Identification of a Novel Cortactin SH3 Domain-Binding Protein and Its Localization to Growth Cones of Cultured Neurons (original) (raw)
Abstract
Cortactin is an actin-binding protein that contains several potential signaling motifs including a Src homology 3 (SH3) domain at the distal C terminus. Translocation of cortactin to specific cortical actin structures and hyperphosphorylation of cortactin on tyrosine have been associated with the cortical cytoskeleton reorganization induced by a variety of cellular stimuli. The function of cortactin in these processes is largely unknown in part due to the lack of information about cellular binding partners for cortactin. Here we report the identification of a novel cortactin-binding protein of approximately 180 kDa by yeast two-hybrid interaction screening. The interaction of cortactin with this 180-kDa protein was confirmed by both in vitro and in vivo methods, and the SH3 domain of cortactin was found to direct this interaction. Since this protein represents the first reported natural ligand for the cortactin SH3 domain, we designated it CortBP1 for cortactin-binding protein 1. CortBP1 contains two recognizable sequence motifs within its C-terminal region, including a consensus sequence for cortactin SH3 domain-binding peptides and a sterile alpha motif. Northern and Western blot analysis indicated that CortBP1 is expressed predominately in brain tissue. Immunofluorescence studies revealed colocalization of CortBP1 with cortactin and cortical actin filaments in lamellipodia and membrane ruffles in fibroblasts expressing CortBP1. Colocalization of endogenous CortBP1 and cortactin was also observed in growth cones of developing hippocampal neurons, implicating CortBP1 and cortactin in cytoskeleton reorganization during neurite outgrowth.
Cells undergo rearrangement of the cortical cytoskeleton, a submembranous actin filament (F-actin)-based network, during a variety of cellular processes including differentiation, proliferation, migration, and oncogenic transformation (9, 10, 43, 67). The cortical cytoskeleton not only controls cell morphology but is also involved in transmitting signals between the plasma membrane and intracellular compartments (7, 8, 38). A large body of evidence indicates that small GTP-binding proteins, tyrosine kinases, and serine/threonine kinases play a pivotal role in regulating the dynamic structure of the cortical cytoskeleton (30, 33). The molecular mechanisms by which these enzymes regulate cortical actin polymerization and reorganization are currently unclear. Identification of actin-associated targets of these enzymes is important for unveiling signaling pathways correlated with the cortical F-actin remodeling.
Cortactin, an F-actin-binding substrate for the nonreceptor tyrosine kinase pp60_src_ (39, 78), is distinguished by the presence of several potential signaling sequence motifs (63, 77, 79). The N-terminal half of the protein contains six and a half tandem repeats of a 37-amino-acid sequence. The repeat region is required and sufficient for efficient association with F-actin as assessed by in vitro cosedimentation assays (78). The role of this region in mediating the interaction with F-actin has been further confirmed by the blockage of the cosedimentation of cortactin with F-actin by a cortactin-specific monoclonal antibody (MAb) whose epitope is located in the repeats (78). Since this region does not show significant sequence similarities with other actin-binding proteins, cortactin represents a distinctive family of F-actin-binding proteins. The C-terminal half of cortactin consists of a predicted α-helix of 50 to 60 residues, a region enriched in proline, serine, and threonine, and a Src homology 3 (SH3) domain that has been found in numerous signaling proteins (15). The cortactin SH3 domain has significant sequence and topological similarity with the SH3 domains of the cortactin-related protein HS1 (76%; discussed below), the yeast actin-binding protein 1 (53% [24]), and the c-Abl tyrosine kinase substrate Abi2 (53% [17]). All SH3 domains characterized so far mediate protein-protein interactions via recognition of polyproline motifs with a left-handed helical conformation (49). The SH3 domain-mediated interactions are implicated in regulation of enzyme activities, targeting of proteins to specific subcellular compartments, and coupling of signaling pathways (15). Little is known about the function of the cortactin SH3 domain except for its selective interaction with peptides containing a consensus sequence of +PPΨPXKP (66). It is interesting to note that a cortactin-related protein termed HS1, which is expressed exclusively in hematopoietic cells (40), contains three and a half copies of a 37-amino-acid motif in the N-terminal region and an SH3 domain at the distal C terminus. Each unit of the repeats and the SH3 domain of HS1 display approximately 70% sequence identity to analogous regions in cortactin, whereas sequence similarity in other regions is limited (40, 41). Proliferative responses and clonal deletion of B cells and T cells upon antigen receptor cross-linking are significantly impaired in mice lacking HS1, implicating important roles of HS1 in antigen receptor-initiated signaling processes (70).
Studies of tyrosine phosphorylation and intracellular distribution of cortactin have suggested involvement of cortactin in signaling pathways induced by oncogenic transformation and cell surface activation. In a number of nontransformed adherent cells grown in culture, cortactin localizes to poorly defined punctate structures in the cytoplasm and within F-actin-based lamellipodia and membrane ruffles at the cell cortex (73, 78). Lamellipodia and membrane ruffles are veil-like membrane structures that have been implicated in guiding the migration of motile fibroblasts and of the neuronal growth cones (59). Cortactin is normally phosphorylated on serine and threonine, but it becomes heavily phosphorylated on tyrosine in oncogenic pp60_src_-transformed fibroblasts (39, 77). Src-induced transformation causes accumulation of cortactin and F-actin in podosomes (77), which are labile cell substratum adhesion sites in the transformed cells (71). Overexpression of cortactin and redistribution of cortactin into podosome-like structures (56, 61, 63) have been observed in human carcinoma cells with amplification of the chromosome 11q13 region in which the human cortactin gene is located (61). Since podosomes contain a substratum-degrading proteolytic activity (12) and amplification of the 11q13 region is correlated with poor tumor prognosis (50, 62), cortactin is suggested to be involved in upregulating an invasive and metastatic behavior associated with tumors with 11q13 amplification (63). In addition, a transient increase in tyrosyl-phosphorylated cortactin is induced by a variety of types of cell surface activation, including treatment of fibroblasts with growth factors (23, 79), activation of platelets with thrombin (76), activation of intercellular adhesion molecule 1 in brain endothelial cells (25), and invasion of epithelial cells by Shigella flexneri (19). All these cellular processes involve dramatic changes in the cortical cytoskeleton, and translocation of cortactin to specific cortical cytoskeleton structures has been observed in response to some of these external stimuli. For example, during _S. flexneri_-mediated cell invasion, which requires assembly of F-actin at the bacterial entry sites (14), cortactin coaccumulates with F-actin in the membrane ruffles at the sites of bacterial entry and in the periphery of the early phagosomes (19).
Given its multidomain structure and regulated subcellular localization, cortactin has been suggested to recruit other signaling molecules to the cortical cytoskeleton (25, 53, 76, 78). In this study, we have identified a novel cortactin-binding protein of approximately 180 kDa, which is designated CortBP1 for cortactin-binding protein 1. A consensus sequence for cortactin SH3 domain-binding peptides (66) is present in the CortBP1 sequences, and the cortactin SH3 domain is required and sufficient for the interaction with CortBP1. The C terminus of CortBP1 contains a sterile alpha motif (SAM), which has been found in a number of signaling proteins involved in developmental regulation (60). CortBP1 is expressed predominately in brain tissue as determined by both Northern and Western blot analysis. Immunofluorescence studies revealed colocalization of CortBP1 with cortactin and cortical F-actin in lamellipodia and membrane ruffles in fibroblasts overexpressing CortBP1. Colocalization of endogenous CortBP1 and cortactin was observed in growth cones in differentiating hippocampal neurons. These data place CortBP1 and cortactin in the same subcellular compartment and implicate a possible involvement of CortBP1 and cortactin in the dynamic actin reorganization during neuronal growth cone extension.
MATERIALS AND METHODS
DNA constructs.
To generate the construct pPC97-SR5, expressing the cortactin SH3 domain fused to the GAL4 DNA-binding domain (GAL4 BD), the mouse cortactin cDNA clone mp85.L7 (51) was amplified by PCR with primers 1597 (5′-GTCCCCCGGGCATCACAGCCATCGCC-3′) and 1786 (5′-CCGGAATTCTGGGTGGCAGCCCTACT-3′). The resultant PCR product was inserted as an _Sma_I/_Eco_RI fragment into pPC97 (13). The expression construct for GAL4 BD-cortactinSH3W525K was generated by subcloning the _Bgl_II/_Not_I fragment (amino acids 506 to 546) of pFlag-cortactinW525K (described below) into _Bgl_II/_Not_I-digested pPC97-SR5.
To express full-length CortBP1 in mammalian cells, pcDNA3.6 and pcDNA3.2 were generated by subcloning the _Not_I-digested inserts of phage DNA clones 6.b and 2.b (see Fig. 2A) into _Not_I-excised pcDNA3.1(−) (Invitrogen). The full-length CortBP1 cDNA, pcDNA3.62, was then made by subcloning the 2.4-kb _Eco_RV/_Bam_HI fragment of pcDNA3.2 into _Eco_RV/_Bam_HI-digested pcDNA3.6 construct.
FIG. 2.
Amino acid sequence and homology domains of CortBP1. (A) Structure of CortBP1 cDNA clones. The CortBP1 cDNA, assembled from the sequences of partial CortBP1 cDNA clones, is illustrated as a line with the positions of the predicted translation initiation and termination codons indicated. Also shown are the positions of restriction sites used for analysis and subcloning, including _Eco_RI (RI), _Bgl_II (Bg), _Eco_RV (RV), and _Bam_HI (Ba). The relative positions of the 5′ ends of nine partial cDNA clones are indicated at the left. Clones 15.a, 25.a, and 10.a were isolated from a rat hippocampus cDNA library, whereas clones 17.b, 14.b, 2.b, 7.b, and 6.b were from a rat brain 5′-stretch-plus cDNA library. (B) Deduced amino acid sequence of CortBP1. The residues encoded by the two-hybrid cDNA clone 2H are shaded. The SAM domain is underlined, and the ppI motif is indicated by dots. (C) Alignment of the predicted SAM domain of CortBP1 with related SAM domains. The boxed amino acid residues are identical or conserved in more than 70% of the SAM domains compared. The gaps in the alignment are denoted by dots. Ψ, aliphatic residues; Φ, aromatic residues. (D) Comparison of the CortBP1 ppI motif, the consensus for cortactin SH3 domain-binding peptides, and the consensus for class II ligands of SH3 domains. Predicted SH3-contacting residues are capitalized. +, R or K; Ψ, aliphatic residues; x, any amino acid residue.
The glutathione _S_-transferase (GST)–CortBP1 construct pGEX-CortBP1cte was generated by subcloning the _Sal_I/_Not_I insert of pPC86-2H (amino acids 941 to 1252) into pGEX4T-2 (Pharmacia Biotech). For expression of CortBP1cte in mammalian cells, pRK5-CortBP1cte was engineered by subcloning the _Bam_HI fragment of pGEX-CortBP1cte into pRK5myc (54). The resultant construct encoded Myc-CortBP1cte recombinant protein that contained the 10-amino-acid Myc epitope tag (EQKLISEEDL) fused to the N terminus of CortBP1cte.
pFlag-cortactin, the Flag-tagged cortactin construct, was generated by PCR with primers 17676 (5′-GGTACCATGTGGAAAGCCTCTGCAGGC-3′) and 17677 (5′-GAATTCCTACTGCCGCAGCTCCACATAG-3′), which flank the open reading frame of the mouse cortactin cDNA clone mp85.L7 (51). The resultant PCR product was subcloned into pCR-Script (Stratagene), digested with _Kpn_I and _Eco_RI, and subcloned into pcDNA3Flag2AB (20). pFlag-cortactin W525K was then generated by mutagenizing pFlag-cortactin with primer pairs (5′-GAAATGATTGACGATGGCAAGTGGCGTGGGGTGTGCAAG-3′ and 5′-CTTGCACACCCCACGCCACTTGCCATCGTCAATCATTTC-3′) with the QuickChange site-directed mutagenesis kit (Stratagene). The coding regions of all constructs were confirmed by direct DNA sequence analysis.
Yeast two-hybrid analysis, cDNA cloning, and sequence analysis.
Transformation of yeast strain Y190 (34) was performed by the lithium acetate method (29), and β-galactosidase activity in individual transformants was measured by the filter lift assay (4). Initially, five variants of cortactin fused to GAL4 BD were generated as baits. However, Y190 cells harboring each of four constructs which encoded full-length proteins or the C-terminal regions (amino acids 329 to 546, 396 to 546, or 473 to 546) exhibited readily detectable β-galactosidase activity. A construct, pPC97-SR5, lacking the 17-amino-acid sequence (473-GHYQAEDDTYDGYESDL-489) common to the four transactivating constructs did not induce any β-galactosidase activity and was used to screen a rat brain library. Yeast transformants containing pPC97-SR5 were subsequently transformed with a rat hippocampus cDNA library in pPC86 (13) and selected for growth on Leu− Trp− His− plates with 25 mM 3-aminotriazole. Three independent positive clones that showed detectable β-galactosidase activity within 12 h of analysis were identified from 5 × 105 yeast transformants and purified by three rounds of colony purification. Loss of pPC97-SR5 from purified positive colonies was induced by Trp− selection, and positive library plasmids were isolated. To verify the interaction, purified positive plasmids were retransformed into Y190 cells together with pPC97, pPC97-SR5, or pPC97-SR5W525K, and resultant transformants were analyzed for β-galactosidase activity.
To obtain the full-length cDNA sequence for CortBP1, a λgt11 rat brain cDNA library (Clontech) was screened with an [α-32P]CTP-labeled 0.87-kb _Sal_I/_Bgl_II fragment of insert 2H (see Fig. 2A). Subsequently, a 5′ 0.88-kb _Eco_RI-digested fragment derived from clone 10.a (see Fig. 2A) was labeled with [α-32P]CTP and used as probe to screen a 5′-plus-stretch rat hippocampus library in λgt11 (Clontech) until clones (clones 6.b and 7.b [Fig. 2A]) containing 5′ in-frame stop codons were isolated. Throughout the complete coding region, at least two independent clones of the same region were sequenced on both strands. DNA sequence was obtained by the dideoxy chain termination method. Both DNA and amino acid sequences were analyzed with Genetics Computer Group sequence analysis software.
Northern blot analysis.
A 0.87-kb _Sal_I-_Bgl_II fragment of insert 2H, a 1.25-kb _Eco_RI-_Sac_I fragment of the mouse cortactin cDNA clone mp85.L7 (51), and a β-actin cDNA probe (Clontech) were labeled with [α-32P]CTP by primer extension with random hexamernucleotide primer (Pharmacia Biotech). A multiple-tissue Northern blot (Clontech), each lane of which contains approximately 2 μg of poly(A)+ RNA from specific adult mouse tissues, was incubated with each probe (2 × 108 to 5 × 108 cpm/μg) overnight at 42°C. The filter was then washed twice (5 min) in washing buffer 1 (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS]) at room temperature, twice (5 min) in washing buffer 2 (0.2× SSC, 0.1% SDS) at room temperature, and then twice (15 min) in prewarmed washing buffer 2 at 42°C. After each hybridization, the probe was stripped by incubating the filter in boiled 0.5% SDS for 10 min and then in 2× SSC for 5 min at room temperature.
GST fusion proteins and in vitro binding analysis.
GST fusion proteins were expressed and purified as described elsewhere (65). Briefly, 5 ml of clarified lysates from Escherichia coli BL21 expressing GST fusion proteins in NETN (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 50 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride, 0.05 U of aprotinin per ml) was incubated with 200 μl of 50% glutathione-Sepharose bead slurry (Pharmacia Biotech) at 4°C for 1 h with constant rocking. The beads were then washed three times with 1 ml of NETN. To make antigen for CortBP1-specific antiserum, purified GST-CortBP1cte proteins were eluted by incubating the beads with glutathione elution buffer (20 mM reduced glutathione, 120 mM NaCl, 100 mM Tris-HCl, pH 8.0) at room temperature for 10 min with constant rocking. The eluates were then dialyzed against phosphate-buffered saline (PBS) and concentrated to approximately 1 mg/ml.
For in vitro binding experiments, immobilized GST fusion proteins were mixed with 1 ml of 1-mg/ml cell lysates in modified radioimmunoprecipitation assay buffer (RIPA) (50 mM HEPES [pH 7.2], 150 mM NaCl, 2 mM EDTA, 0.5% sodium deoxycholate, 1% Nonidet P-40, 50 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride, 0.05 U of aprotinin per ml, 1 mM sodium vanadate, 40 mM sodium fluoride) at 4°C for 2 to 3 h. After being washed once with 1 ml of modified RIPA and three times with 1 ml of HNTG (20 mM HEPES [pH 7.0], 150 mM NaCl, 0.1% Triton X-100, 10% glycerol), precipitated proteins were subjected to Western blot analysis with the cortactin-specific MAb 4F11.
Antibodies and immunoprecipitation analysis.
Rabbit polyclonal antiserum (anti-CortBP1) was raised against purified GST-CortBP1cte (amino acids 941 to 1252). Both anti-CortBP1 and preimmune serum were protein G purified. The specificity of anti-CortBP1 against CortBP1 was confirmed as follows: 500 μg of lysates from NIH 3T3 cells overexpressing Myc epitope-tagged CortBP1cte was immunoprecipitated with anti-CortBP1, preimmune serum, or the Myc-specific MAb 9E10. Precipitated proteins were resolved on SDS–12% polyacrylamide gel electrophoresis (PAGE) gels and subjected to Western blot analysis with anti-CortBP1. Anti-CortBP1 recognized a species present in the immunocomplex of anti-CortBP1 but not in preimmune immunocomplex that comigrated with the recombinant protein present in the immunocomplex of MAb 9E10. Direct Western blot analysis of 100 μg of rat brain lysate revealed a major polypeptide of approximately 180 to 200 kDa (see Fig. 8A) and a minor peptide species of approximately 75 kDa (data not shown).
FIG. 8.
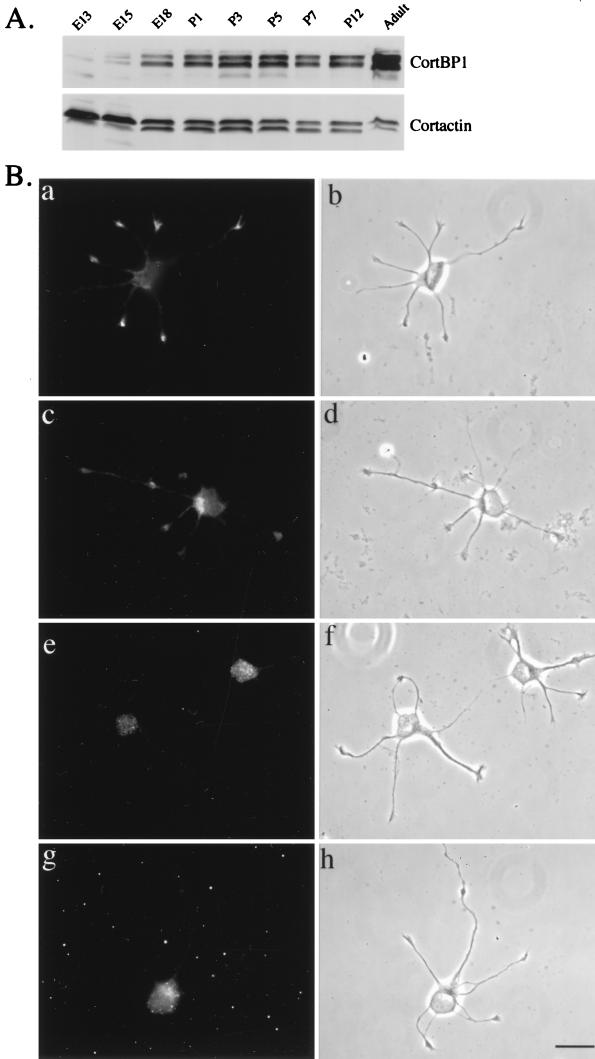
(A) Expression pattern of CortBP1 and cortactin in rat brain lysates at different developmental stages. One hundred micrograms of lysates prepared from rat brains at the indicated developmental stages was resolved on SDS–10% PAGE, transferred to a nitrocellulose filter, and blotted with anti-CortBP1 (top panel). The filter was then stripped and reblotted with the anticortactin MAb 4F11 (bottom panel). E13, E15, and E18, embryonic days 13, 15, and 18; P1, P3, P5, P7, and P12, postnatal days 1, 3, 5, 7, and 12. (B) Localization of cortactin and CortBP1 in rat hippocampal neurons. Cultured hippocampal neurons were immunostained with the anticortactin MAb 4F11 (a and b), anti-CortBP1 (c and d), anti-CortBP1 blocked with purified antigen (g and h), or preimmune serum (e and f). Panels b, d, f, and h are phase-contrast images of the cells shown in panels a, c, e, and g, respectively. Bar, 20 μm.
The cortactin-specific polyclonal antibody, anti-Cterm, was generated by purifying rabbit antiserum raised against a GST-cortactin fusion protein (GST.p80 [78]) by using a cyanogen bromide-Sepharose 4B column (Pharmacia) coupled with a GST fusion protein of the C-terminal half of mouse cortactin (amino acids 329 to 546 [73]). The cortactin-specific MAb 4F11 has been previously described (39). Mouse MAbs specific for NCK (anti-NCK), for the Myc epitope (9E10), and for the Flag epitope (M5) were purchased from Transduction Laboratories, Santa Cruz Biotechnology, and Sigma, respectively.
For immunoprecipitating CortBP1, 5 to 10 μg of protein G-purified anti-CortBP1 or preimmune serum was incubated with 500 μl of modified RIPA containing 50 μl of protein A-Sepharose beads (Sigma) at 4°C for 1 h, followed by three washes with 1 ml of modified RIPA. Tissue extracts prepared by homogenizing adult male rat (Sprague-Dawley) tissues in modified RIPA containing 0.05% SDS or cell lysates were incubated with immobilized anti-CortBP1 or preimmune antiserum at 4°C for 2 to 3 h. After three washes with 1 ml of modified RIPA, immunoprecipitates were subjected to Western blot analysis with 1 μg of the MAb 4F11 per ml, 5 μg of anti-CortBP1 per ml, or 250 ng of the MAb anti-NCK per ml.
Cell culture, transfection, and immunofluorescence microscopy.
NIH 3T3 cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and penicillin-streptomycin. Both 10T1/2neo cells, which are a stable transfectant of C3H-10T1/2 murine fibroblasts with pSV2neo, and 5Hd47 cells (c-Src overexpressors), which are a stable transfectant of C3H-10T1/2 cells with pSV2neo and a plasmid encoding chicken c-Src (11), were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, penicillin-streptomycin, and G418. PC12 cells were cultured as described elsewhere (32). Primary cultures of hippocampal neurons prepared from embryonic day-18 rats (5) were generously provided by Gary Banker.
Cells were transiently transfected with SuperFect (Qiagen). Thirty-six hours posttransfection, NIH 3T3 cells transfected with expression constructs were lysed and subjected to further analysis. For detection of CortBP1 localization in fibroblasts, 10T1/2neo or 5Hd47 cells were seeded on glass coverslips, grown to approximately 60% confluence, and transfected with pRK5-CortBP1cte. Sixteen hours posttransfection, cells were fixed and processed for immunofluorescence analysis.
For immunofluorescence experiments, cultured fibroblasts were fixed with 4% paraformaldehyde in PBS for 20 min and permeabilized with 0.5% Triton X-100 in PBS for 3 min. Cultured hippocampal neurons were fixed and permeabilized with methanol for 20 min. After three washes with PBS, cells were blocked with 20% goat serum–2% bovine serum albumin in PBS for 1 h and incubated with primary antibodies (2.0 to 5.0 μg of anti-CortBP1 per ml, 5 μg of preimmune serum per ml, 0.2 μg of 9E10 per ml, 1 to 2 μg of 4F11 per ml, or 1.0 μg of anti-Cterm per ml) in PBS containing 10% goat serum and 1% bovine serum albumin for 1 h. After three washes with PBS, cells were then incubated with 1.5 μg of fluorescein isothiocyanate- or Texas Red-conjugated secondary antibodies (goat anti-rabbit or goat anti-mouse [Jackson ImmunoResearch]) per ml for 45 min. To visualize F-actin, 0.4 U of Texas Red-conjugated phalloidin (Molecular Probes) per ml was coincubated with secondary antibodies. For the antigen blocking experiment, 5 μg of anti-CortBP1 per ml was preincubated with 5 μg of purified GST-CortBP1cte per ml for 2 h at room temperature prior to labeling. Cells were viewed with a Leitz DMR fluorescence microscope and photographed with a cooled charge-coupled device camera controlled by the ISee software program (Inovision). Images were processed on a Macintosh computer with Adobe Photoshop.
Nucleotide sequence accession number.
The nucleotide sequence of the CortBP1 cDNA (nucleotides 1 to 4636) has been given the GenBank accession no. AF060116.
RESULTS
Identification and molecular cloning of cortactin-binding protein 1.
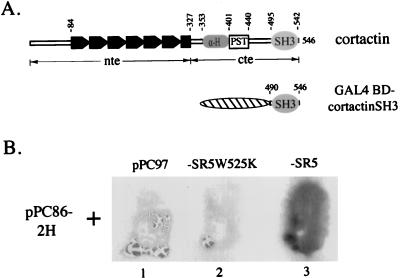
To better define the role of cortactin in cell signaling events, we utilized the yeast two-hybrid system (28) to isolate cDNA clones encoding cortactin-binding proteins. With GAL4 BD-cortactinSH3 as the bait (Fig. 1A), three unique positive clones were isolated from a rat hippocampus cDNA library. The clone pPC86-2H, which reproducibly exhibited the highest affinity for the bait (data not shown), was further characterized. The identified interaction was dependent upon the SH3 domain since the GAL4 BD alone or GAL4 BD-cortactinSH3W525K, in which a tryptophan residue highly conserved in SH3 domains (69) was replaced with a lysine residue, did not display any detectable interaction with the fusion protein encoded by pPC86-2H (Fig. 1B).
FIG. 1.
Identification of cortactin SH3 domain-interacting proteins. (A) Diagram of cortactin domain structure and the cortactin SH3 domain expressed as a GAL4 BD (denoted by hatched oval) hybrid. The tandem repeats in the N-terminal half are denoted by solid boxes; the predicted α-helical region, the proline/serine/threonine-rich region, and the SH3 domain are denoted by α-H, PST, and SH3, respectively. The numerical positions of amino acid residues of mouse cortactin are also indicated. The N-terminal half is designated nte, and the C-terminal half is designated cte. (B) Analysis of the CortBP1 and cortactin SH3 domain interaction by the two-hybrid assay. Y190 cells were cotransformed with pPC86-2H and pPC97 (lane 1), pPC86-2H and pPC97-SR5W525K (lane 2), or pPC86-2H and pPC97-SR5 (lane 3). The resultant transformants were subjected to the filter lift assay for 12 h.
Sequence analysis revealed that pPC86-2H contained an insert (2H) of approximately 2.5 kb with a predicted open reading frame of 312 amino acids contiguous with the GAL4 BD (Fig. 2A and B). Neither the DNA nor the deduced amino acid sequence had significant overall homology to any sequences in current databases. However, a proline-rich sequence (KPPVPPKP, hereafter referred to as the ppI motif) was found by inspection of the predicted amino acid sequence. The ppI motif showed striking similarity to the consensus sequence for cortactin SH3 domain-binding peptides (66), matching at all seven positions (Fig. 2D) that are predicted to be SH3 domain-contacting–scaffolding residues (27). In addition, the last 67 amino acids of this polypeptide showed extensive similarity to SAM domains (Fig. 2C), 60- to 70-amino-acid sequence motifs with a predicted secondary structure consisting of four short α-helices linked by loops (58). SAM domains have been identified in a variety of proteins (60) including EPH family receptor tyrosine kinases, yeast sterile proteins including the serine/threonine kinases Byr2p and Ste11p, the β subunit of trimeric G protein Ste4p, and the cytoskeleton-associated proteins Boi1p and Boi2p, as well as several Drosophila proteins involved in developmental regulation. As the first reported natural ligand for the cortactin SH3 domain, this protein was designated CortBP1 for cortactin-binding protein 1.
To obtain the complete cDNA sequence of CortBP1, two rat brain cDNA libraries were sequentially screened. Eight overlapping partial cDNAs were isolated, and selected regions of these cDNAs were sequenced (Fig. 2A). The 5′-most cDNA clones, 6.b and 7.b (Fig. 2A), contained an in-frame methionine codon (nucleotides 425 to 427) that was preceded by translational termination codons in all three reading frames. The first predicted ATG conformed to the canonical Kozak sequence (42), matching at positions −3, −4, and −5. The predicted termination codon (nucleotides 4181 to 4183) was present in two additional clones (clone 15.a and clone 2.b [Fig. 2A]). The 3′-most cDNA clone, clone 2.b, contained a 3′ untranslated region of approximately 2.7 kb. However, neither a polyadenylation signal (75) nor a polyadenosine sequence was found at the 3′ end of clone 2.b, implicating an even longer 3′ noncoding sequence.
The CortBP1 open reading frame, deduced from sequences of the partial cDNA clones, encoded a 1,252-amino-acid protein with a calculated molecular mass of 134 kDa (Fig. 2B). The deduced N-terminal 940 residues of CortBP1 5′ cDNA did not show significant overall similarity to any identified proteins in the existing databases. Amino acid composition analysis revealed that the CortBP1 protein is considerably rich in proline and serine residues (12 and 10%, respectively).
Tissue distribution analysis of the CortBP1 transcript.
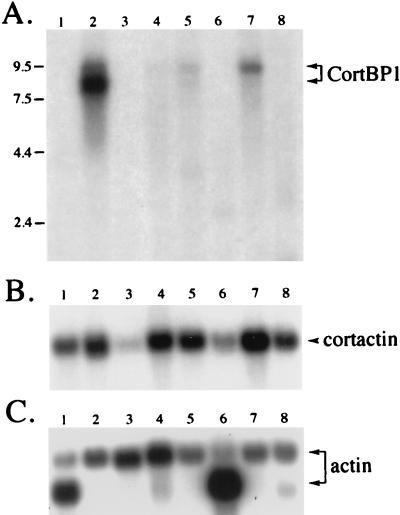
To investigate the tissue distribution of the CortBP1 expression, a CortBP1 cDNA fragment (nucleotides 3245 to 4111) derived from the clone 2H was used to probe a mouse multiple-tissue Northern blot. A major mRNA species of approximately 8 kb was detected exclusively in brain tissue, whereas a much less abundant species of approximately 9.5 kb was detected in brain, kidney, liver, and lung tissue (Fig. 3A). No hybridization to mRNAs from heart, spleen, skeletal muscle, and testis tissue was detected. Northern blot analysis using two other independent probes (from cDNA 2H and clone 10.a) that represented different regions of CortBP1 cDNA yielded identical results (data not shown).
FIG. 3.
Tissue distribution of CortBP1 mRNA and cortactin mRNA. A blot containing poly(A)+ RNA from multiple mouse tissues was hybridized with a CortBP1 cDNA fragment (A), a mouse cortactin cDNA fragment (B), or a β-actin cDNA fragment (C) as described in Materials and Methods. Lanes: 1, heart; 2, brain; 3, spleen; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; 8, testis. The positions of markers of known size (kilobases) are indicated at the left.
Previous studies have shown that the cortactin gene is widely expressed in murine tissues (51). In order to compare the expression pattern of the cortactin gene with that of the CortBP1 gene, the same RNA blot was rehybridized with a mouse cortactin cDNA probe (Fig. 3B). Both cortactin and CortBP1 mRNAs were expressed at high levels in brain tissue, whereas little or no expression of both proteins was observed in spleen or skeletal muscle tissue. On the other hand, cortactin was expressed in heart, lung, liver, and testis tissues, in which little CortBP1 mRNA expression was detected.
Expression of the CortBP1 protein.
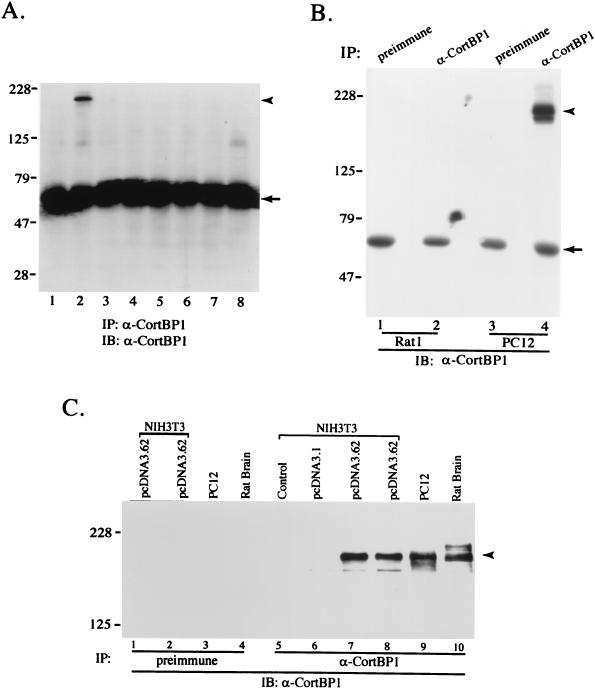
To characterize the endogenous CortBP1 protein, a rabbit polyclonal serum (anti-CortBP1) was raised against the C-terminal region (amino acids 941 to 1252) of CortBP1 (CortBP1cte) as described in Materials and Methods. CortBP1 expression in a number of adult rat tissues was then assessed by immunoprecipitation and Western blot analysis with anti-CortBP1. As shown in Fig. 4A, a protein of approximately 180 kDa was detected in brain lysates while preimmune serum showed no reactivity with this protein (data not shown). Interestingly, no immunoreactive bands were detected in the other tissues examined, including heart, spleen, lung, liver, skeletal muscle, kidney, and testis. Analysis of several cell lines revealed expression of the CortBP1 protein in the rat adrenal pheochromocytoma cell line PC12, whereas expression of p180 CortBP1 was undetectable in the rat fibroblast cell line RAT1 (Fig. 4B) or in mouse NIH 3T3 cells (Fig. 4C, lane 5).
FIG. 4.
Expression of the CortBP1 protein. (A and B) Distribution of the CortBP1 protein in lysates from rat tissues and cell lines. Lysate (1.0 mg) derived from eight adult rat tissues (A) or RAT1 or PC12 cells (B) was incubated with anti-CortBP1 or preimmune serum. Cellular proteins in the immunocomplexes were subjected to Western blot analysis with anti-CortBP1. (A) Lane 1, heart; lane 2, brain; lane 3, spleen; lane 4, lung; lane 5, liver; lane 6, skeletal muscle; lane 7, kidney; lane 8, testis. (C) Expression of full-length CortBP1 in NIH 3T3 cells. Five hundred micrograms of lysates from NIH 3T3 cells (lane 5), NIH 3T3 cells transfected with pcDNA3.1 (lane 6), or NIH 3T3 cells transfected with pcDNA3.62 (lanes 1, 2, 7, and 8) or 1.0 mg of lysates from PC12 cells (lanes 3 and 9) or rat brain (lanes 4 and 10) was incubated with anti-CortBP1 (lanes 5 to 10) or preimmune serum (lanes 1 to 4). Precipitated proteins were subjected to Western blot analysis with anti-CortBP1. The positions of molecular mass markers are indicated in kilodaltons at the left of each panel. The positions of p180 CortBP1 and the heavy chain of immunoglobulin G are indicated with arrowheads and arrows, respectively. IP, immunoprecipitation; IB, immunoblotting.
Since the relative molecular mass of CortBP1 determined by SDS-PAGE was much greater than the molecular mass calculated from the predicted amino acid sequence, a construct (pcDNA3.62) comprising the entire predicted coding sequence was generated from clone 6.b and clone 2.b, subcloned into the expression vector pcDNA3.1, and transfected into NIH 3T3 cells. As shown in Fig. 4C, lysates from cells transfected with pcDNA3.62 contained a major species that comigrated with the major CortBP1-immunoreactive band in PC12 and rat brain lysates. No immunoreactive band of the appropriate size was detected in lysates from nontransfected or pcDNA3.1-transfected cells. Based on this result, we conclude that the open reading frame deduced from the isolated CortBP1 cDNA clones contains the complete coding region for p180 CortBP1.
Characterization of the interaction between CortBP1 and cortactin.
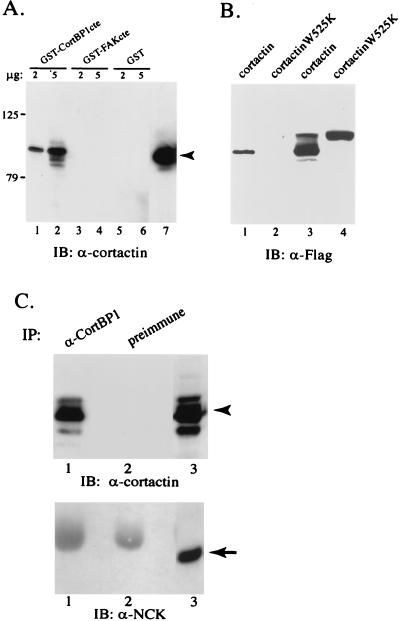
The identification of CortBP1 by two-hybrid screening indicated an interaction between the cortactin SH3 domain and the CortBP1cte containing the ppI motif. To further explore the interaction between CortBP1 and cortactin, we examined the ability of GST-CortBP1cte to associate with endogenous cortactin in NIH 3T3 cell lysates. GST, GST-CortBP1cte, or GST-FAKcte (amino acids 687 to 1054 [35]) fusion proteins were coupled to glutathione-Sepharose beads and incubated with NIH 3T3 cell lysates. GST-FAKcte was used as a control to specify the interaction since the FAKcte polypeptide contains two class II SH3 ligand motifs that have been shown to serve as the binding sites for the SH3 domains of p130_cas_ and Graf (35, 37). The amount of cortactin bound to immobilized GST fusion proteins was then determined by Western blot analysis with the cortactin-specific MAb 4F11. As shown in Fig. 5A, cortactin coprecipitated with immobilized GST-CortBP1cte fusion proteins in a dose-dependent manner whereas little cortactin coprecipitated with beads coated with either GST alone or GST-FAKcte.
FIG. 5.
Interaction between CortBP1 and cortactin. (A) Specific interaction of GST-CortBP1cte with cortactin in NIH 3T3 cell lysates. Two (lanes 1, 3, and 5) or five (lanes 2, 4, and 6) micrograms of each immobilized GST fusion protein was incubated with 600 μg of NIH 3T3 cell lysates. Precipitated cellular proteins were subjected to Western blot analysis with the anti-cortactin MAb 4F11. Lane 7 contains 30 μg of total NIH 3T3 cell lysate. The arrowhead indicates the position of cortactin. (B) Requirement for a functional cortactin SH3 domain for CortBP1 interaction. Five micrograms of GST-CortBP1cte protein was mixed with 700 μg of lysates from NIH 3T3 cells expressing Flag-tagged wild-type cortactin (lane 1) or cortactin SH3 mutant (lane 2). The protein complexes were collected by centrifugation and analyzed by Western blot analysis with the Flag-specific MAb M5. As loading controls, 50 μg of cell lysates expressing wild-type and mutant cortactin was loaded in lanes 3 and 4, respectively. (C) In vivo interaction of CortBP1 and cortactin in PC12 cells. Seven hundred micrograms of PC12 cell lysates was incubated with anti-CortBP1 (lanes 1) or preimmune serum (lanes 2). Immunocomplexes were subjected to Western blot analysis with 4F11 (top panel) and subsequently with an anti-NCK MAb (bottom panel). Lanes 3 were loaded with 100 μg of total PC12 cell lysates. IB, immunoblotting; IP, immunoprecipitation. Arrowhead, position of cortactin; arrow, position of NCK.
To assess the requirement of the cortactin SH3 domain for interacting with CortBP1, we compared the ability of GST-CortBP1cte to bind to an epitope (Flag)-tagged full-length cortactin and to a Flag-tagged cortactinW525K mutant expressed in NIH 3T3 cells. Lysates from cells transfected with pFlag-cortactin contained two major species that were reactive with the Flag epitope-specific MAb M5 (Fig. 5B). In contrast, pFlag-cortactinW525K-transfected cell lysates contained only one major species comigrating with the slower-migrating form of the wild-type cortactin (Fig. 5B, lanes 3 and 4), implicating a conformational change resulting from the point mutation in the SH3 domain. Little cortactinW525K was pulled down by immobilized GST-CortBP1cte fusion proteins, whereas wild-type cortactin was readily detected in the precipitated complex (Fig. 5B, lanes 1 and 2), indicating the requirement for SH3 domain integrity for mediating interaction with CortBP1.
To assess the ability of endogenous CortBP1 to bind cortactin, the in vivo interaction between CortBP1 and cortactin was examined in undifferentiated PC12 cells in which both proteins are expressed. PC12 cell lysates were incubated with either anti-CortBP1 or preimmune serum, and cortactin present in the immunocomplexes was detected by Western blot analysis with the MAb 4F11. Approximately 5 to 15% of the cortactin present in PC12 cell lysates was reproducibly recovered in the anti-CortBP1 immunocomplex, whereas no detectable cortactin was present in preimmune complex (Fig. 5C). When the same protein blot was probed with a MAb specific for NCK (an unrelated SH3-containing protein [44]), no NCK protein could be detected in the CortBP1 immunocomplex (Fig. 5C). Similar results were obtained with PC12 cells differentiated by treatment with nerve growth factor for 1 to 60 min or 1 to 9 days (data not shown). Thus, both in vitro and in vivo protein interaction analysis confirmed the association of cortactin with CortBP1.
Subcellular localization of ectopically expressed CortBP1 in fibroblasts.
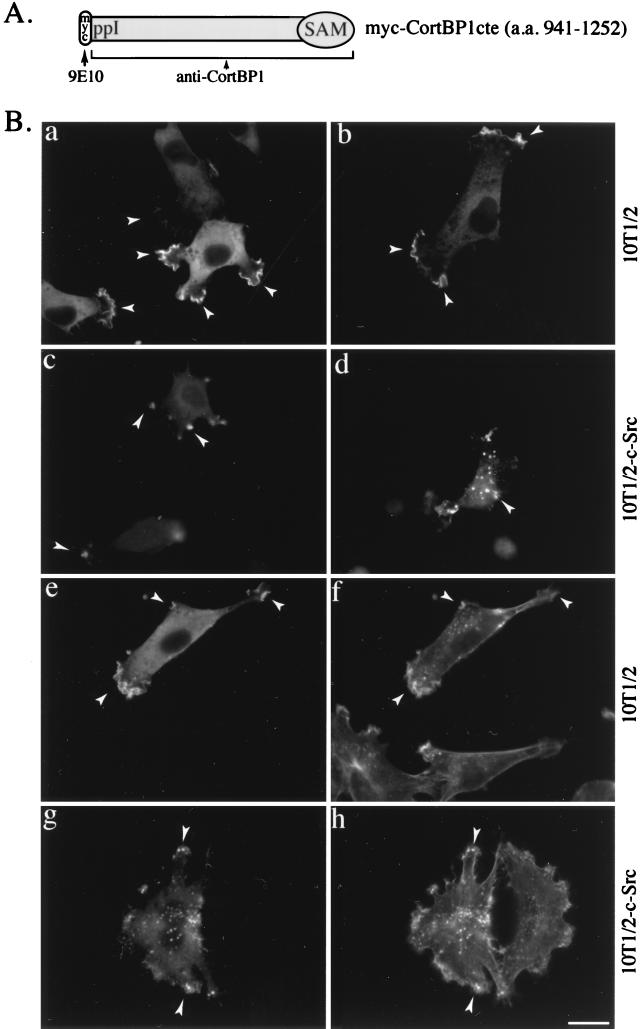
Previous studies have shown that a significant portion of cortactin colocalizes with F-actin-containing structures including lamellipodia and membrane ruffles in fibroblasts as well as in podosomes in Src-transformed fibroblasts (77, 78). To assess whether CortBP1 would colocalize with cortactin present in the cortical cytoskeleton, Myc epitope-tagged CortBP1cte (Fig. 6A) was expressed in 10T1/2 cells and 10T1/2 cells overexpressing c-Src (10T1/2-c-Src). Localization of CortBP1 was determined by immunofluorescence staining with either anti-CortBP1 or Myc epitope-specific MAb 9E10. In 10T1/2 cells overexpressing CortBP1, both antibodies revealed intense staining of CortBP1 within lamellipodia and membrane ruffles (Fig. 6B [a and b]). Accumulation of CortBP1 at the cell cortex was most prominent at the leading and trailing edges of motile cells (Fig. 6B and 7). These lamellipodia and membrane ruffles were enriched with F-actin as shown by costaining experiments with phalloidin (Fig. 6B [e and f]). Little background staining was detected in untransfected cells with either anti-CortBP1 (Fig. 7a and b) or MAb 9E10 (Fig. 6B [e and f]). Similar staining patterns of CortBP1 were also observed in two other fibroblast cell lines, REF52 and Swiss 3T3, overexpressing CortBP1cte (data not shown). When expressed in 10T1/2-c-Src cells, CortBP1 accumulated in dot-like structures in the peripheral extensions in some cells as well as throughout the cytoplasm in others (Fig. 6B [c and d]). As shown in Fig. 6B (g and h), these dot-like structures were enriched with F-actin and are likely membrane-substratum contact sites similar to podosomes (77).
FIG. 6.
Colocalization of CortBP1cte and cortical F-actin in 10T1/2 cells. (A) Diagram of Myc epitope-tagged CortBP1cte. The MAb 9E10 is directed to the Myc epitope, and anti-CortBP1 recognizes CortBP1cte. (B) Intracellular distribution of CortBP1 in 10T1/2 and 10T1/2-c-Src cells. 10T1/2 (a, b, e, and f) or 10T1/2-c-Src (c, d, g, and h) cells were transfected with the Myc-CortBP1cte expression vector and fixed 16 to 18 h posttransfection. Localization of CortBP1 in individual cells was detected with either anti-CortBP1 (a and c) or 9E10 (b and d). Transfected cells were also stained with 9E10 (e and g) in combination with fluorescently conjugated phalloidin (f and h) to visualize the colocalization of CortBP1 and F-actin. Arrowheads indicate lamellipodia and membrane ruffles in 10T1/2 cells and podosome-like structures in 10T1/2-c-Src cells. Bar, 20 μm.
FIG. 7.
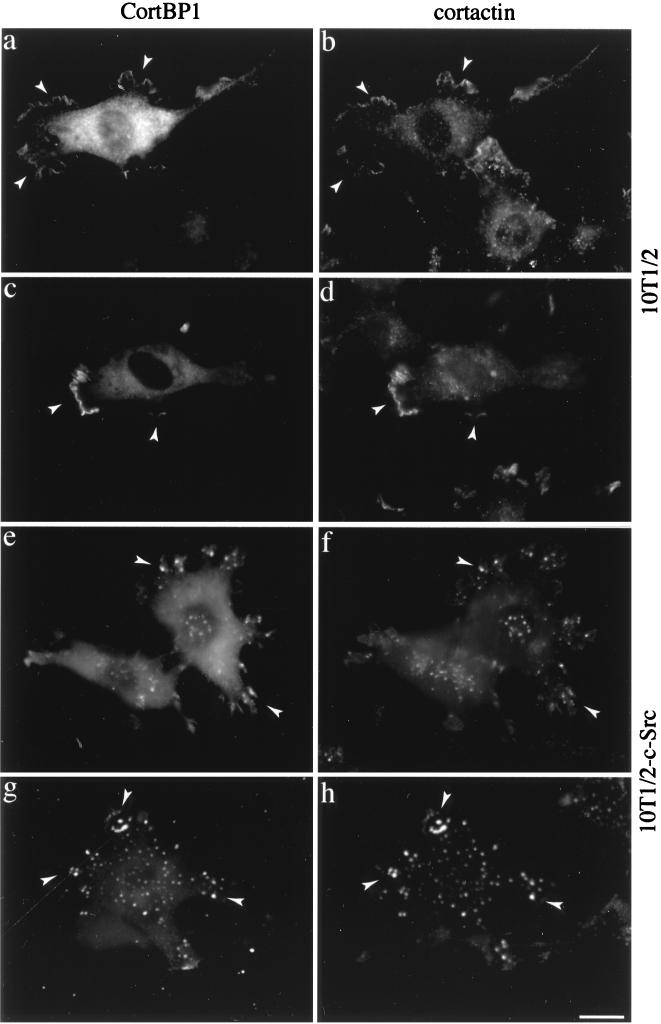
Colocalization of CortBP1 and cortactin in 10T1/2 cells. 10T1/2 (a to d) and 10T1/2-c-Src (e to h) cells were transfected with the Myc-CortBP1cte expression vector and fixed 16 to 18 h posttransfection. Cells were then immunostained either with anti-CortBP1 in combination with the anti-cortactin MAb 4F11 (a, b, e, and f) or with 9E10 in combination with the cortactin-specific antibody anti-Cterm (c, d, g, and h). Arrowheads indicate lamellipodia and membrane ruffles in 10T1/2 cells and podosome-like structures in 10T1/2-c-Src cells. Bar, 20 μm.
To confirm the colocalization of CortBP1 with cortactin, cells overexpressing CortBP1 were stained with anti-CortBP1 in combination with the MAb 4F11 (Fig. 7a, b, e, and f) or, alternatively, with 9E10 in combination with the cortactin-specific polyclonal antibody anti-Cterm (Fig. 7c, d, g, and h). As shown in Fig. 7, significant costaining of CortBP1 and cortactin was observed in lamellipodia and membrane ruffles in 10T1/2 cells and in podosome-like structures in 10T1/2-c-Src cells. These data confirmed the colocalization of CortBP1 and cortactin within subcellular compartments enriched with cortical F-actin.
Analysis of CortBP1 and cortactin in differentiating hippocampal neurons.
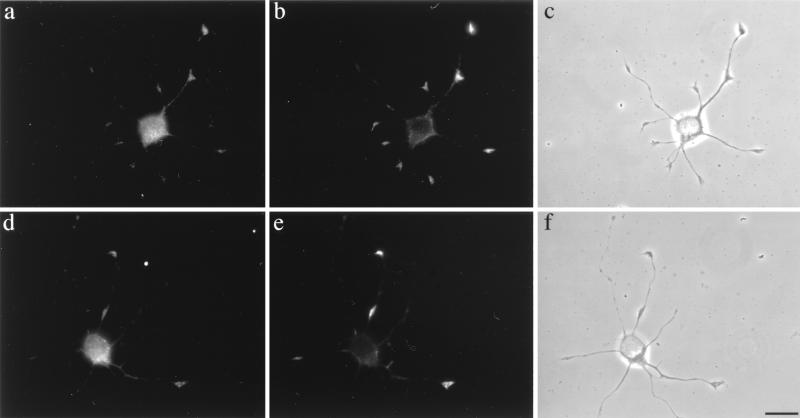
The intense costaining of CortBP1 and cortactin within the lamellipodia and membrane ruffles prompted our studies of the intracellular distribution of these two proteins in differentiating neurons which contain similar F-actin cortical structures in growth cones (45). To find neurons expressing both proteins, we first examined the expression levels of cortactin and CortBP1 in brain lysates isolated from rats at different developmental stages. Western blot analysis of brain lysates revealed that the expression levels of CortBP1, the major bands recognized by anti-CortBP1 (data not shown), increased during the course of embryonic development and remained at high levels during postnatal development (Fig. 8A). In contrast, the expression level of cortactin in rat brain was relatively constant at all of the nine developmental stages examined (Fig. 8A). Since expression of both CortBP1 and cortactin is readily detected in the embryonic 18-day rat brain lysates, in vitro-cultured neurons explanted from embryonic 18-day rat hippocampus were used to assess the subcellular localization of CortBP1 and cortactin by indirect immunofluorescence with the CortBP1-specific anti-CortBP1 and the cortactin-specific MAb 4F11. After 24 h in culture, a majority of cells displayed extended neurites with defined growth cone structures as previously described (31). At this stage, cortactin displayed intense staining within growth cones and weaker staining in the periphery of the cell body (Fig. 8B [a and b]). Little staining of cortactin was observed in neurite shafts. Interestingly, CortBP1 was observed within growth cones and the cell body (Fig. 8B [c and d]). Preincubation of anti-CortBP1 with purified antigen effectively blocked the staining in growth cones, leaving residual fluorescence within the cell body (Fig. 8B [g and h]). Staining of the cell body but not growth cones was seen with preimmune serum (Fig. 8B [e and f]). The staining patterns of cortactin and CortBP1 within growth cones are very similar in that they primarily localized at the central portion of growth cones. Coimmunostaining analysis further suggested the colocalization of CortBP1 and cortactin within growth cones (Fig. 9). These experiments clearly indicate that endogenous cortactin and CortBP1 reside in the same intracellular compartment in outgrowing neurites, supporting a direct interaction of these two proteins in vivo.
FIG. 9.
Colocalization of CortBP1 and cortactin in growth cones of rat hippocampal neurons. Cultured rat hippocampal neurons were immunostained with the anticortactin MAb 4F11 (b and e) in combination with anti-CortBP1 (a and d). Panels c and f are phase-contrast images of the cells shown in panels a and b and d and e, respectively. Bar, 20 μm.
DISCUSSION
Previous studies have suggested that the actin-binding protein cortactin may mediate aspects of cell signaling associated with the cortical cytoskeleton. The present study provides evidence that cortactin forms an SH3 domain-dependent protein complex with a novel 180-kDa protein. This protein represents the first identified natural ligand for the cortactin SH3 domain and is thus referred to as CortBP1. Sequence analysis of CortBP1 revealed the presence of two readily identifiable sequence motifs in the C-terminal region: a sequence virtually identical to a consensus sequence defined for the cortactin SH3 domain-binding peptides (66) and a sequence similar to recently identified SAM domains (58). The expression pattern of CortBP1 is restricted, being expressed predominantly in brain tissue. The stable interaction of cortactin and CortBP1 was demonstrated by coimmunoprecipitation of cortactin with CortBP1 in PC12 cell lysates. In addition, CortBP1, overexpressed in fibroblasts, colocalizes with cortactin and cortical F-actin within lamellipodia and membrane ruffles in normal cells and within podosome-like structures in c-Src overexpressors. Colocalization of endogenous cortactin and CortBP1 was also observed in growth cones of differentiating rat hippocampal neurons. These data indicate that endogenous CortBP1 and cortactin reside within the same subcellular compartment and suggest a possible involvement of cortactin and CortBP1 in the dynamic actin reorganization during neuritogenesis.
Data obtained by yeast two-hybrid and in vitro interaction analysis indicate that the SH3 domain of cortactin is responsible for mediating the observed interaction with CortBP1. Mutation of the highly conserved tryptophan residue to lysine in the cortactin SH3 domain efficiently blocked the interaction of full-length cortactin with CortBP1, as evidenced by the inability of GST-CortBP1 to pull down mutant cortactin from cell lysates. These data, coupled with the observation that mutant SH3 domain fusions fail to bind CortBP1 in the yeast two-hybrid assay, indicate the dependence of the interaction with CortBP1 on the structural integrity of the cortactin SH3 domain. The critical role of the cortactin SH3 domain for interacting with CortBP1 is further supported by the presence of the ppI motif in CortBP1, which displays a precise match with the consensus sequence (Fig. 2D) for cortactin SH3 domain-binding peptides. The sequence of this consensus motif, identified by screening a biased X6PXXPX6 peptide library with GST-cortactin SH3 fusion proteins, is related to class II ligands for SH3 domains (66). Interestingly, the cortactin SH3 domain does not show detectable interaction with peptides preferred by SH3 domains of other proteins including Src, Yes, Abl, Crk, Grb2, and phospholipase Cγ (66). The apparent specificity of the cortactin SH3 domain may derive from two unusual residue selections in cortactin SH3 domain-binding peptides. First, whereas an aliphatic residue is present at the −1 position (Fig. 2D) in most class II ligands (27), a positively charged residue (lysine or arginine) is present at the analogous position in cortactin SH3 domain-binding peptides. Second, the cortactin SH3 domain-binding peptides show a strict preference for lysine over arginine at the 5 position (Fig. 2D). We have examined the contribution of the CortBP1 ppI motif to interaction with cortactin by mutational analysis. Mutation of proline 948 and proline 950 to alanine within the ppI motif (e.g., …KPA948VA950PKP…) reduced the ability of CortBP1 to bind the cortactin SH3 domain in two-hybrid interaction analysis and with endogenous cortactin in in vitro GST pull-down experiments (data not shown). While these results underscore the importance of the CortBP1 ppI motif in binding the cortactin SH3 domain, we cannot rule out the contribution of other CortBP1 sequences in mediating the stable interactions with the cortactin SH3 domain.
The C-terminal 67 amino acids of CortBP1 display extensive similarity to SAM domains, recently identified protein modules present in diverse eukaryotic organisms from yeasts to humans (60). SAM-containing proteins have been implicated in developmental regulation and signal transduction (60). For example, SAM-containing proteins are essential for pheromone-induced sexual differentiation in yeast (Byr2p, Ste11p, and Ste4p [16]) and for regulating anterior-posterior patterning in Drosophila oocytes (polyhomeotic protein and Bicaudal-C [18, 46]). Two yeast SAM domain-containing proteins, Boi1p and Boi2p, are associated with cytoskeleton and play an important role in the maintenance of cell polarity during bud formation (6). While the molecular function of SAM domains remains largely unclear, recent studies of several SAM-containing proteins suggest a possible role of SAM domains in mediating protein-protein interactions (3, 48, 55, 64). Given the apparent association of CortBP1 with cortactin–F-actin complexes, it will be interesting to further investigate whether the CortBP1 SAM domain is responsible for recruiting other signaling proteins to the dynamic cortical cytoskeleton in growth cones.
In contrast to the cortactin gene, which is expressed in a wide variety of tissues, the expression pattern of the CortBP1 gene appears restricted. Northern blot analysis has shown that the major species of CortBP1 transcript (∼8 kb) is present exclusively in brain tissue and a less abundant species (∼9.5 kb) is present in brain, kidney, lung, and liver tissues. Whereas the CortBP1 protein is readily detected in brain tissues by Western blot analysis, we have failed to detect p180 CortBP1 in kidney, lung, and liver tissues. The difficulty of detecting the CortBP1 protein in these tissues may be due to the lower sensitivity of the Western blot analysis. The restricted expression pattern of CortBP1 suggests that the cortactin-CortBP1 interaction may have physiological significance in neurons and that in other cell types cortactin may interact with either CortBP1-like protein(s) or another tissue-specific binding partner(s).
Previous studies have shown that cortactin displays an intense staining within lamellipodia as well as a punctate staining within the cytoplasm in adherent cells cultured in the presence of serum (77, 78). Recent studies from our laboratory (73) have shown that the distribution of cortactin between the cortical cytoskeleton and cytoplasmic structures is regulated. Sequestration of cortactin to the cytoplasmic pool increases upon serum starvation. Translocation of cortactin into lamellipodia and membrane ruffles is dramatically enhanced by activation of Rac1, a small GTP-binding protein of the Rho family. The cortical cytoskeleton-targeting signal is located within the N-terminal half of cortactin, whereas sequences within the C-terminal half appear to be dispensable for cortical actin localization (74). Based on these data and the documented function of SH3 domains in recruiting their ligands to certain subcellular compartments (15), we propose that the cortactin SH3 domain plays a role in targeting its binding partner(s) to the cortical cytoskeleton. In support of this, we have shown here that a significant portion of overexpressed CortBP1 colocalizes with cortactin and F-actin in lamellipodia and membrane ruffles of cultured murine fibroblast cells. The colocalization of CortBP1 with cortactin and F-actin was also observed in podosome-like structures in 10T1/2 cells overexpressing c-Src. These observations strongly suggest that cortactin mediates formation of multiprotein complexes within the cortical cytoskeleton, by the concomitant interaction with cortical F-actin via its N-terminal repeats and with other cellular proteins via the SH3 domain at its C terminus. Thus, cortactin would serve as a cortical actin-specific docking protein for binding partners such as CortBP1.
We have further shown in this study that endogenous cortactin and CortBP1 colocalize within the cortical F-actin-containing subcellular compartment in primary cultures of differentiating rat neurons. Indirect immunofluorescence analysis suggests that both cortactin and CortBP1 are components of growth cones in cultured neurons isolated from embryonic 18-day rat hippocampus. These cells, when placed in culture, extend neurites mimicking their developmental morphology in vivo (22). The immunostaining of CortBP1 within growth cones appeared to be specific, since preimmune serum and antigen-blocked antibody failed to stain growth cones. The cortactin staining is essentially confined to growth cones, similar to the staining pattern of cortactin observed within growth cones of cultured Xenopus spinal cord neurons (57). Both CortBP1 staining and cortactin staining in growth cones of rat hippocampal neurons are primarily localized within the central portion of growth cones, very similar to the previously described staining pattern of F-actin in these cells (31). Additionally, costaining experiments support the colocalization of cortactin and F-actin in growth cones of the rat hippocampal neurons (data not shown). Previous studies have suggested that the F-actin remodeling within neuronal growth cones, in response to extracellular attraction or repulsion cues, plays an important role in regulating directional neurite extension (7, 45). Many proteins, including members of the Src family of tyrosine kinases (36, 47); actin-binding proteins such as profilin (26), gelsolin (68), myosin-V (72), and neurabin (52); and membrane-associated protein GAP-43 (2), have been identified as components of growth cones. Of note, the recently identified neural tissue-specific F-actin-binding protein neurabin has been implicated in neurite formation (52). The localization of cortactin and CortBP1 within growth cones, together with the brain-specific expression of CortBP1, suggests that the cortactin-CortBP1 interaction may also be involved in signaling pathways associated with the formation and migration of growth cones during neurite outgrowth.
A database search for CortBP1 homologs in other organisms reveals two human brain cDNA fragments in the expressed sequence tag database (accession no. m86079 and h41098 [1]) and one human DNA segment in the sequence-tagged site database (accession no. z51760 [21]). Both display high DNA sequence identity (81, 90, and 88%, respectively) to rat CortBP1 cDNA. The similarity between rat CortBP1 and the predicted open reading frames of these DNA sequences spans the N-terminal region (amino acids 294 to 405, 27 to 171, and 1 to 57, respectively). The existence of a putative human CortBP1 homolog is an indication of the conservation of CortBP1 function in other organisms and an involvement of CortBP1 in common biological processes in diverse organisms.
ACKNOWLEDGMENTS
We thank G. Banker and H. Asmussen for in vitro-cultured rat hippocampal neurons, S. J. Parsons for 10T1/2neo and 5Hd47 cells, A. A. Lanahan for pPC86 and pPC97 vectors and the rat hippocampus cDNA library in pPC86, A. Hall for the pRK5myc vector, J. S. Morrow for the pcDNA3Flag2AB vector, and M. T. Harte for the GST-FAKcte expression vector. We also thank M. E. Cox for helpful discussions.
This work was supported by grants CA29243 and CA40042 from the DHHS-NCI and grant 4491 from the Council for Tobacco Research, Inc., to J. T. Parsons; S. A. Weed is supported by NIH postdoctoral fellowship 1 F32 CA75695-01; W.-C. Xiong is supported by NIH NRSA fellowship NS09918.
ADDENDUM IN PROOF
CortBP1 contains a single PDZ domain (amino acids 38 to 133) that has sequence similarity to the PDZ domains of PSD95, Disc large-1, and Z0-1.
REFERENCES
- 1.Adams M D, Dubnick M, Kerlavage A R, Moreno R, Kelley J M, Utterback T R, Nagle J W, Fields C, Venter J C. Sequence identification of 2,375 human brain genes. Nature (London) 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- 2.Aigner L, Caroni P. Absence of persistent spreading, branching, and adhesion in GAP-43-depleted growth cones. J Cell Biol. 1995;128:647–660. doi: 10.1083/jcb.128.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr M M, Tu H, Aelst L V, Wigler M. Identification of Ste4 as a potential regulator of Byr2 in the sexual response pathway of Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:5597–5603. doi: 10.1128/mcb.16.10.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel P L, Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett W P, Banker G A. An electron microscopic study of the development of axons and dendrites by hippocampal neurons in culture. I. Cells which develop without intercellular contacts. J Neurosci. 1984;4:1944–1953. doi: 10.1523/JNEUROSCI.04-08-01944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender L, Lo H S, Lee H, Kokojan V, Peterson J, Bender A. Associations among PH and SH3 domain-containing proteins and Rho-type GTPases in yeast. J Cell Biol. 1996;133:879–894. doi: 10.1083/jcb.133.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentley D, O’Connor T P. Cytoskeleton events in growth cone steering. Curr Opin Neurobiol. 1994;4:43–48. doi: 10.1016/0959-4388(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 8.Bretscher A. Microfilament structure and function in the cortical cytoskeleton. Annu Rev Cell Biol. 1991;7:337–374. doi: 10.1146/annurev.cb.07.110191.002005. [DOI] [PubMed] [Google Scholar]
- 9.Button E, Shapland C, Lawson D. Actin, its associated proteins and metastasis. Cell Motil Cytoskelet. 1995;30:247–251. doi: 10.1002/cm.970300402. [DOI] [PubMed] [Google Scholar]
- 10.Cao L G, Wang Y L. Mechanism of the formation of contractile ring in dividing culture animal cells. J Cell Biol. 1990;110:1089–1095. doi: 10.1083/jcb.110.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang J-H, Gill S, Settleman J, Parsons S J. c-Src regulates the simultaneous rearrangement of actin cytoskeleton, p190RhoGAP, and p120RasGAP following epidermal growth factor stimulation. J Cell Biol. 1995;130:355–368. doi: 10.1083/jcb.130.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W-T, Chen J-M, Parsons S J, Parsons J T. Local degradation of fibronectin at sites of expression of the transforming gene product pp60src. Nature (London) 1985;316:156–158. doi: 10.1038/316156a0. [DOI] [PubMed] [Google Scholar]
- 13.Chevray P M, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clerc P, Sansonetti P J. Entry of Shigella flexneri into HeLa cells: evidence for directed phagocytosis involving actin polymerization and myosin accumulation. Infect Immun. 1987;55:2681–2688. doi: 10.1128/iai.55.11.2681-2688.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen G B, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 16.Cook S, McCormick F. Ras blooms on sterile ground. Science. 1994;369:361–362. doi: 10.1038/369361a0. [DOI] [PubMed] [Google Scholar]
- 17.Dai Z, Pendergast A M. Abi2, a novel SH3-containing protein, interacts with the c-Ab1 tyrosine kinase and modulates c-Ab1 transforming activity. Genes Dev. 1995;9:2526–2528. doi: 10.1101/gad.9.21.2569. [DOI] [PubMed] [Google Scholar]
- 18.DeCamillis M, Cheng N, Pierre D, Brock H W. The polyhomeotic gene of Drosophila encodes a chromatin protein that shares polytene chromosome-binding sites with Polycomb. Genes Dev. 1992;6:223–232. doi: 10.1101/gad.6.2.223. [DOI] [PubMed] [Google Scholar]
- 19.Dehio C, Prevost M-C, Sansonetti P J. Invasion of epithelial cells by Shigella flexneri induces tyrosine phosphorylation of cortactin by a pp60c-src-mediated signaling pathway. EMBO J. 1995;14:2471–2482. doi: 10.1002/j.1460-2075.1995.tb07244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devarajan P, Stabach P R, Dematteis M A, Morrow J S. Na, K-ATPase transport from endoplasmic reticulum to Golgi requires the Golgi spectrin-ankyrin G119 skeleton in Madin-Darby canine kidney cells. Proc Natl Acad Sci USA. 1997;94:10711–10716. doi: 10.1073/pnas.94.20.10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature (London) 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 22.Dotti C G, Sullivan C A, Banker G A. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Downing J R, Reynolds A B. PDGF, CSF-1 and EGF induce tyrosine phosphorylation of p120, a pp60src transformation-associated substrate. Oncogene. 1991;6:607–613. [PubMed] [Google Scholar]
- 24.Drubin D G, Mulholland J, Zhu Z, Botstein D. Homology of a yeast actin-binding protein to signal transduction proteins and myosin-1. Nature (London) 1990;343:288–290. doi: 10.1038/343288a0. [DOI] [PubMed] [Google Scholar]
- 25.Durieu-Trautmann O, Chaverot N, Cazaubon S, Strosberg A D, Couraud P-O. Intercellular adhesion molecule 1 activation induces tyrosine phosphorylation of the cytoskeleton-associated protein cortactin in brain microvessel endothelial cells. J Biol Chem. 1994;269:12536–12540. [PubMed] [Google Scholar]
- 26.Faivre-Sarrailh C, Lena J Y, Had L, Vignes M, Lindberg U. Localization of profilin at presynaptic sites in the cerebellar cortex; implication for the regulation of the actin-polymerization state during axonal elongation and synaptogenesis. J Neurocytol. 1993;22:1060–1072. doi: 10.1007/BF01235749. [DOI] [PubMed] [Google Scholar]
- 27.Feng S, Chen J K, Yu H, Simon J A, Schreiber S L. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 28.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 29.Gietz R D, Schiestl R H. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 30.Goodman C S. Mechanisms and molecules that control growth cone guidance. Annu Rev Neurosci. 1996;19:341–377. doi: 10.1146/annurev.ne.19.030196.002013. [DOI] [PubMed] [Google Scholar]
- 31.Goslin K, Birgbauer E, Banker G, Solomon F. The role of cytoskeleton in organizing growth cones: a microfilament-associated growth cone component depends upon microtubules for its localization. J Cell Biol. 1989;109:1621–1631. doi: 10.1083/jcb.109.4.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greene L A, Aletta J M, Rukenstein A, Green S H. PC12 pheochromocytoma cells: culture, nerve growth factor treatment, and experimental exploitation. Methods Enzymol. 1987;147:207–216. doi: 10.1016/0076-6879(87)47111-5. [DOI] [PubMed] [Google Scholar]
- 33.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 34.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 35.Harte M T, Hildebrand J D, Burnham M R, Bouton A H, Parsons J T. p130Cas, a substrate associated with v-Src and v-Crk, localizes to focal adhesions and binds to focal adhesion kinase. J Biol Chem. 1996;271:13649–13655. doi: 10.1074/jbc.271.23.13649. [DOI] [PubMed] [Google Scholar]
- 36.Helmke S, Pfenninger K H. Growth cone enrichment and cytoskeletal association of non-receptor tyrosine kinases. Cell Motil Cytoskelet. 1995;30:194–207. doi: 10.1002/cm.970300304. [DOI] [PubMed] [Google Scholar]
- 37.Hildebrand J D, Taylor J M, Parsons J T. An SH3 domain-containing GTPase-activating protein for Rho and Cdc42 associates with focal adhesion kinase. Mol Cell Biol. 1996;16:3169–3178. doi: 10.1128/mcb.16.6.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hitt A, Luna E J. Membrane interactions with the actin cytoskeleton. Curr Opin Cell Biol. 1994;6:120–130. doi: 10.1016/0955-0674(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 39.Kanner S B, Reynolds A B, Vines R R, Parsons J T. Monoclonal antibodies to individual tyrosine-phosphorylated protein substrates of oncogene-encoded tyrosine kinases. Proc Natl Acad Sci USA. 1990;87:3328–3332. doi: 10.1073/pnas.87.9.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitamura D, Kaneko H, Miyagoe Y, Ariyasu T, Watanabe T. Isolation and characterization of a novel human gene expressed specifically in the cells of hematopoietic lineage. Nucleic Acids Res. 1989;17:9367–9379. [PMC free article] [PubMed] [Google Scholar]
- 41.Kitamura D, Kaneko H, Taniuchi I, Akagi K, Yamamura K-I, Watanabe T. Molecular cloning and characterization of mouse HS1. Biochem Biophys Res Commun. 1995;208:1137–1146. doi: 10.1006/bbrc.1995.1452. [DOI] [PubMed] [Google Scholar]
- 42.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larabell C A. Cortical cytoskeleton of the Xenopus oocyte, egg, and early embryo. Curr Top Dev Biol. 1995;31:433–453. doi: 10.1016/s0070-2153(08)60235-5. [DOI] [PubMed] [Google Scholar]
- 44.Lehmann J M, Riethmuller G, Johnson J P. NCK, a melanoma cDNA encoding a cytoplasmic protein consisting of the src homology units SH2 and SH3. Nucleic Acids Res. 1990;18:1048. doi: 10.1093/nar/18.4.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin C-H, Thompson C, Forscher P. Cytoskeletal reorganization underlying growth cone motility. Curr Opin Neurobiol. 1994;4:640–647. doi: 10.1016/0959-4388(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 46.Mahone M, Saffman E E, Lasko P F. Localized Bicaudal-C RNA encodes a protein containing a KH domain, the RNA binding motif of FMR1. EMBO J. 1995;14:2043–2055. doi: 10.1002/j.1460-2075.1995.tb07196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maness P F, Aubry M, Shores C G, Frame L, Pfenninger K H. c-Src gene product in developing rat brain is enriched in nerve growth cone membranes. Proc Natl Acad Sci USA. 1988;85:5001–5005. doi: 10.1073/pnas.85.14.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marcus S, Polverino A, Barr M, Wigler M. Complexes between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc Natl Acad Sci USA. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayer B J, Eck M J. Minding your p’s and q’s. Curr Biol. 1995;5:364–367. doi: 10.1016/s0960-9822(95)00073-x. [DOI] [PubMed] [Google Scholar]
- 50.Meredith S D, Levine P A, Burns J A, Gaffey M J, Boyd J C, Weiss L M, Erickson N L, Williams M E. Chromosome 11q13 amplification in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 1995;121:790–794. doi: 10.1001/archotol.1995.01890070076016. [DOI] [PubMed] [Google Scholar]
- 51.Miglarese M R, Mannion-Henderson J, Wu H, Parsons J T, Bender T P. The protein tyrosine kinase substrate cortactin is differentially expressed in murine B lymphoid tumors. Oncogene. 1994;9:1989–1998. [PubMed] [Google Scholar]
- 52.Nakanishi H, Obaishi H, Satoh A, Wada M, Mandai K, Satoh K, Nishioka H, Matsuura Y, Mizoguchi A, Takai Y. Neurabin: a novel neural tissue-specific actin filament-binding protein involved in neurite formation. J Cell Biol. 1997;139:951–961. doi: 10.1083/jcb.139.4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okamura H, Resh M D. p80/85 cortactin associates with the Src SH2 domain and colocalizes with v-Src in transformed cells. J Biol Chem. 1995;270:26613–26618. doi: 10.1074/jbc.270.44.26613. [DOI] [PubMed] [Google Scholar]
- 54.Olson M F, Pasteris N G, Gorski J L, Hall A. Faciogenital dysplasia protein (FGD1) and Vav, two related proteins required for normal embryonic development, are upstream regulators of Rho GTPases. Curr Biol. 1996;6:1628–1633. doi: 10.1016/s0960-9822(02)70786-0. [DOI] [PubMed] [Google Scholar]
- 55.Pandey A, Duan H, Dixit V M. Characterization of a novel Src-like adaptor protein that associates with the Eck receptor tyrosine kinase. J Biol Chem. 1995;270:19201–19204. doi: 10.1074/jbc.270.33.19201. [DOI] [PubMed] [Google Scholar]
- 56.Patel A M, Incognito L S, Schechter G L, Wasilenko W J, Somers K D. Amplification and expression of EMS-1 (cortactin) in head and neck squamous cell carcinoma cell lines. Oncogene. 1996;12:31–35. [PubMed] [Google Scholar]
- 57.Peng H B, Xie H, Dai Z. Association of cortactin with developing neuromuscular specializations. J Neurocytol. 1997;26:637–650. doi: 10.1023/a:1018545827369. [DOI] [PubMed] [Google Scholar]
- 58.Ponting C P. SAM: a novel motif in yeast sterile and Drosophila polyhomeotic proteins. Protein Sci. 1995;4:1928–1930. doi: 10.1002/pro.5560040927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ridley A J. Membrane ruffling and signal transduction. Bioessays. 1994;16:321–327. doi: 10.1002/bies.950160506. [DOI] [PubMed] [Google Scholar]
- 60.Schultz J, Ponting C P, Hofmann K, Bork P. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 1997;6:249–253. doi: 10.1002/pro.5560060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schuuring E, Verhoeven E, Mooi W J, Michalides R J A M. Identification and cloning of two overexpressed genes, U21B31/PRAD1 and EMS1, within the amplified chromosome 11q13 region in human carcinomas. Oncogene. 1992;7:355–361. [PubMed] [Google Scholar]
- 62.Schuuring E, Verhoeven E, Tinteren H V, Peterse J L, Nunnink B, Thunnissen F B J M, Devilee P, Cornelisse C J, Vijver M J V D, Mooi W J, Michalides R J A M. Amplification of genes within the chromosome 11q13 region is indicative of poor prognosis in patients with operable breast cancer. Cancer Res. 1992;52:5229–5234. [PubMed] [Google Scholar]
- 63.Schuuring E, Verhoeven E, Litvinov S, Michalides R J A M. The product of the EMS1 gene, amplified and overexpressed in human carcinomas, is homologous to a v-src substrate and is located in cell-substratum contact sites. Mol Cell Biol. 1993;13:2891–2898. doi: 10.1128/mcb.13.5.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Serra-Pages C, Kedersha N L, Fazikas L, Medley Q, Debant A, Streuli M. The LAR transmembrane protein tyrosine phosphatase and a coiled-coil LAR-interacting protein co-localize at focal adhesions. EMBO J. 1995;14:2827–2838. doi: 10.1002/j.1460-2075.1995.tb07282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 66.Sparks A B, Rider J E, Hoffman N G, Fowlkes D M, Quilliam L A, Kay B K. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Ab1, Cortactin, p53bp2, PLCγ, Crk, and Grb2. Proc Natl Acad Sci USA. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stossel T P. On the crawling of animal cells. Science. 1993;260:1086–1094. doi: 10.1126/science.8493552. [DOI] [PubMed] [Google Scholar]
- 68.Tanaka J, Kira M, Sobue K. Gelsolin is localized in neuronal growth cones. Dev Brain Res. 1993;76:268–271. doi: 10.1016/0165-3806(93)90217-x. [DOI] [PubMed] [Google Scholar]
- 69.Tanaka M, Gupta R, Mayer B J. Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adaptor protein. Mol Cell Biol. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taniuchi I, Kitamura D, Maekawa Y, Fukuda T, Kishi H, Watanabe T. Antigen-receptor induced clonal expansion and deletion of lymphocytes are impaired in mice lacking HS1 protein, a substrate of the antigen-receptor-coupled tyrosine kinases. EMBO J. 1995;14:3664–3678. doi: 10.1002/j.1460-2075.1995.tb00036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tarone G, Cirillo D, Giancotti F G, Comoglio P M, Marchisio P C. Rous sarcoma virus-transformed fibroblasts adhere primarily at discrete protrusions of the ventral membrane celled podosomes. Exp Cell Res. 1985;159:141–157. doi: 10.1016/s0014-4827(85)80044-6. [DOI] [PubMed] [Google Scholar]
- 72.Wang F-S, Wolenski J S, Cheney R E, Mooseker M S, Jay D G. Function of myosin-V in filopodial extension of neuronal growth cones. Science. 1996;273:660–663. doi: 10.1126/science.273.5275.660. [DOI] [PubMed] [Google Scholar]
- 73.Weed S A, Du Y, Parsons J T. Translocation of cortactin to the cell periphery is mediated by the small GTPase Rac1. J Cell Sci. 1998;111:2433–2443. doi: 10.1242/jcs.111.16.2433. [DOI] [PubMed] [Google Scholar]
- 74.Weed, S. A., Y. Du, and J. T. Parsons. Unpublished data.
- 75.Wilusz J, Pettine S M, Shenk T. Functional analysis of point mutations in the AAUAAA motif of the SV40 late polyadenylation signal. Nucleic Acids Res. 1989;17:3899–3908. doi: 10.1093/nar/17.10.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wong S, Reynolds A B, Papkoff J. Platelet activation leads to increased c-src kinase activity and association of c-src with an 85-kDa tyrosine phosphoprotein. Oncogene. 1992;7:2407–2415. [PubMed] [Google Scholar]
- 77.Wu H, Reynolds A B, Kanner S B, Vines R R, Parsons J T. Identification and characterization of a novel cytoskeleton-associated pp60src substrate. Mol Cell Biol. 1991;11:5113–5124. doi: 10.1128/mcb.11.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu H, Parsons J T. Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J Cell Biol. 1993;120:1417–1426. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhan X, Hu X, Hampton B, Burgess W H, Friesel R, Maciag T. Murine cortactin is phosphorylated in response to fibroblast growth factor-1 on tyrosine residues late in the G1 phase of the BALB/c 3T3 cell cycle. J Biol Chem. 1993;268:24427–24431. [PubMed] [Google Scholar]