (9). Unless otherwise indicated, cells were cultured for 5 days before analysis. Basement membrane ECM either was purified from EHS tumors by the method of Taub et al. (42) or was supplied as Matrigel from Collaborative Research.
SCp2 cells were transfected with the murine Id-1 cDNA driven by the mouse mammary tumor virus promoter as previously described (9). The transfected cells were initially pooled. Single cell-derived clones were subsequently derived by plating cells at limiting dilutions in 24-well plates. After 10 days, wells with visible colonies were trypsinized and replated onto 35-mm-diameter dishes. When nearly confluent, the cells were replated onto 100-mm-diameter dishes. The population was expanded by subculturing at a ratio of 1:4, and cells were used after 5 to 8 passages after the first 1:4 subculture.
The human breast cancer cell lines T47D, MCF-7, Hs578T, BT-549, MDA-MB-231, ZR75-1, and SKBR-3 were purchased from the American Type Culture Collection. The MDA-MB-436 cell line was originally purchased from the American Type Culture Collection and was given to us by R. Lupu (Berkeley National Laboratory). MDA-MB-435 cells were derived from the original cell line by selection in nude mice for the highly aggressive subpopulation (37a). Cells were passaged in DMEM containing 10% FBS and insulin (5 μg/ml; Sigma). For serum-free conditions, FBS was omitted from the medium.
DNA synthesis and autoradiography.
Cells plated on coverslips were labeled with [3H]methylthymidine (10 μCi/ml; 60 to 70 Ci/mmol) for 24 h, washed twice with PBS, then fixed for 5 min with a 1:1 (vol/vol) mixture of acetone and methanol at −20°C. Where indicated, cell nuclei were stained for 2 min with DAPI diluted 1:10,000 in PBS. The coverslips were air dried, coated with Kodak NTB2 emulsion (1:2 dilution), and exposed for 16 to 24 h. The coverslips were developed with D-19, fixed with Kodak Rapid-Fix, and viewed by phase-contrast microscopy.
Boyden chamber invasion assays.
Invasion assays were performed in modified Boyden chambers with 8-μm-pore-size filter inserts for 24-well plates (Collaborative Research). Filters were coated with 10 to 12 μl of ice-cold basement membrane ECM at 8 to 12 mg of protein/ml. Cells (0.5 × 105 to 1 × 105) were added to the upper chamber in 200 μl of DMEM-F12. The lower chamber was filled with 300 μl of NIH 3T3 cell-conditioned medium. Where indicated, GM6001 was added at 0.2 mM to both chambers immediately after cell plating. After a 16- to 20-h incubation, the cells were fixed with 2.5% glutaraldehyde in PBS and stained with 0.5% toluidine blue in 2% Na2CO3. Cells that remained in the basement membrane or attached to the upper side of the filter were removed with paper towels. Cells on the lower side of the filter were examined by light microscopy and counted.
Anchorage-dependent growth assays.
Liquefied 2% agarose was mixed with an equal volume of 2× DMEM-F12 growth medium lacking serum and supplemented with insulin (10 μg/ml) and gentamicin (100 μg/ml) (2× medium). One milliliter of the mixture was layered onto 35-mm-diameter dishes to create a 1% agarose base. Liquefied 0.6% agarose was mixed with an equal volume of 2× medium, and 10 ml of this solution was mixed with 1 ml of growth medium containing 105 cells to yield 104 cells/ml in 0.27% agarose; 1 ml of this cell suspension was layered on top of the 1% agarose base, and 1 ml of DMEM-F12 containing 5% FBS was added. The cells were incubated for 14 days, after which representative fields were photographed under phase-contrast microscopy.
Tumorigenicity assays.
Cells were injected subcutaneously into nude mice at 4 × 106 cells per site, two sites per animal, and two animals for each cell type (TCL1, SCg6, SCp2, SCp2–antisense Id-1, and SCp2–Id-1). Animals injected with TCL1 and SCg6 cells developed easily detectable tumors (at least 1 cm3) within 3 weeks and were sacrificed after 4 weeks. The remaining animals remained tumor negative for a minimum of 5 months.
Immunofluorescence.
Cells cultured on coverslips were washed with PBS, fixed for 5 min with acetone-methanol (1:1, vol/vol) at −20°C, permeabilized for 5 min with 1% Triton X-100 in PBS, and washed with PBS. A rabbit polyclonal antiserum raised against bovine keratins (Dako, Carpinteria, Calif.) was diluted 1:10 in 0.2% BSA in PBS and applied for 60 min at 37°C, followed by three washes in PBS. The coverslips were then incubated with biotin-conjugated anti-rabbit antibody (1:100 dilution; Amersham Corp.) for 30 min at 37°C and were washed three times in PBS. Finally, the coverslips were incubated with fluorescein isothiocyanate-conjugated streptavidin (1:100 dilution; Amersham Corp.) for 30 min at 37°C and were washed in PBS. Cell nuclei were stained with DAPI, as described above, and the coverslips were mounted in glycerol-gelatin (Sigma) for viewing by epifluorescence.
RNA isolation and analysis.
Total cellular RNA was isolated and purified as described by Chomczynski and Sacchi (5). The RNA (10 μg) was size fractionated by electrophoresis through formaldehyde-agarose gels and transferred to a nylon membrane (Hybond N from Amersham Corp.). The membrane was hybridized to 32P-labeled probes prepared by random oligonucleotide priming, washed, and exposed to XAR-5 film for autoradiography as described by Maniatis et al. (31). The β-casein probe was the 540-bp mouse cDNA (from J. Rosen, Baylor College of Medicine, Houston, Tex.), and the Id-1 probe was either the murine Id-1 cDNA (4) or the human Id-1 cDNA (14).
RT-PCR and Southern analysis.
Transcripts for murine gelatinases A (72-kDa MMP) and B (92-kDa MMP) were detected by RT-PCR. cDNA was synthesized from total RNA by using SuperscriptII Reverse TranscriptaseII (Gibco-BRL), and 100 ng was used for PCR. The 5′ and 3′ PCR primers were TTGAGAAGGATGGCAAGTATGG and ACACCTTGCCATCGTTGC for gelatinase A, GGCGTGTCTGGAGATTCGA and AGGGTCCACCTTGGTCACC for gelatinase B, and ACCACAGTCCATGCCATCAC and TCCACCACCCTGTTGCTGTA for GAPDH. PCR was performed in 20 mM Tris-HCl (pH 8.8)–2 mM MgSO4–10 mM KCl–10 mM (NH4)2SO4–0.1% Triton X-100–100 μg of BSA/ml–0.125 mM deoxynucleoside triphosphates–0.8 μM each PCR primer–0.05 U of Pfu DNA polymerase/μl by using 35 cycles for amplification of gelatinase cDNAs and 25 cycles for amplification of GAPDH cDNA. The cycle conditions were 1 min of denaturation at 94°C, 1 min of annealing at 58°C, and 30 s of extension at 72°C. For Southern analysis, one-fifth of the PCR reaction product was separated on a 2.1% agarose gel, transferred to a nylon membrane (Hybond N+), and hybridized with cDNA inserts labeled with 32P by random priming. cDNAs encoding murine gelatinase A or B (33, 34) were a gift from Z. Werb, University of California, San Francisco, and the GAPDH cDNA was obtained from Clontech (Palo Alto, Calif.). Hybridization was carried out in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt’s solution, 0.5% sodium dodecyl sulfate, and 50% formamide at 42°C overnight. The membranes were washed at a final stringency of 0.2× SSC and 0.1% sodium dodecyl sulfate at 68°C and were exposed to XAR-5 film for autoradiography.
Zymography.
Proliferating cells (106 in 100-mm-diameter dishes) were shifted to serum-free medium for 2 to 3 days, after which they were given 10 ml of fresh serum-free medium. Forty-eight hours later, the conditioned medium was collected and concentrated 10- to 15-fold by using 10-kDa-cutoff filters (Millipore, Bedford, Mass.). The concentrated medium was analyzed on casein and gelatin substrate gels, as described by Fisher and Werb (10) and Talhouk et al. (41). Briefly, gels consisted of 8 to 10% polyacrylamide and 3 mg of α-casein or gelatin (Sigma)/ml. Concentrated conditioned medium was mixed with nonreducing Laemmli sample buffer and incubated at 37°C for 15 min. After electrophoresis, the gels were incubated for 1 h in 2.5% Triton X-100 at room temperature, followed by 24 to 48 h in substrate buffer (100 mM Tris-HCl [pH 7.4]–15 mM CaCl2) in the absence or presence of GM6001 (0.2 mM in DMSO; supplied by Glycomed Corporation and obtained from Z. Werb [12]), EDTA (10 mM), _ortho_-phenanthroline (1 mM in DMSO; Sigma), PMSF (5 mM), or AEBSF (0.5 mM; Calbiochem). Where appropriate, control gels were incubated with buffer containing solvent only. The gels were stained with Coomassie blue for 30 min and were destained with 30% methanol–10% acetic acid. Caseinase and gelatinase activities were visible as clear bands, indicative of proteolysis of the substrate protein.
RESULTS
Id-1 induces an invasive, migratory phenotype in mammary epithelial cells.
SCp2 mammary epithelial cells grow as a monolayer in 5% serum. When given lactogenic hormones and basement membrane ECM in serum-free medium, they arrest growth, form three-dimensional alveolar structures, and express the milk protein β-casein (8). Alveoli formed by SCp2 cells are stable, maintaining their structure and β-casein expression for more than 2 weeks. Under these conditions, Id-1 is not expressed. By contrast, SCp2 cells that constitutively express Id-1 form poorly compacted alveoli that become increasingly disorganized; after 6 to 8 days, cells at the periphery detach from the structure and synthesize DNA (9).
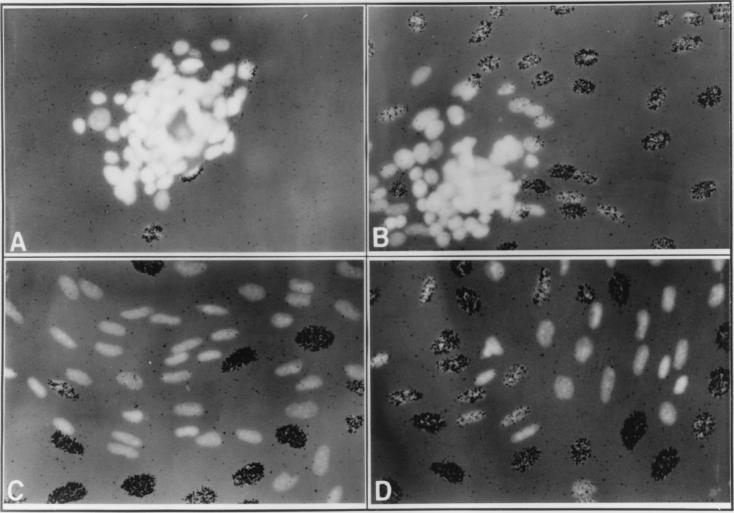
Using a pooled population of SCp2 cells that constitutively express a murine Id-1 transgene (SCp2–Id-1 cells) (9), we more precisely monitored the fate of cells that detached from the alveolar structure. Within 10 days, approximately 30 to 40% of the SCp2–Id-1 alveolar structures showed substantial disintegration. Following detachment from the alveolar structure, SCp2–Id-1 cells actively invaded and migrated through the surrounding ECM (Fig. 1). The migrating cells had an elongated nuclear morphology, compared to the rounded nuclei of cells in the early stages of disaggregation. Initial detachment and invasion occurred in the absence of cell proliferation (Fig. 1A). However, 2 to 4 days after initial detachment, SCp2–Id-1 cells that had migrated extensively through the ECM were abundant, and many of these cells synthesized DNA (Fig. 1B to D). For the most part, DNA synthesis was evident only in cells that had migrated some distance from the alveolar structure. Thus, the initiation of invasion and migration was not due to resumption of growth; rather, cells resumed proliferation only after they had detached and migrated from the three-dimensional structure. As previously described (9), spheres comprised of control cells transfected with the vector alone were very stable, remaining viable and morphologically unchanged even after more than 10 days on basement membrane ECM.
FIG. 1.
Instability of the three-dimensional organization and loss of growth arrest of SCp2–Id-1 cells. A pooled population of SCp2–Id-1 cells was induced to differentiate for 8 (A), 10 (B), or 12 (C and D) days, [3H]thymidine was added for 24 h preceding fixation, and the cells were then stained with DAPI and processed for autoradiography as described in Materials and Methods. Shown are the DAPI fluorescence and autoradiography. Depending on the batch of EHS ECM or Matrigel, disaggregation of the three-dimensional structures and resumption of DNA synthesis occurred 1 to 2 days earlier or later than in the experiments for which results are shown here. Magnification, ×300.
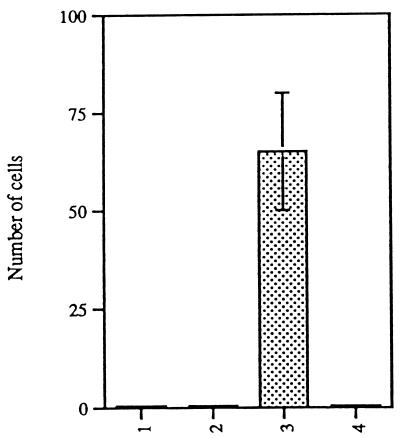
To quantify the invasion and migration of SCp2–Id-1 cells, they and control cells were assayed in Boyden chambers (2). Cells were added to the upper portion of the chamber; conditioned medium from mouse fibroblasts, used as a source of chemoattractants (2), was added to the lower compartment. The porous filter separating the two compartments was coated with basement membrane ECM. After a 16- to 20-h incubation, cells that had migrated through the ECM to the lower surface of the filter were fixed, stained, and counted (Fig. 2). The 16- to 20-h incubation time ensured that only a small fraction of invasive cells migrated through the filter, which in turn ensured that the fraction of migratory cells was small enough to score reliably.
FIG. 2.
SCp2–Id-1 cells invade the ECM and migrate in a Boyden chamber. Parental SCp2 cells (lane 1), SCp2 cells transfected with an insertless vector (lane 2), SCp2–Id-1 cells (lane 3), and SCp2 cells transfected with Id-1 in the antisense orientation (lane 4) were plated on ECM-coated filters in Boyden chambers; the number of cells that migrated through the filter after 16 to 20 h was determined as described in Materials and Methods. Error bars indicate standard deviations from three or four independent fields. The data shown are from one of five independent experiments which showed very similar differences among the cell types.
Four types of cells were compared in this assay: (i) parental SCp2 cells, (ii) SCp2 cells transfected with an insertless vector, (iii) SCp2–Id-1 cells, and (iv) SCp2 cells transfected with the Id-1 cDNA in the antisense orientation. Of these cell types, only SCp2–Id-1 cells were invasive. Under these assay conditions, none of the control (parental or insertless-vector) cells and none of the cells expressing antisense Id-1 migrated through the filter. By contrast, 0.7 to 1% of a population of one of the most invasive breast cancer cell lines (MDA-MB-231, previously described [43]) migrated through the filter, although only about 0.05% of the SCg6-transformed cells, which were previously shown to be invasive (28), migrated through the ECM to the lower surface of the filter (data not shown). In the case of the SCp2–Id-1 cells, 0.2 to 0.3% migrated through the filter. Thus, SCp2–Id-1 cells, which were transfected with a single gene, were 20 to 30% as invasive as one of the most aggressive breast cancer cell lines (which harbors multiple mutations) and four- to sixfold more invasive than their SCg6-transformed counterparts.
We conclude that constitutive expression of the Id-1 gene can induce an invasive and migratory phenotype in nontransformed and nontumorigenic SCp2 mammary epithelial cells.
Constitutive Id-1 expression is not sufficient for anchorage-independent growth or tumorigenicity.
In many model systems of malignant transformation, unregulated expression of normal or activated proto-oncogenes drives cell proliferation, and invasiveness often develops subsequent to, or concomitant with, tumorigenicity. Although Id-1 did not appear in this regard to act like a typical oncogene, we nonetheless asked whether constitutive Id-1 expression transformed SCp2 cells, using the criteria of anchorage-independent growth and tumorigenicity in nude mice.
We first tested the ability of the cells to grow in an anchorage-independent manner. As expected, control cells and cells transfected with Id-1 in the antisense orientation failed to grow in soft agar (Fig. 3A and C). Similarly, SCp2–Id-1 cells failed to form colonies in soft agar, remaining as single cells for at least 14 days (Fig. 3B). It is interesting that, in soft agar, SCp2–Id-1 cells appeared twice as large as control cells; the reason for this size increase is not known. Malignant TCL1 cells (isolated from a murine mammary tumor [28]), used as a positive control, formed large colonies after 14 days in soft agar (Fig. 3D). We conclude that constitutive expression of Id-1 does not induce anchorage-independent growth in SCp2 mammary epithelial cells.
FIG. 3.
SCp2–Id-1 cells do not grow in an anchorage-independent manner. Parental SCp2 cells (A), SCp2–Id-1 cells (B), SCp2 cells expressing an Id-1 antisense vector (C), and TCL1 mammary tumor cells (D) were seeded in soft agar as described in Materials and Methods and photographed 14 days later. Magnification, ×50.
We next tested SCp2–Id-1 cells for their ability to form tumors. Cells were injected subcutaneously into nude mice. The positive control, TCL1 cells, formed tumors (at least 1 cm3) within 3 weeks (Table 1). The same was true for SCg6, a cell line with mesenchymal and transformed properties that was isolated from the same population from which SCp2 cells were isolated (8) (Table 1). By contrast, neither parental SCp2 cells, SCp2 cells expressing the Id-1 antisense cDNA, nor SCp2–Id-1 cells formed tumors after 5 months (Table 1).
TABLE 1.
SCp2–Id-1 cells are not tumorigenica
| Cell type | No. of tumors/no. of sites injected (time) |
|---|---|
| TCL1 | 4/4 (after 3 wk) |
| SCg6 | 4/4 (after 3 wk) |
| SCp2 | 0/4 (after 5 mo) |
| SCp2–antisense Id-1 | 0/4 (after 5 mo) |
| SCp2–Id-1 | 0/4 (after 5 mo) |
We conclude that constitutive Id-1 expression in SCp2 mammary epithelial cells is not sufficient to lead to the transformed phenotypes of anchorage-independent growth in culture and in vivo, despite its ability to induce an invasive phenotype.
Isolation and characterization of cloned SCp2–Id-1 cells.
The experiments described thus far used a pooled population of SCp2–Id-1 cells, which is heterogeneous with respect to Id-1 expression. To eliminate this heterogeneity and better define the role of Id-1 in inducing an invasive phenotype, we isolated single-cell-derived SCp2–Id-1 clones that expressed the Id-1 transgene to varying levels. The clones were assessed for cytokeratin filaments (a general characteristic of epithelial cells), morphology in monolayer culture, and ability to form alveolar structures in response to basement membrane ECM. In addition, RNA was isolated 5 days after the cells were exposed to basement membrane and hormones and was analyzed for Id-1 and β-casein mRNA. The Id-1 transgene mRNA was distinguishable from the endogenous Id-1 mRNA by its slightly larger size; the endogenous transcript was barely detectable under these conditions (9).
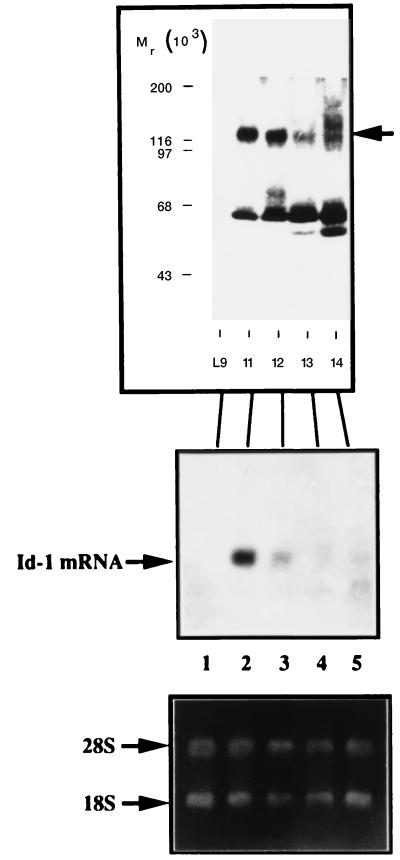
One subclone, SCp2–Id-1A cells, did not express detectable Id-1 transgene mRNA (Fig. 4a, lane 1). These cells grew as compact colonies in monolayer culture and expressed cytokeratin filaments (Fig. 4b, panel B). They also differentiated similarly to untransfected SCp2 cells, as judged by their ability to express high levels of β-casein mRNA (Fig. 4a, lane 1) and form stable alveolar structures (data not shown), like untransfected SCp2 cells. These cells were therefore used as negative controls.
FIG. 4.
Characterization of SCp2 cell clones expressing constitutive Id-1. (a) SCp2–Id-1 cells were plated at limiting dilution, and five independent clones (SCp2–Id-1A through SCp2–Id-1E) were isolated and amplified. Cells from each of these clones were exposed to basement membrane and hormones for 5 days and were analyzed for expression of the Id-1 transgene and β-casein mRNA, as described in Materials and Methods. Shown are the autoradiogram of the Northern blot and the ethidium bromide-stained Northern gel made to confirm RNA integrity and quantitation. Lane 1, SCp2–Id-1A; lane 2, SCp2–Id-1B; lane 3, SCp2–Id-1C; lane 4, SCp2–Id-1D; lane 5, SCp2–Id-1E. (b) SCp2–Id-1A (A and B) and SCp2–Id-1E (C and D) cells were grown in monolayer culture, fixed, and stained with DAPI (A and C) or processed for immunofluorescence by using a pan-keratin antibody (B and D), as described in Materials and Methods.
SCp2–Id-1B and SCp2–Id-1C cells expressed moderate levels of the Id-1 transgene, which were below the levels of Id-1 mRNA expressed by proliferating control cells. These cells expressed lower levels of β-casein mRNA than SCp2–Id-1A cells (Fig. 4a, lanes 2 and 3), but they expressed cytokeratin filaments and formed alveolar structures (data not shown).
Finally, SCp2–Id-1D and SCp2–Id-1E cells expressed high levels of the Id-1 transgene and undetectable levels of β-casein (Fig. 4a, lanes 4 and 5). In monolayer culture, SCp2–Id-1E cells were less cuboidal and grew as more-dispersed entities than SCp2–Id-1A cells (Fig. 4b, panels C and D). Their failure to express β-casein was not due to a loss of epithelial characteristics. SCp2–Id-1E cells, which expressed the highest levels of Id-1, as well as SCp2–Id-1D cells (data not shown), expressed cytokeratin filaments (Fig. 4b, panel D). However, SCp2–Id-1D and SCp2–Id-1E cells, like the pooled SCp2–Id-1 cells, formed only loose alveolar structures, from which they eventually detached and invaded the ECM (see Fig. 7) (data not shown).
FIG. 7.
The invasive phenotype of Id-1-expressing cells is repressed by an MMP inhibitor. SCp2–Id-1A cells in 0.5% DMSO (lane 1), SCp2–Id-1E cells in 0.5% DMSO (lane 2), and SCp2–Id-1E cells in GM6001 (200 μM; 0.5% DMSO) (lane 3) were plated on ECM-coated filters in Boyden chambers, and the numbers of cells that migrated through the membrane after 16 to 20 h were determined as described in Materials and Methods and the legend to Fig. 2. Error bars indicate standard deviations from three to four independent fields; the data shown are from one of three independent experiments.
These results confirm in cloned populations that mammary epithelial cells constitutively expressing Id-1 do not undergo a complete epithelial-to-mesenchymal transition; they retain some epithelial-cell characteristics (such as keratin expression) but fail to functionally differentiate and to maintain three-dimensional organization on the ECM. SCp2–Id-1A and SCp2–Id-1E cells, which express undetectable and high levels of the Id-1 transgene, respectively, were used for further studies.
A potentially novel metalloproteinase is secreted by Id-1-expressing cells.
The ability of SCp2–Id-1 cells to invade the ECM suggested that Id-1 might induce expression of ECM-degrading proteases. The major classes of proteases that degrade ECM are serine, cysteine, and aspartyl proteases, and metalloproteinases (10). The Zn2+-containing, Ca2+-stabilized MMPs are of particular interest because they are implicated in the remodeling of the mammary gland during involution (29, 41) and the initial steps of tumor-cell invasion (25). Of the major MMPs, interstitial collagenase (56 kDa) and gelatinases A (72 kDa) and B (92 kDa) are detectable on gelatin substrate gels, whereas stromelysins (57 kDa for stromelysin-1) and matrilysin (30 kDa) are detectable on casein substrate gels (reviewed by Fisher and Werb [10]; see also reference 22).
We examined the secretion of proteases by SCp2–Id-1A and SCp2–Id-1E cells, using conditioned medium and gelatin or casein substrate gel zymography. Cells were incubated in serum-free medium for 3 days prior to collection of conditioned medium for zymography. Under these conditions, the endogenous Id-1 gene is not expressed (9), and SCp2–Id-1A and SCp2–Id-1E express undetectable and high levels of the Id-1 transgene, respectively.
Gelatin substrate gels showed that SCp2–Id-1A and SCp2–Id-1E cells differed only in the secretion of a high-molecular-mass (approximately 120-kDa) gelatinase. The 120-kDa gelatinase was abundantly expressed by serum-deprived SCp2–Id-1E cells (Fig. 5A, lane 2) as well as SCp2–Id-1D cells (data not shown). Secretion of this 120-kDa gelatinase was not due to clonal variation. Conditioned medium from the uncloned SCp2–Id-1 pooled population also showed a gelatinase of ∼120 kDa (Fig. 5C, lane 2). This gelatinase was undetectable in serum-deprived SCp2–Id-1A (Fig. 5A, lane 1) and control SCp2 (Fig. 5C, lane 1) cells. Thus, secretion of a 120-kDa gelatinase correlated with Id-1 expression.
FIG. 5.
Expression of a 120-kDa gelatinase by Id-1-expressing mammary epithelial cells. (A) Gelatin zymogram of conditioned media from SCp2–Id-1A (lane 1) and SCp2–Id-1E (lane 2) cells. Cells were cultured in serum-free medium, and conditioned media were harvested and analyzed on a gelatin substrate gel, as described in Materials and Methods. (B) Gelatin zymogram of conditioned media from SCp2–Id-1A cells either growth arrested by serum deprivation (lane 1) or growing in 5% serum (lane 2). (C) Gelatin zymogram of conditioned media from control SCp2 cells (lane 1) and an uncloned SCp2–Id-1-transfected pooled population (lane 2). Cells were cultured in serum-free medium. (D) Gelatin zymogram of SCp2–Id-1A (lanes 1, 3, and 5) and SCp2–Id-1E (lanes 2, 4, and 6) cell-conditioned media incubated with DMSO (lanes 1 and 2), the MMP inhibitor GM6001 (0.2 mM in DMSO) (lanes 3 and 4), or the serine proteinase inhibitor PMSF (5 mM in DMSO) (lanes 5 and 6). (E) Casein zymogram of conditioned media from SCp2–Id-1A (lanes 1 and 3) and SCp2–Id-1E (lanes 2 and 4) cells incubated with DMSO (lanes 1 and 2) or GM6001 (lanes 3 and 4). In panels A through D, arrows mark the positions of the 120-kDa MMP.
Secretion of the 120-kDa gelatinase correlated with expression of the endogenous Id-1 gene as well as with that of the Id-1 transgene. Thus, the 120-kDa proteinase was secreted by SCp2–Id-1A cells (in which expression of the Id-1 transgene is undetectable) while they were proliferating in monolayer culture (Fig. 5B, lane 2). Under these conditions, the endogenous Id-1 gene is expressed at high levels (9).
The 120-kDa gelatinase had characteristics of an MMP. It was sensitive to the MMP inhibitors GM6001 (Fig. 5D, lane 4), EDTA, and _ortho_-phenanthroline (data not shown). By contrast, it was insensitive to the serine protease inhibitors PMSF (Fig. 5D, lane 6) and AEBSF (data not shown). The 120-kDa MMP appeared to be the only MMP secreted by Id-1-expressing cells. The two gelatinases with apparent molecular sizes of 70 and 90 kDa, which were expressed by both SCp2–Id-1A and SCp2–Id-1E cells, were not inhibited by any of the MMP inhibitors GM6001 (Fig. 5D, lane 4), _ortho_-phenanthroline, and EDTA (data not shown), and therefore neither is likely to be gelatinase A or B.
Casein substrate gels showed one major caseinase of approximately 50 kDa that was expressed by both SCp2–Id-1A and SCp2–Id-1E cells. This protease was not inhibited by the metalloproteinase inhibitor GM6001 (Fig. 5E). Therefore, it is most likely not the metalloproteinase stromelysin-1.
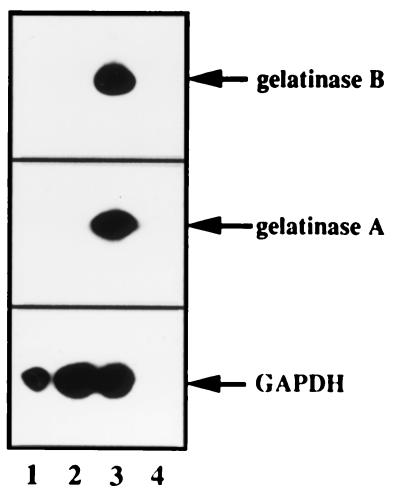
To definitively rule out the possibility that gelatinases A and B were expressed in SCp2 cells, as well as the possibility that the 120-kDa MMP was a complex between gelatinase B and its carrier protein (21), we analyzed RNA by PCR and Southern blotting for gelatinase-A and -B mRNAs. SCp2–Id-1A cells, which do not express the Id-1 transgene, SCp2–Id-1E cells, which express high levels of the Id-1 transgene, and the mesenchyme-like mammary SCg6 cells were deprived of serum for 3 days before RNA was extracted and synthesized into cDNA for PCR analysis (Fig. 6). The 326-bp PCR product expected from the gelatinase-A cDNA and the 190-bp product expected for the gelatinase-B cDNA were detected only in SCg6 cells (Fig. 6, lane 3). We conclude that SCp2 cells, whether or not they express Id-1, do not express gelatinase A or B and that therefore the 120-kDa gelatinase is not a gelatinase B-containing complex (21).
FIG. 6.
SCp2 cells do not express gelatinase A or B. SCp2 cells were serum deprived for 3 days before RNA was extracted, transcribed into cDNA, and analyzed by PCR for gelatinase-A and -B cDNA sequences, as described in Materials and Methods. Arrows indicate the positions of the amplified products for gelatinases A and B and the control gene, encoding GAPDH. Lane 1, SCp2–Id-1A cells; lane 2, SCp2–Id-1E cells; lane 3, SCg6 cells; lane 4, no cDNA control.
We conclude that SCp2 mammary epithelial cells secrete a single detectable MMP, having an apparent molecular size of 120 kDa, when they express Id-1. This MMP does not belong to the stromelysin subclass of MMPs, which degrades casein, but rather is a type IV collagenase MMP family member and thus degrades gelatin, a denatured collagen.
The Id-1-related MMP is essential for the invasive phenotype of SCp2 cells.
Because the 120-kDa MMP appears to be the only proteinase whose secretion correlates with Id-1 expression, and constitutive Id-1 expression renders cells invasive, we explored the possibility that this MMP is critical for the invasive phenotype of mammary epithelial cells.
We first tested the abilities of SCp2–Id-1A and SCp2–Id-1E cells to invade basement membrane ECM in a Boyden chamber invasion assay (Fig. 7). SCp2–Id-1A cells, like untransfected SCp2 cells (Fig. 2), were not invasive, or only minimally invasive, in this assay (Fig. 7). Under the assay conditions, the endogenous Id-1 gene is not expressed and SCp2–Id-1A cells express undetectable levels of the Id-1 transgene. By contrast, SCp2–Id-1E cells, like uncloned SCp2–Id-1 cells (Fig. 2), were demonstrably invasive under the same conditions, consistent with the high levels of the Id-1 transgene expressed by these cells.
To test the role of the 120-kDa MMP in the invasive phenotype induced by Id-1, we used MMP inhibitors in the invasion assay. We first tested the toxicities of two compounds, GM6001 and phenanthroline. SCp2 cells were treated with either compound, the solvent (DMSO), or nothing for the duration of the invasion assays (20 h), and viability was assessed by trypan blue exclusion. There were no differences in viability among untreated, DMSO-treated, and GM6001-treated cells (data not shown). By contrast, all the phenanthroline-treated cells died within 20 h of treatment. We therefore used GM6001 in the invasion assay. GM6001 reduced the invasiveness of SCp2–Id-1E cells about fourfold (Fig. 7, lane 3). Because the 120-kDa gelatinase is the only detectable MMP expressed by these cells, this result suggests that much of the invasive phenotype induced by Id-1 can be attributed to the 120-kDa MMP.
Id-1 and the 120-kDa gelatinase are expressed during mammary gland involution.
In studying proteases during mouse mammary-gland development, Talhouk et al. (41) described a gelatinase having a molecular size greater than 110 kDa that was not expressed during lactation (Fig. 8, top panel, lane 1) but was expressed during the early stages of involution (days 1 and 2 [lanes 2 and 3], declining by day 3 [lane 4]). The identity or function of this gelatinase was not determined or discussed. To explore the possibility that this gelatin-degrading proteinase may be the 120-kDa MMP expressed by Id-1-expressing cells, we isolated RNA from lactating and involuting mouse mammary glands and measured Id-1 expression by Northern analysis (Fig. 8, lower panels). Id-1 mRNA was undetectable in the lactating gland (lane 1) but was highly expressed early in involution (day 1 and 2 [lanes 2 and 3]); Id-1 expression began to decline by the 3rd day of involution (lane 4). Thus, the correlation between the expression of Id-1 and a 120-kDa gelatinase observed in mammary epithelial-cell cultures is also seen in the intact mammary gland during involution.
FIG. 8.
Correlation between expression of the 120-kDa gelatinase and Id-1 in vivo. Cell extracts were prepared from lactating and involuting glands (as described by Talhouk et al. [41]) and analyzed by gelatin zymography. In the upper panel, the arrow marks the position of the 120-kDa gelatinase. RNA was isolated from mammary glands at the same stages and analyzed on Northern blots for Id-1 mRNA (middle panel). Lane 1, day 9 of lactation; lanes 2 through 5, days 1, 2, 3, and 4 of involution, respectively. The ethidium bromide-stained gel is shown in the lower panel to confirm RNA integrity and quantitation.
Id-1 and 120-kDa gelatinase expression in invasive human breast cancer cells.
Our finding that ectopic Id-1 expression induced a 120-kDa gelatinase and an invasive phenotype in mouse mammary epithelial cells suggested that Id-1 and its associated gelatinase could, at least in some instances, contribute to human breast cancer progression. To begin to explore this possibility, we examined human breast cancer cell lines exhibiting varying degrees of invasiveness in culture and in vivo.
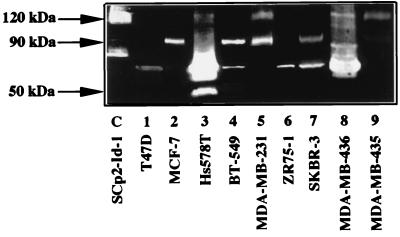
We examined four differentiated, essentially noninvasive breast cancer cell lines, T47D, MCF-7, ZR75-1, and SKBR-3 (43), and five poorly differentiated and invasive cell lines, Hs578T, BT-549, MDA-MB-231, MDA-MB-436, and MDA-MB-435 (27, 30, 43, 50) (Fig. 9). These cell lines have been evaluated for invasiveness in culture, by using the Boyden chamber assay (2), and in vivo, by using metastatic tumor formation in nude mice (43). By both assays (under serum-free and/or estrogen-free conditions), T47D, MCF-7, ZR75-1, and SKBR-3 cells were noninvasive. By contrast, Hs578T, BT-549, and particularly MDA-MB-231, MDA-MB-436, and MDA-MB-435 cells were highly invasive by both assays. We confirmed the reported invasive potentials of these cells, using the Boyden chamber assay (data not shown).
FIG. 9.
Id-1 expression in nine human breast cancer cell lines. Cells were cultured in serum-free medium for 2 days before RNA was extracted and subjected to Northern blotting. The blots were then hybridized with a human Id-1 cDNA probe. Hybridization to the 28S rRNA is also indicated.
When cells were cultured in serum-free medium for 2 days, Id-1 mRNA was undetectable in the noninvasive T47D, MCF-7, ZR75-1, and SKBR-3 cells (Fig. 9, lanes 1, 2, 6, and 7) but was easily detectable in the highly invasive MDA-MB-231, MDA-MB-436, and MDA-MB-435 cells (Fig. 9, lanes 5, 8, and 9). Of the other invasive cells, Hs578T expressed low levels of Id-1 mRNA (Fig. 9, lane 3), whereas Id-1 mRNA was undetectable in BT-549 (Fig. 9, lane 4). Thus, the invasive potential of the human breast cancer cell lines MDA-MB-231, MDA-MB-436, MDA-MB-435, and, to a lesser extent, Hs578T could, at least in part, derive from unregulated expression of Id-1 and its associated 120-kDa gelatinase.
Consistent with this idea, a 120-kDa gelatinase was detected in conditioned media from the invasive cells that expressed Id-1 (Hs578T, MDA-MB-231, MDA-MB-436, and MDA-MB-435; Fig. 10, lanes 3, 5, 8, and 9, respectively). This gelatinase was not detected in conditioned media from the noninvasive cell lines T47D, MCF-7, ZR75-1, and SKBR-3 (Fig. 10, lanes 1, 2, 6, and 7) or from the invasive cell line that did not express Id-1 (BT-549; Fig. 10, lane 4). The 120-kDa gelatinase expressed by the human breast cancer cells comigrated with the 120-kDa gelatinase expressed by Id-1-transfected SCp2 cells (Fig. 10, lane C). As previously reported (1), the 72- and/or 92-kDa gelatinases were detected in most of these human cell lines, whether or not they were invasive. Despite the secretion of these gelatinases by the cells, only the 120-kDa gelatinase-expressing cells were invasive in the Boyden chamber invasion assay (reference 43 and data not shown). The exception was the invasive BT-549 cell line, which neither expressed Id-1 mRNA nor secreted the 120-kDa gelatinase. BT-549 cells express many MMPs (by zymography), including high levels of membrane type 1 MMPs (11).
FIG. 10.
Expression of a 120-kDa gelatinase in Id-1-positive cells. Serum-free conditioned media from SCp2–Id-1 transfected cells (lane C [control]) and nine human breast cancer cell lines (lanes 1 to 9) were analyzed by gelatin zymography.
Thus, among nine human breast tumor cells examined, only Id-1-expressing cells also expressed the 120-kDa gelatinase, and all Id-1-negative cells failed to express the 120-kDa gelatinase. Moreover, the Id-1- and 120-kDa gelatinase-expressing cells were all invasive in culture and in vivo.
DISCUSSION
The mammary gland is one of the few organs that undergo striking morphological and functional changes during adult life, particularly during pregnancy, lactation, and involution. In both humans and mice, fetal, virgin adult, and pregnant mammary glands undergo extensive temporal and spatial remodeling, which entails invasion, migration, and relocation of cells to generate the ductal and alveolar structures of the gland. Once lactation is terminated, there is additional and extensive tissue remodeling as the gland returns to its resting state. In recent years, progress has been made in elucidating the mechanisms that regulate mammary gland-specific gene expression and the transformation of mammary epithelial cells to malignancy (3, 39). However, much less is known about the mechanisms, particularly the transcriptional mechanisms, that regulate the development and remodeling of the normal mammary gland.
SCp2 cells as a model for normal mammary epithelial cells.
SCp2 is an immortal murine cell line that nonetheless expresses many characteristics of epithelial cells in the pregnant and lactating mammary gland. SCp2 cells proliferate in monolayer culture in response to serum growth factors but arrest growth, form alveolar structures, and express milk proteins in response to lactogenic hormones and basement membrane components. Arrested growth is necessary, but not sufficient, for differentiation. The differentiation of SCp2 cells in culture is remarkably similar to the differentiation of mammary epithelial cells in vivo (8). Here, we extend this similarity to expression of a 120-kDa MMP that appears to be controlled by Id-1, a negative regulator of bHLH transcription factors (4).
Id-1 as a negative regulator of mammary epithelial-cell differentiation.
During proliferation, but not during arrested growth or differentiation, SCp2 cells express Id-1. The expression of Id-1 and that of the milk protein β-casein are inversely correlated in cultured SCp2 cells (9), as well as in the mammary gland in virgin, pregnant, and lactating mice (9a). Indeed, Id-1 is a negative regulator of the functional differentiation of SCp2 cells. When constitutively expressed, Id-1 prevents the strong cell-cell contacts typical of differentiated cells and blocks milk protein expression. Although the precise mechanism by which Id-1 inhibits differentiation is not known, it is clear that it does not act by preventing the growth arrest induced by hormones and ECM (9).
Id-1 is presumed to repress differentiation by inhibiting one or more bHLH transcription factors. By analogy with the role of bHLH proteins in the differentiation of muscle, neuronal, and lymphoid cells (24, 40, 47), bHLH transcription factors may be required for differentiation-specific gene expression in the mammary gland. However, our results suggest an additional role for bHLH proteins in the mammary gland: repression of a 120-kDa MMP, whose activity permits the epithelial cells to migrate and invade the ECM.
An Id-1-regulated gelatinase expressed by mammary epithelial cells.
Id-1 expression, whether originating from the endogenous gene or a transgene, correlated strongly with the expression of a 120-kDa gelatinase having the characteristics of an MMP. This protease appeared to be the only metalloproteinase expressed by SCp2 mammary epithelial cells. The well-characterized MMPs stromelysin and gelatinases A and B (72- and 92-kDa type IV collagenases) were not expressed by SCp2 cells. By contrast, gelatinases A and B were expressed by SCg6, a stroma-like cell line derived from the same culture from which SCp2 cells were cloned (8). SCg6 cells also express stromelysin-1 (28). These findings suggest that the expression of stromelysin and gelatinases A and B during involution of the mammary gland may derive from the nonepithelial cells in the tissue (29).
The epithelial cells of the mammary gland, on the other hand, may express the 120-kDa MMP. Talhouk et al. (41) described a gelatinase with an apparent molecular size exceeding 110 kDa that was expressed during the early stages of involution. We found that Id-1 mRNA was not expressed during lactation, when the 120-kDa gelatinase is undetectable, but was expressed early in involution (days 1 and 2). We suggest that this gelatinase may be the 120-kDa MMP identified in Id-1-expressing SCp2 cells. Thus, there is a correlation between Id-1 expression and secretion of a 120-kDa gelatinase in vivo, as well as in cultured cells.
The Id-1-regulated gelatinase is critical for epithelial-cell invasiveness.
SCp2 cells arrest growth when in contact with basement membrane ECM. Under these conditions, Id-1 is not expressed, the cells maintain strong contacts, and they do not invade the surrounding ECM (9). Constitutive Id-1 expression did not prevent the growth arrest but conferred an invasive phenotype on the cells. Only after Id-1-expressing SCp2 cells had invaded the ECM did they resume proliferation. Thus, Id-1 appeared to be a regulator of the invasive phenotype rather than a stimulator of cell proliferation per se. This invasive phenotype, in turn, appeared to depend primarily on the 120-kDa gelatinase (MMP). This MMP was the only detectable target of GM6001, a nontoxic MMP inhibitor (12), and GM6001 effectively inhibited the invasive phenotype of Id-1-expressing cells. Thus, Id-1 and its related 120-kDa MMP were key regulators of the invasive phenotype of SCp2 cells. During involution, the Id-1-associated MMP may participate in remodeling the gland in vivo. We suggest that Id-1 and its related MMP may be key regulators of the transient invasive phenotype acquired by the epithelial cells during certain stages of normal mammary-gland development and remodeling.
Id-1 and the 120-kDa gelatinase in tumor cell invasion.
The invasive phenotype induced by Id-1 was not the result of malignant transformation. Id-1-expressing SCp2 cells did not grow in an anchorage-independent manner and did not form detectable tumors in nude mice. Thus, Id-1 differs from oncogenes such as v-Ha-ras, which converts mouse mammary epithelial cells into invasive but also tumorigenic cells (13). Furthermore, Id-1 did not induce an invasive phenotype by converting cells to a stromal or mesenchymal phenotype. Id-1-expressing SCp2 cells maintained their epithelial characteristics, such as keratin expression, and did not express stromal MMPs. Thus, the action of Id-1 differs from that of genes of the ets family. c-Ets, a transcription factor expressed by stromal fibroblasts, promotes epithelial tumor cell invasion (48) by inducing stromal MMPs such as stromelysin-1. E1AF, a new member of the ets family, induces an invasive and migratory phenotype in human MCF-7 breast cancer cells (19), presumably by inducing gelatinase B as well as stromelysin-1.
Although the Id-1-induced invasive phenotype was not a consequence of malignant transformation, our results with human breast cancer cells suggest that constitutive Id-1 expression, and its associated 120-kDa gelatinase, may play a role in the invasive phenotype of at least some aggressive human breast tumors. We hypothesize that Id-1 and the 120-kDa gelatinase may constitute a thus far unrecognized pathway for tumor cell invasion. A very recent report (32) suggests that the Id-1–120-kDa gelatinase pathway we describe here may be of substantial clinical importance. In that report, a gelatinase of approximately the same size as the one described here was detected in urine from metastatic breast cancer patients but not in urine specimens from patients with other types of cancer. The authors acknowledge that the identity of this gelatinase is as yet unknown but suggest that it might serve as a predictor of metastatic breast cancer. By contrast, the 72- and 92-kDa gelatinases detected in urine were suggested to serve as predictors of organ-confined cancers. These suggestions are consistent with our results showing that the 72- and/or 92-kDa gelatinase is expressed by differentiated and noninvasive human breast cancer cells, whereas the 120-kDa gelatinase is expressed only in invasive breast cancer cells.
In conclusion, we propose that Id-1 regulates the invasive phenotype of breast epithelial cells, in part through the activity of a 120-kDa gelatinase, during normal mammary-gland development and remodeling. Although this phenotype is not necessarily linked to tumorigenesis, it may well be reactivated during progression toward malignancy in some breast cancers, for example, during the transition from an in situ to an invasive carcinoma. We do not yet know whether Id-1 induces the 120-kDa gelatinase by directly inactivating a bHLH repressor of the gene or whether it acts indirectly by altering the expression of other genes. We are currently attempting to clone the 120-kDa gelatinase in order to answer these questions.
ACKNOWLEDGMENTS
We thank Y. Jen (Memorial Sloan-Kettering Cancer Center, New York, N.Y.) for the murine Id-1 cDNA, J. Rosen for the β-casein cDNA, and Z. Werb for the gelatinase-A and -B cDNAs and the GM6001. We also thank S. Liang, A. Lochter, R. Lupu, and Z. Werb for assistance with some of the experiments and for helpful discussions.
P.-Y.D. and C.Q.L. contributed equally to this work.
This work was supported by grants from the U.S. Department of Energy (contract DE-AC03-76SF00098) to M.J.B. and J.C. and by a New Investigator Award from the University of California Breast Cancer Research Program (1KB0274) to P.-Y.D.
REFERENCES
- 1.Abbas Abidi S M, Howard E W, Dmytryk J J, Pento J T. Differential influence of antiestrogens on the in vitro release of gelatinases (type IV collagenases) by invasive and non-invasive breast cancer cells. Clin Exp Metastasis. 1997;15:432–439. doi: 10.1023/a:1018458406797. [DOI] [PubMed] [Google Scholar]
- 2.Albini A, Iwamoto Y, Kleinman H K, Martin G R, Aaronson S A, Kozlowski J M, McEwan R N. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 1987;47:3239–3245. [PubMed] [Google Scholar]
- 3.Band V. Preneoplastic transformation of human mammary epithelial cells. Semin Cancer Biol. 1995;6:185–192. doi: 10.1006/scbi.1995.0015. [DOI] [PubMed] [Google Scholar]
- 4.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Cross J C, Flannery M L, Blanar M A, Steingrimsson E, Jenkins N A, Copeland N G, Rutter W J, Werb Z. Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development. 1995;121:2513–2523. doi: 10.1242/dev.121.8.2513. [DOI] [PubMed] [Google Scholar]
- 7.Danielson K G, Oborn C J, Durban E M, Buetel J S, Medina D. Epithelial mouse mammary cell line exhibiting normal morphogenesis in vivo and functional differentiation in vitro. Proc Natl Acad Sci USA. 1984;81:3756–3760. doi: 10.1073/pnas.81.12.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desprez P Y, Roskelley C, Campisi J, Bissell M J. Isolation of functional cell lines from a mouse mammary epithelial cell strain: the importance of basement membrane and cell-cell interaction. Mol Cell Differ. 1993;1:99–110. [Google Scholar]
- 9.Desprez P Y, Hara E, Bissell M J, Campisi J. Suppression of mammary epithelial cell differentiation by the helix-loop-helix protein Id-1. Mol Cell Biol. 1995;15:3398–3404. doi: 10.1128/mcb.15.6.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Desprez, P. Y., C. Sympson, J. Campisi, and M. J. Bissell. Unpublished data.
- 10.Fisher S J, Werb Z. The catabolism of extracellular matrix components. In: Haralson M A, Hassell J R, editors. Extracellular matrix: a practical approach. Oxford, United Kingdom: IRL Press Ltd.; 1995. pp. 261–287. [Google Scholar]
- 11.Gilles C, Polette M, Seiki M, Birembaut P, Thompson E W. Implication of collagen type I-induced membrane type 1-matrix metalloproteinase expression and matrix metalloproteinase-2 activation in the metastatic progression of breast carcinoma. Lab Investig. 1997;76:651–660. [PubMed] [Google Scholar]
- 12.Grobelny D, Poncz L, Galardy R E. Inhibition of human skin fibroblast collagenase, thermolysin, and Pseudomonas aeruginosa elastase by peptide hydroxamic acids. Biochemistry. 1992;31:7152–7154. doi: 10.1021/bi00146a017. [DOI] [PubMed] [Google Scholar]
- 13.Gunzburg W H, Salmons B, Schlaeffli A, Moritz-Legrand S, Jones W, Sarkar N H, Ullrich R. Expression of the oncogenes mil and ras abolishes the in vivo differentiation of mammary epithelial cells. Carcinogenesis. 1988;9:1849–1856. doi: 10.1093/carcin/9.10.1849. [DOI] [PubMed] [Google Scholar]
- 14.Hara E, Yamaguchi T, Nojima H, Ide T, Campisi J, Okayama H, Oda K. Id-related genes encoding HLH proteins are required for G1 progression and are repressed in senescent human fibroblasts. J Biol Chem. 1994;269:2139–2145. [PubMed] [Google Scholar]
- 15.Hara E, Uzman J A, Dimri G P, Nehlin J O, Testori A, Campisi J. The helix-loop-helix Id-1 and a retinoblastoma protein binding mutant of SV40 T antigen synergize to reactivate DNA synthesis in senescent human fibroblasts. Dev Genet. 1996;18:161–172. doi: 10.1002/(SICI)1520-6408(1996)18:2<161::AID-DVG9>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Haslam S Z. The ontogeny of mouse mammary gland responsiveness to ovarian steroid hormones. Endocrinology. 1989;125:2766–2772. doi: 10.1210/endo-125-5-2766. [DOI] [PubMed] [Google Scholar]
- 17.Iavarone A, Garg P, Lasorella A, Hsu J, Israel M A. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 18.Jen Y, Weintraub H, Benezra R. Overexpression of Id protein inhibits the muscle differentiation program: in vivo association of Id with E2A proteins. Genes Dev. 1992;6:1466–1479. doi: 10.1101/gad.6.8.1466. [DOI] [PubMed] [Google Scholar]
- 19.Kaya M, Yoshida K, Higashino F, Mitaka T, Ishii S, Fujinaga K. A single ets-related transcription factor, E1AF, confers invasive phenotype on human cancer cells. Oncogene. 1996;12:221–227. [PubMed] [Google Scholar]
- 20.Kingston R E. Transcription control and differentiation: the HLH family, c-myc and C/EBP. Curr Opin Cell Biol. 1989;1:1081–1087. doi: 10.1016/s0955-0674(89)80054-7. [DOI] [PubMed] [Google Scholar]
- 21.Kjeldsen L, Johnsen A H, Sengelov H, Borregaard N. Isolation and primary structure of NGAL, a novel protein associated with human neutrophil gelatinase. J Biol Chem. 1993;268:10425–10432. [PubMed] [Google Scholar]
- 22.Kleiner D E, Stetler-Stevenson W G. Quantitative zymography: detection of picogram quantities of gelatinases. Anal Biochem. 1994;218:325–329. doi: 10.1006/abio.1994.1186. [DOI] [PubMed] [Google Scholar]
- 23.Kreider B L, Benezra R, Rovera G, Kadesch T. Inhibition of myeloid differentiation by the helix-loop-helix protein Id. Science. 1992;255:1700–1702. doi: 10.1126/science.1372755. [DOI] [PubMed] [Google Scholar]
- 24.Lee J E, Hollenberg S M, Snider L, Turner D L, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by neuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 25.Liotta L A, Steeg P S, Stetler-Stevenson W G. Cancer metastasis and angiogenesis: an imbalance of positive and negative regulation. Cell. 1991;64:327–336. doi: 10.1016/0092-8674(91)90642-c. [DOI] [PubMed] [Google Scholar]
- 26.Lister J, Forrester W C, Baron M H. Inhibition of an erythroid differentiation switch by the helix-loop-helix protein Id1. J Biol Chem. 1995;270:17939–17946. doi: 10.1074/jbc.270.30.17939. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z, Brattain M G, Appert H. Differential display of reticulocalbin in the highly invasive cell line, MDA-MB-435, versus the poorly invasive cell line, MCF-7. Biochem Biophys Res Commun. 1997;231:283–289. doi: 10.1006/bbrc.1997.6083. [DOI] [PubMed] [Google Scholar]
- 28.Lochter A, Srebrow A, Sympson C J, Terracio N, Werb Z, Bissell M J. Misregulation of stromelysin-1 expression in mouse mammary tumor cells accompanies acquisition of stromelysin-1-dependent invasive properties. J Biol Chem. 1997;272:5007–5015. doi: 10.1074/jbc.272.8.5007. [DOI] [PubMed] [Google Scholar]
- 29.Lund L R, Romer J, Thomasset N, Solberg H, Pyke C, Bissell M J, Dano K, Werb Z. Two distinct phases of apoptosis in mammary gland involution: proteinase-independent and -dependent pathways. Development. 1996;122:181–193. doi: 10.1242/dev.122.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maemura M, Akiyama S K, Woods V L, Jr, Dickson R B. Expression and ligand binding of alpha 2 beta 1 integrin on breast carcinoma cells. Clin Exp Metastasis. 1995;13:223–235. doi: 10.1007/BF00133478. [DOI] [PubMed] [Google Scholar]
- 31.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Moses M A, Wiederschain D, Loughlin K R, Zurakowski D, Lamb C C, Freeman M R. Increased incidence of matrix metalloproteinases in urine of cancer patients. Cancer Res. 1998;58:1395–1399. [PubMed] [Google Scholar]
- 33.Reponen P, Sahlberg C, Huhtala P, Hurskainen T, Thesleff I, Tryggvason K. Molecular cloning of murine 72-kDa type IV collagenase and its expression during mouse development. J Biol Chem. 1992;267:7856–7862. [PubMed] [Google Scholar]
- 34.Reponen P, Sahlberg C, Munaut C, Thesleff I, Tryggvason K. High expression of 92-kD type IV collagenase (gelatinase B) in the osteoclast lineage during mouse development. J Cell Biol. 1994;124:1091–1102. doi: 10.1083/jcb.124.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riechmann V, van Cruchten I, Sablitzky F. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res. 1994;22:749–755. doi: 10.1093/nar/22.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roskelley C D, Desprez P Y, Bissell M J. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc Natl Acad Sci USA. 1994;91:12378–12382. doi: 10.1073/pnas.91.26.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidhauser C, Bissell M J, Myers C A, Casperson G F. Extracellular matrix and hormones transcriptionally regulate bovine beta-casein 5′ sequences in stably transfected mouse mammary cells. Proc Natl Acad Sci USA. 1990;87:9118–9122. doi: 10.1073/pnas.87.23.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Shalaby, R., and G. Colbern (California Pacific Medical Center). Personal communication.
- 38.Stetler-Stevenson W G. Type IV collagenases in tumor invasion and metastasis. Cancer Metastasis Rev. 1990;9:289–303. doi: 10.1007/BF00049520. [DOI] [PubMed] [Google Scholar]
- 39.Streuli C H, Schmidhauser C, Bailey N, Yurchenco P, Skubitz A P, Roskelley C, Bissell M J. Laminin mediates tissue-specific gene expression in mammary epithelia. J Cell Biol. 1995;129:591–603. doi: 10.1083/jcb.129.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun X H. Constitutive expression of the Id1 gene impairs mouse B cell development. Cell. 1994;79:893–900. doi: 10.1016/0092-8674(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 41.Talhouk R S, Chin J R, Unemori E N, Werb Z, Bissell M J. Proteinases of the mammary gland: developmental regulation in vivo and vectorial secretion in culture. Development. 1991;112:439–449. doi: 10.1242/dev.112.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taub M, Wang Y, Szczesny T M, Kleinman H K. Epidermal growth factor or transforming growth factor α is required for kidney tubulogenesis in Matrigel cultures in serum free medium. Proc Natl Acad Sci USA. 1990;87:4002–4006. doi: 10.1073/pnas.87.10.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson E W, Paik S, Brunner N, Sommers C L, Zugmaier G, Clarke R, Shima T B, Torri J, Donahue S, Lippman M E, Martin G R, Dickson R B. Association of increased basement membrane invasiveness with absence of estrogen receptor and expression of vimentin in human breast cancer cell lines. J Cell Physiol. 1992;150:534–544. doi: 10.1002/jcp.1041500314. [DOI] [PubMed] [Google Scholar]
- 44.Traurig H H. Cell proliferation in the mammary gland during late pregnancy and lactation. Anat Rec. 1967;157:489–504. [Google Scholar]
- 45.Traurig H H. A radioautographic study of cell proliferation in the mammary gland of the pregnant mouse. Anat Rec. 1967;159:239–248. doi: 10.1002/ar.1091590213. [DOI] [PubMed] [Google Scholar]
- 46.Wang S, Counterman L J, Haslam S Z. Progesterone action in normal mouse mammary gland. Endocrinology. 1990;127:2183–2189. doi: 10.1210/endo-127-5-2183. [DOI] [PubMed] [Google Scholar]
- 47.Weintraub H, Davis R, Tapscott S J, Thayer M, Krause R, Benezra R, Blackwell T K, Turner D, Rupp R, Hollenberg S, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 48.Wernert N, Gilles F, Fafeur V, Bouali F, Raes M B, Pyke C, Dupressoir T, Seitz G, Vandenbunder B, Stehelin D. Stromal expression of c-Ets1 transcription factor correlates with tumor invasion. Cancer Res. 1994;54:5683–5688. [PubMed] [Google Scholar]
- 49.Witty J P, Wright J H, Matrisian L M. Matrix metalloproteinases are expressed during ductal and alveolar mammary morphogenesis, and misregulation of stromelysin-1 in transgenic mice induces unscheduled alveolar development. Mol Biol Cell. 1995;6:1287–1303. doi: 10.1091/mbc.6.10.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang R D, Fidler I J, Price J E. Relative malignant potential of human breast carcinoma cell lines established from pleural effusions and a brain metastasis. Invasion Metastasis. 1991;11:204–215. [PubMed] [Google Scholar]