CD4 Glycoprotein Degradation Induced by Human Immunodeficiency Virus Type 1 Vpu Protein Requires the Function of Proteasomes and the Ubiquitin-Conjugating Pathway (original) (raw)
Abstract
The human immunodeficiency virus type 1 (HIV-1) vpu gene encodes a type I anchored integral membrane phosphoprotein with two independent functions. First, it regulates virus release from a post-endoplasmic reticulum (ER) compartment by an ion channel activity mediated by its transmembrane anchor. Second, it induces the selective down regulation of host cell receptor proteins (CD4 and major histocompatibility complex class I molecules) in a process involving its phosphorylated cytoplasmic tail. In the present work, we show that the Vpu-induced proteolysis of nascent CD4 can be completely blocked by peptide aldehydes that act as competitive inhibitors of proteasome function and also by lactacystin, which blocks proteasome activity by covalently binding to the catalytic β subunits of proteasomes. The sensitivity of Vpu-induced CD4 degradation to proteasome inhibitors paralleled the inhibition of proteasome degradation of a model ubiquitinated substrate. Characterization of CD4-associated oligosaccharides indicated that CD4 rescued from Vpu-induced degradation by proteasome inhibitors is exported from the ER to the Golgi complex. This finding suggests that retranslocation of CD4 from the ER to the cytosol may be coupled to its proteasomal degradation. CD4 degradation mediated by Vpu does not require the ER chaperone calnexin and is dependent on an intact ubiquitin-conjugating system. This was demonstrated by inhibition of CD4 degradation (i) in cells expressing a thermally inactivated form of the ubiquitin-activating enzyme E1 or (ii) following expression of a mutant form of ubiquitin (Lys48 mutated to Arg48) known to compromise ubiquitin targeting by interfering with the formation of polyubiquitin complexes. CD4 degradation was also prevented by altering the four Lys residues in its cytosolic domain to Arg, suggesting a role for ubiquitination of one or more of these residues in the process of degradation. The results clearly demonstrate a role for the cytosolic ubiquitin-proteasome pathway in the process of Vpu-induced CD4 degradation. In contrast to other viral proteins (human cytomegalovirus US2 and US11), however, whose translocation of host ER molecules into the cytosol occurs in the presence of proteasome inhibitors, Vpu-targeted CD4 remains in the ER in a transport-competent form when proteasome activity is blocked.
The human immunodeficiency virus type 1 (HIV-1)-specific accessory protein Vpu performs two distinct functions in the viral life cycle (11, 12, 29, 34, 46, 47, 50–52; reviewed in references 31 and 55): enhancement of virus particle release from the cell surface, and the selective induction of proteolysis of newly synthesized membrane proteins. Known targets for Vpu include the primary virus receptor CD4 (63, 64) and major histocompatibility complex (MHC) class I molecules (28). Vpu is an oligomeric class I integral membrane phosphoprotein (35, 48, 49) with a structurally and functionally defined domain architecture: an N-terminal transmembrane anchor and C-terminal cytoplasmic tail (20, 34, 45, 47, 50, 65). Vpu-induced degradation of endoplasmic reticulum (ER) membrane proteins involves the phosphorylated cytoplasmic tail of the protein (50), whereas the virion release function is mediated by a cation-selective ion channel activity associated with the membrane anchor (19, 31, 45, 47).
CD4 is a 55-kDa class I integral membrane glycoprotein that serves as the primary coreceptor for HIV entry into cells. CD4 consists of a large lumenal domain, a transmembrane peptide, and a 38-residue cytoplasmic tail. It is expressed on the surface of a subset of T lymphocytes that recognize MHC class II-associated peptides, and it plays a pivotal role in the development and maintenance of the immune system (reviewed in reference 30). Down regulation of CD4 in HIV-1-infected cells is mediated through several independent mechanisms (reviewed in references 5 and 55): intracellular complex formation of CD4 with the HIV envelope protein gp160 (8, 14), endocytosis of cell surface CD4 induced by the HIV-1 nef gene product (1, 2), and ER degradation induced by the HIV-1 vpu gene product (63, 64).
Vpu-induced degradation of CD4 is an example of ER-associated protein degradation (ERAD). ERAD is a common outcome when proteins in the secretory pathway are unable to acquire their native structure (4). Although it was thought that ERAD occurs exclusively inside membrane vesicles of the ER or other related secretory compartments, this has gained little direct experimental support. Indeed, there are several recent reports that ERAD may actually represent export of the target protein to the cytosol, where it is degraded by cytosolic proteases. It was found that in yeast, a secreted protein, prepro-α-factor (pαF), is exported from microsomes and degraded in the cytosol in a proteasome-dependent manner (36). This process was dependent on the presence of calnexin, an ER-resident molecular chaperone that interacts with N-linked oligosaccharides containing terminal glucose residues (3). In mammalian cells, two human cytomegalovirus (HCMV) proteins, US2 and US11, were found to cause the retranslocation of MHC class I molecules from the ER to the cytosol, where they are destroyed by proteasomes (61, 62). In the case of US2, class I molecules were found to associate with a protein (Sec61) present in the channel normally used to translocate newly synthesized proteins into the ER (termed the translocon), leading to the suggestion that the ERAD substrates are delivered to the cytosol by retrograde transport through the Sec61-containing pore (61). Fujita et al. (24) reported that, similar to these findings, the proteasome-specific inhibitor lactacystin (LC) partially blocked CD4 degradation in transfected HeLa cells coexpressing CD4, Vpu, and HIV-1 Env glycoproteins. In the present study, we show that Vpu-induced CD4 degradation can be completely blocked by proteasome inhibitors, does not require the ER chaperone calnexin, but requires the function of the cytosolic polyubiquitination machinery which apparently targets potential ubiquitination sites within the CD4 cytoplasmic tail. Our findings point to differences between the mechanism of Vpu-mediated CD4 degradation and ERAD processes induced by the HCMV proteins US2 and US11 (61, 62).
MATERIALS AND METHODS
Cell culture and transfection.
HeLa cells (ATCC CCL2; American Type Culture Collection, Rockville, Md.) were propagated and transfected as previously described (63, 64). A3.01 cells, a CD4+ human T-cell line, were cultivated as described previously (23, 51). _tsA_1S9 cells were maintained and thermally challenged as described previously (13). The identity of the E1 mutation was routinely confirmed by shifting cells overnight to 39°C, resulting in gross morphological alteration obvious in the inverted microscope. Calnexin-deficient T-lymphoblastoid cell line NKR(cal−) (26) and its calnexin-transfected derivative NKR(cal+) were cultivated as described previously (43). Plasmid pHIV-CD4 (63), which allows the expression of wild-type CD4 under the control of the HIV-1 long terminal repeat, and the subgenomic HIV-1 expression vectors pNL-A1 (51) have been described previously.
Proteasome inhibitors.
The peptide aldehyde inhibitors _N_-carbobenzoxyl-l-leucinyl-l-leucinyl-l-leucinal (zLLL) and _N_-carbobenzoxyl-l-leucinyl-l-leucinal (zLL) were synthesized as described by Vinitsky et al. (58). LC was provided by E. J. Corey, Department of Chemistry, Harvard University (Cambridge, Mass.).
Construction of pHIVCD4KRcyto.
The four Lys residues in the CD4 cytosolic domain were converted to Arg by site-directed mutagenesis using a two-step PCR strategy. In the first step, two fragments, A and B, were amplified by using primers A1 (5′-GACATCGTGGTGCTAGC) and A2 (5′-GAGGGCACTGGCAGGTCCTCCTCTCACTGAGGAGTCTCCTGATCTGAG) or B1 (5′-GGAGGACCTGCCAGTGCCCTCACCGGTTTCAGAGGACATGTAG) and B2 (5′-GAGAGGGGATCCTCATGAGCAGTGGGG), respectively, for amplification of pHIV-CD4 template DNA (63). Resulting PCR products were purified by standard techniques, mixed, and used as templates for second-round amplification using the external primers A1 and B2. The final PCR product was purified, digested with _Nhe_I and _Bam_HI, and cloned into the _Nhe_I/_Bam_HI sites of pHIV-CD4. The presence of the mutations was confirmed by sequencing. In addition to the Lys-to-Arg substitutions, clone pHIVCD4KRcyto contains an additional amino acid change (W to R) at position 241, located in the CD4 ectodomain. Inasmuch as the ectodomain of CD4 is not critical for Vpu-mediated degradation of the protein, we assume that this substitution has no effect on the sensitivity of CD4KRcyto to Vpu.
rVVs.
The following recombinant vaccinia viruses (rVVs) have been previously described: VV-Vpu and VV-UDEL1 (28), v-CB3 (7), VV-E1 (13), NP (nucleoprotein)-Vac (54), and Ub (ubiquitin)-Arg-NP-Vac (54). The human Ub gene was altered at amino acid position 48 (creating Ub48) by changing lysine to arginine as follows. A _Bsm_I-_Xho_I fragment was isolated from plasmid pUC-Ub (16), and an Arg codon was introduced in position 48. The UbR48 gene was cloned into vector pSC11 (17), and rVVs were generated in CV-1 cells as described previously (10). All rVVs were generated in CV-1 cells, plaque purified, and propagated in thymidine kinase-deficient human 143B osteosarcoma cells as described previously (10).
Antisera and antibodies.
A polyclonal anti-Vpu antiserum (sheep), directed against Vpu residues 41 to 58, and a polyclonal anti-Vpu serum (rabbit), directed against Vpu residues 32 to 81, were used to detect Vpu as described previously (35, 45, 47, 48). For immunocollection of CD4, a mixture of the following antibodies was used: a polyclonal rabbit antiserum directed against glycosylated CD4 (15), monoclonal antibody OKT4a (Ortho Diagnostic Systems, Raritan, N.J.), a rabbit polyclonal antiserum (T4-Cy) raised to keyhole limpet hemocyanin coupled to a synthetic peptide corresponding to CD4 residues 394 to 422 (located in the cytosolic domain), and a polyclonal antiserum directed against recombinant nonglycosylated CD4 produced in sheep (40) or in rabbit. NP was detected by using monoclonal antibody H16-L10-4R5 (ATCC HB65).
Pulse-chase metabolic labeling, immunocollection, and fluorography.
HeLa cells were infected with rVVs at a multiplicity of infection (MOI) of 3 to 10 PFU per cell as indicated in the text. At 2.5 h postinfection (p.i.) cells were starved for 30 min in serum-free and methionine-free medium, and pulse-chase experiments were performed as described previously (28, 45, 63, 64). Immunoprecipitation of CD4 from CHAPS [(3-chloramidopropyl)-dimethyl-ammonio]-1-propanesulfonate]-deoxycholate-treated cell lysates was described before (47). Immunoprecipitates were separated in 10% AcrylAide gels (FMC Bioproducts, Rockland, Maine), and radioactive bands were visualized by fluorography. Quantitation of fluorograms was performed by exposing gels to PhosphorImager screens (Molecular Dynamics, Sunnyvale, Calif.), which were imaged and quantitated by using the PhosphorImager software. Endo-β-_N_-acetylglucosaminidase H (endo H) from Streptomyces plicatus (New England Biolabs, Beverly, Mass.) was used to analyze the oligosaccharide composition of immunoprecipitated CD4 as described previously (28). For Vpu binding studies, cell lysates were prepared in 1% digitonin buffer as described previously (6), and immunoprecipitates collected with CD4 antibodies were denatured in 2% sodium dodecyl sulfate (SDS) for 10 min at 95°C, diluted 1:500 in 0.1% Triton buffer (50 mM Tris-HCl [pH 7.4], 300 mM NaCl, 0.1% Triton X-100), and subjected to a second round of immunocollection with anti-Vpu antibodies.
Assay for CD4 ubiquitination.
HeLa cells expressing CD4 and Vpu were treated with 5 μM zLLL or left untreated. After 10 min of pulse-labeling cells were subjected to CHAPS-deoxycholate lysis. Insoluble proteins were solubilized by incubation in 2% SDS containing 50 μg of DNase I per ml for 30 min at 95°C followed by dilution (1:500) in Triton buffer (50 mM Tris-HCl [pH 7.4], 300 mM NaCl, 0.1% Triton X-100). Both fractions were either immunoprecipitated with anti-CD4 antibodies first, followed by a second round of immunoprecipitation with anti-Ub serum (Sigma, St. Louis, Mo.), or vice versa, with anti-Ub followed by anti-CD4. To detect ubiquitinated CD4 by Western blotting, anti-CD4 antibodies were chemically cross-linked to GammaBind-Plus–Sepharose beads (Pharmacia LKB, Piscataway, N.J.), using dimethyl pimelimidate as described previously (44), and used for immunocollection of CD4. Immunoprecipitates were separated in 10% AcrylAide gels, transferred to Immobilon membranes (Millipore Corp., Bedford, Mass.), and probed with biotinylated anti-Ub antiserum followed by streptavidin-peroxidase.
RESULTS
Proteasome-specific inhibitors block Vpu-induced CD4 proteolysis.
In most of the experiments described below, we used rVVs to express CD4, Vpu, and proteins involved in cytosolic protein degradation. The broad host range of rVV, its transient nature as an expression vector, and high levels of expression of rVV-encoded recombinant proteins facilitated biochemical analysis of Vpu-mediated CD4 degradation in a variety of cell lines under controlled conditions.
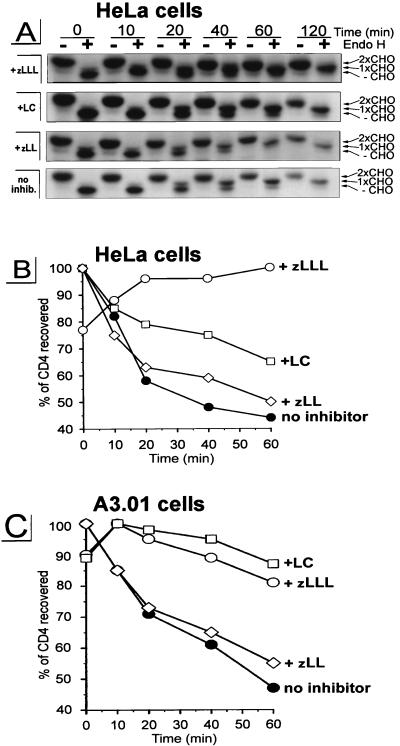
In the first experiment, HeLa cells were coinfected with v-CB3, expressing human CD4 (7), and VV-Vpu, expressing wild-type Vpu (28), or a control rVV, VV-UDEL1, expressing a mutated vpu transcript that is unable to direct translation of Vpu-specific sequences (28). Cells were radiolabeled for 5 min with [35S]methionine and chased for up to 120 min (Fig. 1A). Detergent extracts were immunoprecipitated with a mixture of CD4-specific antibodies and analyzed by SDS-polyacrylamide gel electrophoresis (Fig. 1A). The material recovered was quantitated with a PhosphorImager, and the kinetics of CD4 recovery were determined by calculating the levels of CD4 collected during the chase period relative to the highest level of CD4 recovered (defined as 100%) (Fig. 1B). In the absence of Vpu, rVV-expressed CD4 exhibits a half-life (_t_1/2) of >240 min (not shown). By contrast, coexpression of Vpu resulted in a rapid initial degradation of CD4 (_t_1/2 of ∼30 min) followed by a slower rate of decay (Fig. 1B). This bimodal kinetics of CD4 loss (Fig. 1A and B) is similar to results previously obtained using HeLa cells transfected with HIV-1 subgenomic expression vectors (45, 50, 61, 62) or infected with rVV expressing CD4 and Vpu (28).
FIG. 1.
Treatment with proteasome-specific inhibitors rescues CD4 from Vpu-induced proteolysis. HeLa cells (A and B) or the CD4+ T-cell line A3.01 (C) were coinfected with rVVs expressing either human CD4 (v-CB3) at an MOI of 2 or Vpu (VV-Vpu) at an MOI of 5; 2.5 h p.i., the culture was split and cells were either pretreated with 5 μM zLLL, 5 μM zLL, or 10 μM LC or left untreated. After a 5-min pulse-labeling, cells were chased in the presence or absence of inhibitor for up to 2 h. CD4 molecules were recovered from detergent lysates by using a mixture of anti-CD4 antibodies. The immunoprecipitates recovered from HeLa cell lysates were split and either not treated (−) or treated with endo H (+), separated in a 10% AcrylAide gel, and analyzed by fluorography (A). Only parts of the fluorograms demonstrating CD4-specific bands are shown. The three CD4-specific bands in the endo H-treated samples represent CD4 molecules either partially resistant (1xCHO), completely resistant (2xCHO), or sensitive (−CHO) to endo H treatment as indicated on the right. (B) Relative amounts of CD4 detected in the untreated samples shown in panel A were quantitated with an Image Analyzer, and the stability of CD4 present at different times during the chase period was calculated, using as 100% the highest value for each treatment.
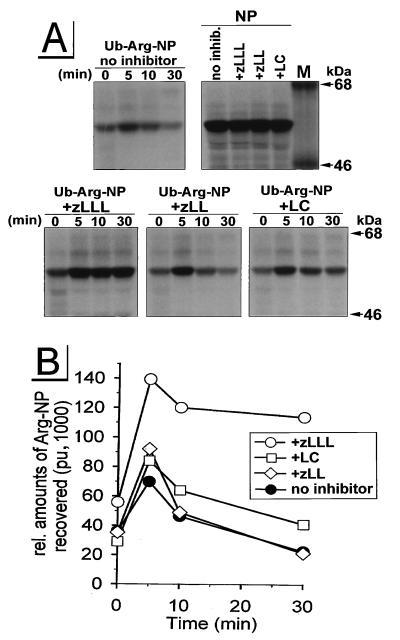
Incubation of cells with the peptide aldehyde zLLL (also known as MG132), which blocks all of the enzymatically defined activities of the proteasome (33, 58), resulted in the complete stabilization of CD4 in Vpu-expressing cells (Fig. 1, +zLLL). To control for the effects of zLLL on nonproteasomal proteases, we also incubated rVV-infected HeLa cells with zLL, which has a spectrum of inhibitory activity on other cytosolic and lysosomal nonproteasomal proteases similar to that of zLLL but does not detectably affect proteasome function at the concentrations used (56, 58). zLL had little effect on CD4 degradation in rVV-infected HeLa cells (Fig. 1, +zLL), supporting the role of proteasomes in the Vpu-mediated CD4 degradation. This was further shown in assays using LC, the most specific proteasome inhibitor available. LC irreversibly inactivates the chymotryptic- and tryptic-like activities of the proteasome by binding covalently to the catalytic β subunits (21). Incubation of HeLa cells with 10 μM LC partially rescued CD4 from Vpu-induced degradation (Fig. 1A and B, +LC). This partial effect of LC on CD4 stability is consistent with recent findings of Fujita et al. (24) for stably transfected HeLa cell clones. Since it is known that proteasome composition and also uptake of inhibitors are cell type dependent (38), we analyzed the effectiveness of LC and zLLL to block CD4 degradation in a human CD4+ T-cell line. We infected parallel cultures of A3.01 cells (23) with v-CB3 and VV-Vpu and conducted a pulse-chase experiment similar to that described for Fig. 1A and B. By contrast to the situation in HeLa cells, in A3.01 cells (Fig. 1C) LC was completely effective, blocking CD4 degradation to a similar extent as zLLL, while the control inhibitor zLL, again, had almost no effect on the Vpu-induced degradation of CD4. From this result, we assumed that the difference between LC (Fig. 1B and C and reference 24) and zLLL (Fig. 1C) in the rescue of CD4 from degradation reflects the degree of inhibition of proteasome activity. To confirm this assumption and to interpret the partial effect of LC on Vpu-mediated CD4 degradation in HeLa cells, we compared the abilities of LC and zLLL to block degradation of previously characterized (54) model cytosolic target proteins (Fig. 2). For this purpose, we used the rVV Ub-Arg-NP-Vac, expressing a chimeric protein consisting of NH2-terminal Ub fused with influenza virus NP with an extraneous Arg at the Ub-NP junction (54). Following translation, the Ub-Arg-NP fusion protein is rapidly removed by the abundant Ub hydrolases in cells, leaving Arg at the NH2 terminus. According to the N-end rule (57), Arg represents a highly destabilizing residue directing the fusion protein into the proteasomal pathway. Consistent with this rule, in our experiment Ub-Arg-NP is degraded with a _t_1/2 of ∼10 min (Fig. 2, panel no inhibitor) following a ∼5-min folding time period observed in all rVV-infected HeLa cultures (Fig. 2). The increased recovery of antibody-reactive Arg-NP fusion protein during the first 5 min of the chase period may reflect masking of antibody binding sites in the nascent fusion protein. Incubation of VV-Ub-Arg-NP-infected cells with the control inhibitor zLL had no significant effect on Ub-Arg-NP stability (Fig. 2, panel +zLL). In contrast, zLLL stabilized Arg-NP over the 30-min chase period (Fig. 2, panel +zLLL), confirming the involvement of proteasomes in the degradation of the Arg-NP fusion. As observed with Vpu-mediated CD4 degradation in HeLa cells (Fig. 1A and B and reference 24), LC only partially protected Arg-NP from degradation during the chase period. The parallel nature of the partial blockade of LC on the degradation of CD4 and the model proteasome substrate Ub-Arg-NP in HeLa cells is consistent with the involvement of proteasomes in both processes and, in addition, provides an explanation for the partial effect of LC observed in HeLa cells (Fig. 1A and B and reference 24). In addition to cell-type-dependent variations in uptake of proteasome inhibitors, it is known that proteolytic activities of the proteasome complex are redundant and depend on the cell-type-specific proteasome subunit composition (18, 37). It is conceivable that the residual proteasome activity observed in HeLa cells treated with LC (Fig. 1A and B and reference 24) is due to the fact that the proteasome composition in HeLa cells is different to that of T cells, where LC was fully active (Fig. 1C).
FIG. 2.
Effects of zLLL, zLL, and LC on proteasomal degradation of the model substrate Ub-Arg-NP in HeLa cells. HeLa cells that had been incubated for 45 min with or without the protease inhibitors were infected for 5 h with an rVV expressing either Ub-Arg-NP or wild-type NP (MOI = 10) in the presence or the absence of the appropriate inhibitor. Cells were pulse-labeled for 1 min and chased for up to 30 min. In panel NP, cells were lysed immediately after the pulse-labeling. Detergent lysates were immunoprecipitated with an anti-NP antibody and separated in a 9% acrylamide gel (A). Relative amounts of NP were quantitated with a PhosphorImager and plotted against time (pu, PhosphorImager units) (B). M, molecular mass standard protein.
In summary, these findings indicate that proteasome activity is required for Vpu-induced CD4 degradation. In other experiments, we also found that neither zLLL nor LC interferes with the binding of Vpu to CD4 (data not shown), a necessary step during Vpu-mediated CD4 degradation (6), suggesting that the requirement for proteasomes reflects their direct participation in this process.
CD4 protected by proteasome inhibitors is not irreversibly translocated to the cytosol.
The proteasome-dependent degradation of several ER substrates has been shown to involve the translocation of the substrate to the cytosol (36, 61, 62). In contrast to those reports, we have been unable to detect accumulation of breakdown intermediates of CD4 in the cytosol, suggesting that CD4 which was rescued by proteasome inhibitors from Vpu-induced degradation remains in the ER. To further evaluate this hypothesis, we conducted endo H analysis on CD4. Substrates which have been retranslocated to the cytosol cannot further be transported through the Golgi complex and therefore will not demonstrate Golgi complex-mediated alterations in N-linked oligosaccharides. We analyzed the export of CD4 from the ER in HeLa cells coexpressing CD4 and Vpu by monitoring the structure of its N-linked oligosaccharides (Fig. 1A). One half of CD4 immunoprecipitated during the chase period was treated with endo H, which removes N-linked oligosaccharides characteristic of the proximal secretory pathway but not those that have been acted on by enzymes in the medial or trans cisternae of the Golgi complex. CD4 possess two N-linked oligosaccharides in positions 272 and 300 within its ectodomain, one of which acquires endo H resistance as the protein traffics through the Golgi complex (53). In the absence of protease inhibitors, virtually all of the CD4 that escapes Vpu-induced degradation is exported from the ER since it acquires one endo H-resistant oligosaccharide (Fig. 1A, 1xCHO). Most importantly, CD4 rescued completely by zLLL or partially by LC from Vpu-mediated degradation acquires endo H resistance with similar kinetics as CD4 expressed in untreated cells. In summary, these results indicate that in the absence of proteasome activity, CD4 remains in the secretory pathway of Vpu-expressing cells in a transport-competent form and therefore is not irreversibly translocated to the cytosol as was reported for other ERAD substrates (36, 61, 62).
Calnexin is not required for Vpu-induced CD4 proteolysis.
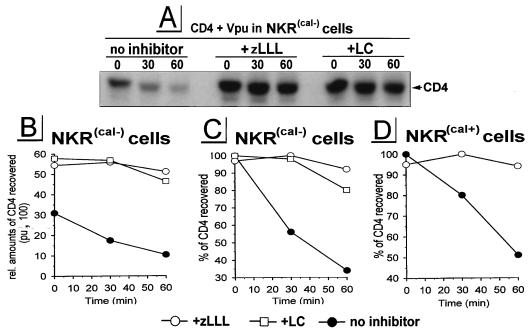
Molecular chaperones like calnexin associate with specific target proteins that define their functions in protein trafficking, protein translocation, gene expression, and proteolysis. The possible involvement of the ER chaperone calnexin in ERAD is supported by observations that ERAD substrates like alpha-1 protease inhibitor (32) and the cystic fibrosis transmembrane conductance regulator (CFTR) mutant DF508 (41) are retained in the ER by interaction with calnexin. More directly, in yeast, calnexin is required for the translocation of pαF to the cytosol (36). To evaluate the potential requirement for calnexin in Vpu-mediated CD4 degradation, we used a mutagenized human T-cell line, NKR(cal−), deficient for calnexin (26) (Fig. 3A to C). A derivative, NKR(cal+) (43), expressing human calnexin from a transfected gene (Fig. 3D) was used as positive control. We infected parallel cultures of NKR cells with v-CB3 and VV-Vpu and performed a pulse-chase protocol as described above. In NKR(cal−) cells, a large initial loss of CD4 occurred immediately after pulse-labeling. This is probably due to rapid degradation of CD4 occurring during the 5-min labeling period, since this would account for the ∼2-fold enhancement of CD4 recovery following treatment of cells with LC or zLLL (Fig. 3A and B). The rate of CD4 degradation with a _t_1/2 of ∼35 min was similar in calnexin-expressing NKR(cal+) cells (Fig. 3D), demonstrating that Vpu-induced CD4 degradation can occur independently of calnexin. As observed for the CD4+ T-cell line A3.01 (Fig. 1C), in both NKR(cal+) and NKR(cal−) cells, LC and zLLL had similar capacities to entirely block Vpu-induced CD4 degradation, supporting the notion that the partial inhibitory effect of LC on proteasome activity is a HeLa cell-type-specific phenomenon which was not observed in T cells.
FIG. 3.
Vpu-induced ERAD of CD4 does not require calnexin. Calnexin-deficient T-cell line NKR(cal−) (A to C) and calnexin-expressing T-cell line NKR(cal+) (D) were coinfected with VV-Vpu and v-CB3, treated with the inhibitors indicated, pulse-labeled for 5 min, and chased for up to 2 h. CD4 molecules recovered were analyzed by fluorography [shown only for NKR(cal−) cells (A)]. Relative amounts of CD4 established by PhosphorImager analysis (B) were used for quantitation of CD4 stability (C and D). pu, PhosphorImager units.
Ub-activating enzyme E1 is involved in Vpu-mediated CD4 proteolysis.
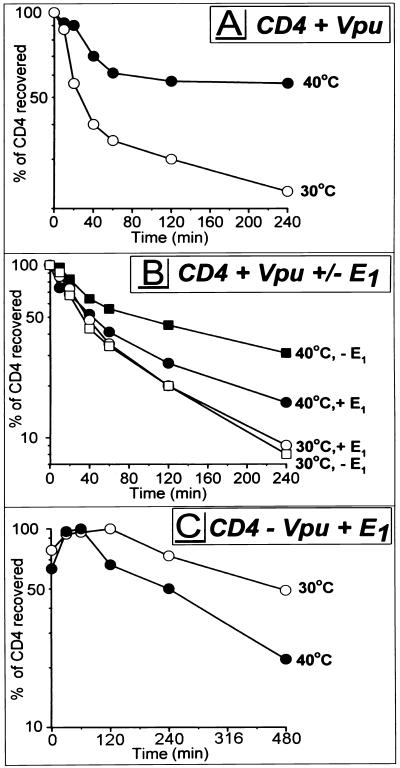
Many of the substrates of the 26S proteasome carry poly-Ub attached to the ɛ-amino group of Lys residues. To examine a potential role of ubiquitination in CD4 degradation, we used _tsA_1S9 cells, a derivative of mouse L929 fibroblasts (66) expressing a temperature-sensitive mutant form of Ub-activating enzyme E1, which is essential for the covalent attachment of Ub to proteins. At or above 39.5°C, protein ubiquitination in _tsA_1S9 cells is reduced and can be restored by infection with an rVV expressing wild-type human E1 (13). _tsA_1S9 cells were coinfected with VV-Vpu and v-CB3, and the standard pulse-chase protocol was followed as described above except that one sample was thermally challenged at 40°C beginning with the initiation of the 30 min Met-free starvation period. Elevating the temperature compromised Vpu-induced CD4 degradation (Fig. 4A). The partial effect is consistent with a residual capacity of _tsA_1S9 cells to ubiquitinate proteins at 40°C (13). Two findings indicate that block of CD4 degradation is a consequence of inactivating E1. First, degradation of CD4 in parental L929 cells was actually greater at 40°C than at 30°C (not shown). Second, coexpression of rVV-encoded wild-type E1 accelerated CD4 degradation at 40°C but not at 30°C (Fig. 4B). rVV-expressed E1 did not completely restore the rate of CD4 degradation to that observed in wild-type cells at 40°C or mutant cells at 30°C. This could be due to differences between the expression level and activity of rVV-expressed E1 and those of endogenous E1. The requirement for Vpu in this process is shown by the much slower decay of CD4 in cells coinfected with v-CB3, VV-E1, and a control virus that does not express Vpu (Fig. 4C), where the _t_1/2s of CD4 were ∼8 h at 30°C and ∼4 h at 40°C. The enhanced turnover of CD4 at 40°C observed in the absence of Vpu (Fig. 4C) could only be counterproductive to the thermal rescue of CD4 degradation observed in _tsA_1S9 cells expressing Vpu (Fig. 4B and C), confirming that the temperature shift to 40°C does not account for the rescue of CD4 from Vpu-induced degradation. In summary, these results indicate that the Ub-activating enzyme E1 is involved in Vpu-mediated CD4 degradation, suggesting that Vpu-induced CD4 degradation depends on the cytosolic ubiquitination machinery.
FIG. 4.
Ub-activating enzyme E1 is important for Vpu-induced CD4 degradation. _tsA_1S9 cells were coinfected with VV-Vpu (MOI = 2) and v-CB3 (MOI = 4) (A), in addition to either VV-E1 (MOI = 4) or VV-UDEL1 (MOI = 4) (B), or with v-CB3, VV-UDEL1, and VV-E1 (C). At 2.5 h p.i., cells were incubated at either permissive (30°C) or nonpermissive (40°C) temperature; 30 min later, standard pulse-chase experiments were conducted. Stability of CD4 recovered during 4 h (A and B) or 8 h (C) of chase periods (not shown) was calculated and plotted against time. Note the difference in time scales between panels A and B (0 to 240 min) and panel C (480 min).
Inhibition of Vpu-mediated CD4 degradation by transdominant mutant UbR48.
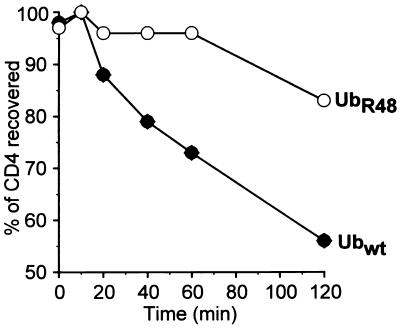
The role of ubiquitination in CD4 degradation was further examined by using an rVV expressing UbR48. The ɛ-amino group of Lys48 is often used for polyubiquitination by forming an isopeptide bond with the COOH terminus of a new Ub molecule. It has been shown that in yeast, multiubiquitination related to proteolysis and cell cycle progression can be blocked by expression of UbR48, which acts in a transdominant negative manner (22). The potential of UbR48 to interfere with Vpu-induced CD4 degradation was analyzed in HeLa cells coinfected with v-CB3, VV-Vpu, and either VV-UbR48 or VV-Ub. To maximize the expression of VV-encoded wild-type and mutant Ub proteins relative to endogenous Ub, we infected cells with relatively high MOIs of rVVs and conducted the pulse-chase experiment at 10 h p.i. Coexpression of UbR48 almost completely blocked CD4 degradation in the initial 60 min following synthesis, which then proceeded at a rate over the next 60 min similar to that observed in cells expressing wild-type Ub, with ∼60% of the pulse-labeled CD4 recovered in the presence of wild-type Ub and ∼80% recovered in the presence of UbR48 after 2 h of chase (Fig. 5). This finding is consistent with the results from Fig. 4 and therefore further supports the involvement of cytosolic protein polyubiquitination in the process of Vpu-mediated CD4 degradation.
FIG. 5.
Vpu-induced CD4 degradation is blocked by coexpression of transdominant mutant UbR48. Parallel HeLa cultures were cotransfected with VV-Vpu (MOI = 4), v-CB3 (MOI = 2), and either VV-UbR48 or VV-Ubwt (MOI = 10). At 10 h p.i., cells were pulse-labeled for 7.5 min, and CD4 molecules immunoprecipitated from chase samples were analyzed by fluorography (not shown). Stability of CD4 molecules recovered was calculated and plotted as a function of time. wt, wild type.
Vpu-induced CD4 degradation requires potential ubiquitination within the CD4 cytoplasmic tail.
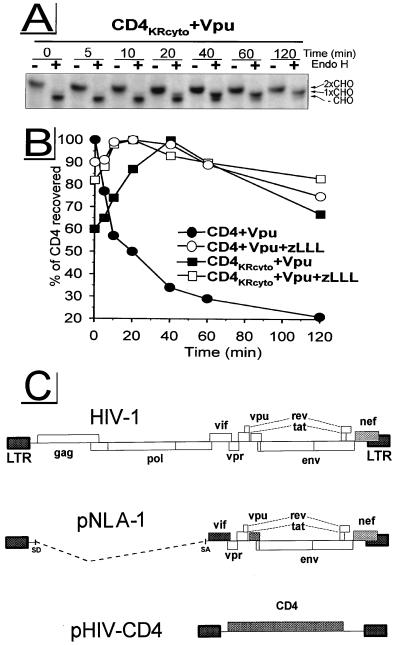
CD4, as a type I integral membrane protein, consists of a 38-amino-acid cytosolic domain that can potentially be exposed to enzymes of the ubiquitin system, while the large ectodomain of CD4 cannot be ubiquitinated but can be exposed to ER lumenal proteases. The cytosolic tail of CD4 possesses four Lys residues. To examine the possibility that one or more of these residues is ubiquitinated during the process of Vpu-induced CD4 degradation, we constructed a mutant, CD4KRcyto, substituting Arg for Lys in positions 411, 417, 418, and 428 of CD4. Although of similar net charge and effect on the folding of the protein backbone, unlike Lys, the Arg side chain has no ɛ-amino group and therefore cannot serve as an anchor for Ub. For the pulse-chase experiments shown in Fig. 6, target molecules (wild-type CD4 and the mutant CD4KRcyto) and the effector Vpu were expressed from HIV-1 subgenomic expression vectors under control of the HIV-1 long terminal repeat promoter (Fig. 6C), the system previously used to study Vpu-induced CD4 degradation (45, 50, 63, 64). Similar to the situation in rVV-infected HeLa cells (Fig. 1 and reference 28), Vpu induced the degradation of wild-type CD4 with a _t_1/2 of ∼20 min; as in the assays described above, degradation was completely blocked by zLLL. In contrast, the tail mutant CD4KRcyto was stable in the presence of wild-type Vpu: the amount of CD4KRcyto recovered increased over the first 40 min of chase and then diminished slowly. As expected, zLLL had no effect on the folding and stability of CD4KRcyto. To analyze the transport of CD4KRcyto, chased samples were subjected in parallel to endo H treatment (Fig. 6A). CD4KRcyto was exported from the ER with kinetics similar to that observed for wild-type CD4 rescued by zLLL (Fig. 1A). Furthermore, binding studies between CD4KRcyto and Vpu by coimmunoprecipitation from digitonin lysates (not shown) demonstrate that mutation of potential ubiquitination sites introduced in the cytoplasmic tail of CD4KRcyto does not detectably modify association of CD4KRcyto with Vpu, a prerequisite for Vpu-induced CD4 degradation (6). Therefore, these findings suggest that polyubiquitination of one or more Lys residues in the cytosolic domain is required for the Vpu-induced CD4 degradation and may therefore occur prior to the proteolysis of CD4 by the 26S proteasome.
FIG. 6.
Potential ubiquitination sites within the CD4 cytoplasmic tail are required for Vpu-induced CD4 proteolysis. HeLa cells were cotransfected with the HIV-1 subgenomic expression vector pNLA-1, expressing wild-type Vpu, in combination with either pHIV-CD4, expressing wild-type CD4, or pHIV-CD4KRcyto, expressing mutant CD4KRcyto carrying Lys-to-Arg mutations within the cytoplasmic tail. At ∼24 h posttransfection, cells were preincubated with 5 μM zLLL or left untreated. Cells were pulse-labeled for 5 min and chased for up to 120 min in the absence or presence of zLLL. CD4 molecules were immunoprecipitated and analyzed by fluorography. Endo H analysis was performed as described for Fig. 1A. Only fluorograms demonstrating samples from experiment CD4KRcyto + Vpu are shown in panel A. Relative amounts of CD4 detected were quantitated and used to calculate the stability of CD4 molecules (B). (C) Schematic structure of HIV-1 subgenomic expression vectors pNLA-1 and pHIV-CD4. LTR, long terminal repeat; SD, splice donor; SA, splice acceptor.
DISCUSSION
Membrane proteins like CD4 are translocated into the ER in an unfolded state and subsequently maintained by various molecular chaperones that promote protein translocation, folding, and oligomerization. Proteins that are misfolded, inappropriately glycosylated, or fail to assemble into multimeric complexes are targeted to the ERAD pathway (4). In addition, proteins that ordinarily fold efficiently like CD4 and MHC class I heavy chain and are normally spared from ERAD can be directed into ERAD by viral factors like the HCMV proteins US2 and US11 (61, 62) or Vpu (63, 64).
Several lines of evidence suggested that Vpu directs CD4 into ERAD. In HIV-1-infected cells Vpu selectively induces destruction of CD4 that is trapped in the ER due to binding to gp160 (8, 10, 63, 64), while Vpu is inactive toward CD4 which has been exported from the ER (63, 64). Also, the activity of Vpu on CD4 can be enhanced by treatment with brefeldin A (63), which blocks ER-to-Golgi transport, is insensitive to inhibitors of lysosomal protein degradation (24), requires ATP consumption (6), and is a membrane-dependent process (11).
Although numerous substrates of the ERAD pathway have been identified, there is little evidence to suggest that proteolysis occurs within the ER lumen or other compartments of the secretory pathway. Indeed, a number of recent findings indicate that degradation of various ERAD substrates is blocked by proteasome inhibitors (24, 25, 36, 60–62). Inasmuch as proteasomes are located strictly in the cytosol and nucleus (note that the subset of proteasomes found in close proximity to the ER membrane [39] are topologically located in the cytosol), this suggests two, non-mutually exclusive possibilities: (i) ERAD substrates are translocated from the ER to the cytosol, where they are degraded by proteasomes; and (ii) proteasomes associated with the cytosolic surface of the ER membrane degrade the cytoplasmic tails of membrane proteins, causing alterations in the membrane and lumenal domains that target the proteins for destruction by proteases in the secretory pathway. As described in the introduction, there is direct evidence for the first possibility (60–62). The second possibility cannot be dismissed but as yet is not directly supported by experimental evidence.
Fujita et al. (24) recently reported that Vpu-induced degradation of CD4 was partially blocked by LC and peptide aldehyde proteasome inhibitors in HeLa cells, while nonproteasomal inhibitors had no effect on CD4 degradation. We confirm and extend these findings, demonstrating that (i) in T cells, LC, the most specific proteasome inhibitor, completely blocks Vpu-induced CD4 degradation; (ii) in contrast to observations with yeast pαF (36), the ER chaperone calnexin is not required for this process; (iii) cytosolic polyubiquitination, which probably targets the 38-amino-acid cytoplasmic tail of CD4, is required for CD4 degradation; and (iv) CD4, which becomes insensitive to Vpu in the presence of proteasome inhibitors, remains in the secretory pathway and is not detected in the cytosol. Together, these findings provide conclusive evidence that Vpu-induced CD4 degradation, previously believed to occur in the ER (63, 64), is at least initiated by the cytosolic proteasome-ubiquitin pathway. However, from the data available so far, we cannot formally rule out the possibility that after initial proteasomal degradation, fragments of CD4 are degraded by undefined ER proteases.
The requirement for ubiquitination in ERAD was first described by Ward et al. (60), who demonstrated that degradation of mutant CFTR is reduced in cells with thermally inactivated E1 or in cells expressing UbR48. CFTR is a multimembrane-spanning protein which is sensitive to proteasome processing (27) but has a large cytosolic domain and limited number of lumenal residues, raising the possibility that the ubiquitination requirement was limited to ER membrane proteins located exclusively or predominantly in the cytosol. Our findings indicate that ubiquitination can also be involved in the degradation of membrane proteins with large lumenal and short cytosolic domains. Furthermore, the requirement for Lys residues in the CD4 cytosolic tail suggests that ubiquitination of these residues is a necessary step in the process of Vpu-stimulated CD4 proteolysis. Based on these observations, we attempted to identify mono- or polyubiquitinated forms of CD4. We were unable, however, to detect ubiquitinated CD4 in the soluble or insoluble fractions of detergent extracts or in total-cell extracts. We were also unable to detect multiple higher-molecular-weight products of CD4 typical for polyubiquitinated protein targets (data not shown, but the procedures used are described in Materials and Methods). It remains to be determined whether this reflects technical limitations in our methodology to detect short-lived intermediates of CD4 or the absence of such Ub-CD4 adducts in general. In the latter case, it is plausible that the requirement for Lys in the cytoplasmic tail of CD4 is unrelated to ubiquitination and that unidentified cellular factors, potentially involved in the Vpu-mediated proteolysis of CD4, are the crucial targets for ubiquitination. Such a model is supported by the recent finding that the LC-sensitive ERAD substrate α1-antitrypsin Z, lacking a transmembrane anchor, associates with calnexin and specifically induces its polyubiquitination (42). Although our results indicate no essential role for calnexin in Vpu-induced CD4 degradation, other ER chaperones may be involved in this process and could be targeted for ubiquitination. It is also noteworthy that no ubiquitinated breakdown intermediates were observed in similar ERAD processes, for instance, the rapid degradation of MHC class I heavy chain in HCMV-infected cells (61, 62).
Retention of CD4 in the ER either by coexpression with the HIV-1 gp160 or by treatment with brefeldin A was reported previously to be required for efficient degradation of CD4 in HeLa cells transfected with HIV-1 subgenomic expression vectors (63, 64). ER retention of CD4, however, is not an absolute requirement for Vpu-induced degradation, since CD4 is degraded in a Vpu-specific manner in the absence of deliberate ER retention in transfected HeLa cells expressing CD4 and Vpu (34). Similarly, we described here and in a previous study (28) that in a variety of cell types, deliberate ER retention is not necessary when CD4 and Vpu are expressed from rVVs. This concurs with the finding that Vpu-induced degradation of chimeric proteins created from vesicular stomatitis virus glycoprotein and CD4 expressed by the vaccinia virus/T7 system occurred without retention of proteins in the ER (9). Taken together, the findings suggest that when large, probably supraphysiological amounts of Vpu are produced, Vpu is able to encounter CD4 with sufficient rapidity that ER retention of CD4 is not needed. Under conditions of HIV-1 infection in at least some cells, however, binding of CD4 to gp160 is needed to retard export of CD4 to enable limiting amounts of Vpu the opportunity to interact (6) and target CD4 for degradation.
Our most important finding is that CD4 rescued from Vpu-mediated degradation by inhibiting proteasome activity is exported normally from the ER. This means either that ERAD of CD4 does not entail the translocation of CD4 from the ER to the cytosol as observed for the HCMV proteins US2 and US11 (61, 62) or that CD4 translocation requires enzymatically active proteasomes. The latter possibility could account for our failure to detect Ub-conjugated CD4 in the presence of proteasome inhibitors. Further, the possible involvement of proteasomes in the control of ER-cytosol translocation is supported by observations of a close association of a subset of proteasomes with the cytosolic face of the ER membrane (39). Alternatively, the translocation process may be inhibited by alterations in cellular physiology associated with proteasome inactivation. In either scenario, the evolutionary significance of such a mechanism may be to forestall the delivery of potentially dangerous unfolded proteins to the cytosol under circumstances when their degradation will be delayed.
It is noteworthy that the Vpu seems to act in a manner distinct from HCMV proteins US2 and US11, which have similar target molecules in MHC class I. It will be of interest in future studies to compare in one system the molecular mechanisms used by these viral membrane proteins to degrade unwanted cellular proteins.
ACKNOWLEDGMENTS
We thank Beth Buschling for excellent technical assistance in preparing rVV stocks and Ronald Willey, Jonathan Silver, Kazunobu Fujita, and Heidi Link Snyder for helpful discussions and critical comments on the manuscript. We thank Peter Cresswell for NKR(cal−) and NKR(cal+) cell lines, Larry Thompson for _tsA_1S9 cells, Ed Berger for rVV v-CB3, Alain Townsend for rVV-Ub-R-NP, Alicia Buckler-White and Bachoti Rao for oligonucleotide synthesis and sequence analysis, and John Coligan for peptide synthesis. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: antiserum (sheep) to CD4 from Michael Phelan and an antiserum to CD4 (T4-4) from R. Sweet, SmithKline Beecham Pharmaceuticals.
REFERENCES
- 1.Aiken C J, Konner J, Landau N R, Lenburg E, Trono D. Nef induces CD4 endocytosis: requirement of a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:503–513. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S J, Lenburg M, Landau N R, Garcia V J. The cytoplasmic domain of CD4 is sufficient for its down-regulation from the cell surface by human immunodeficiency virus type 1 Nef. J Virol. 1994;68:3092–3101. doi: 10.1128/jvi.68.5.3092-3101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergeron J J M, Brenner M B, Thomas D Y, Williams D B. Calnexin: a membrane bound chaperone of the endoplasmic reticulum. Trends Biochem Sci. 1994;19:124–128. doi: 10.1016/0968-0004(94)90205-4. [DOI] [PubMed] [Google Scholar]
- 4.Bonifacino J S, Klausner R D. Degradation of proteins in the endoplasmic reticulum. In: Ciechanover A, Schwartz A L, editors. Cellular proteolytic systems. Vol. 15. New York, N.Y: Wiley-Liss; 1994. pp. 137–160. [Google Scholar]
- 5.Bour S, Boulerice F, Wainberg M A. Inhibition of gp160 and CD4 maturation in U937 cells after both defective and productive infections by human immunodeficiency virus type 1. J Virol. 1991;65:6387–6396. doi: 10.1128/jvi.65.12.6387-6396.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bour S, Schubert U, Strebel K. The human immunodeficiency virus type-1 Vpu protein specifically binds to the cytoplasmic domain of CD4: implications for the mechanism of degradation. J Virol. 1995;69:1510–1520. doi: 10.1128/jvi.69.3.1510-1520.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broder C C, Dimitrov D S, Blumenthal R, Berger E A. The block to HIV-1 envelope glycoprotein-mediated membrane fusion in animal cells expressing human CD4 can be overcome by a human cell component(s) Virology. 1993;193:483–491. doi: 10.1006/viro.1993.1151. [DOI] [PubMed] [Google Scholar]
- 8.Buonocore L, Rose J K. Prevention of HIV-1 glycoprotein transport by soluble CD4 retained in the endoplasmic reticulum. Nature. 1990;345:625–628. doi: 10.1038/345625a0. [DOI] [PubMed] [Google Scholar]
- 9.Buonocore L, Turi T G, Crise B, Rose J K. Stimulation of heterologous protein degradation by the Vpu protein of HIV-1 requires the transmembrane and cytoplasmic domains of CD4. Virology. 1994;204:482–486. doi: 10.1006/viro.1994.1560. [DOI] [PubMed] [Google Scholar]
- 10.Chakrabarti S, Brechling K, Moss B. Vaccinia virus expression vector: coexpression of β-galactosidase provides visual screening of recombinant virus plaques. Mol Cell Biol. 1985;5:3403–3409. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen M Y, Maldarelli F, Karczewski M K, Willey R L, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces degradation of CD4 in vitro: the cytoplasmic domain of CD4 contributes to Vpu sensitivity. J Virol. 1993;67:3877–3884. doi: 10.1128/jvi.67.7.3877-3884.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen E A, Terwilliger E F, Sordroski J G, Haseltine W A. Identification of a protein encoded by the vpu gene of HIV-1. Nature. 1988;334:532–534. doi: 10.1038/334532a0. [DOI] [PubMed] [Google Scholar]
- 13.Cox J H, Galardy P, Bennink J R, Yewdell J W. Presentation of endogenous and exogenous antigens is not affected by inactivation of E1 ubiquitin-activating enzyme in temperature-sensitive cell lines. J Immunol. 1995;154:511–519. [PubMed] [Google Scholar]
- 14.Crise B, Buonocore L, Rose J K. CD4 is retained in the endoplasmic reticulum by the human immunodeficiency virus type 1 glycoprotein precursor. J Virol. 1990;64:5585–5593. doi: 10.1128/jvi.64.11.5585-5593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deen K C, McDougal J S, Inacker R, Folena-Wasserman G, Arthos J, Rosenberg J, Maddon P, Axel J R, Sweet R W. A soluble form of CD4 (T4) protein inhibits AIDS virus infection. Nature. 1988;331:82–84. doi: 10.1038/331082a0. [DOI] [PubMed] [Google Scholar]
- 16.Eckert D J, Khan M I, Marsh J, Butt T R, Crooke S T. Chemical synthesis and expression of a cassette adapted ubiquitin gene. J Biol Chem. 1987;262:3524–3527. [PubMed] [Google Scholar]
- 17.Eisenlohr L C, Bacik I, Bennink J R, Bernstein K, Yewdell J W. Expression of a membrane protease enhances presentation of endogenous antigens to MHC class I-restricted T lymphocytes. Cell. 1992;71:963–972. doi: 10.1016/0092-8674(92)90392-p. [DOI] [PubMed] [Google Scholar]
- 18.Eleuteri A M, Kohanski R A, Cardozo C, Orlowski M. Bovine spleen multicatalytic proteinase complex (proteasome); replacement of X, Y, and Z subunits by LMP7, and MECL1 and changes in properties and specificity. J Biol Chem. 1997;272:11824–11831. doi: 10.1074/jbc.272.18.11824. [DOI] [PubMed] [Google Scholar]
- 19.Ewart G D, Sutherland T, Gage P W, Cox G B. The Vpu protein of human immunodeficiency virus type 1 forms cation-selective ion channels. J Virol. 1996;70:7108–7115. doi: 10.1128/jvi.70.10.7108-7115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Federau T, Schubert U, Floßdorf J, Henklein P, Schomburg D, Wray V. Solution structure of the cytoplasmic domain of the human immunodeficiency virus type 1 encoded virus protein U (Vpu) Int J Peptide Protein Res. 1996;47:297–310. doi: 10.1111/j.1399-3011.1996.tb01359.x. [DOI] [PubMed] [Google Scholar]
- 21.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystein. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 22.Finley D, Sadis S, Monia B P, Boucher P, Ecker D J, Crooke S T, Chau V. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folks T, Benn S, Rabson A, Theodore T, Hoggan D, Martin M A, Lightfoot M, Sell K. Characterization of a continuous T-cell line susceptible to the cytopathic effects of the acquired immunodeficiency syndrome (AIDS)-associated retrovirus. Proc Natl Acad Sci USA. 1985;8:4539–4543. doi: 10.1073/pnas.82.13.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujita K, Omura S, Silver J. Rapid degradation of CD4 in cells expressing human immunodeficiency virus type 1 Env and Vpu is blocked by proteasome inhibitors. J Gen Virol. 1997;78:619–625. doi: 10.1099/0022-1317-78-3-619. [DOI] [PubMed] [Google Scholar]
- 25.Hiller M M, Finger A, Schweiger M, Wolf D H. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 26.Howell D N, Andreotti P E, Dawson J R, Cresswell P. Natural killing target antigens as inducers of interferon: studies with an immunoselected, natural killing-resistant human T lymphoblastoid cell line. J Immunol. 1985;134:971–976. [PubMed] [Google Scholar]
- 27.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A L, Riordan J R. Multiple proteolytic system, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 28.Kerkau T, Bacik I, Yewdell J, Bennink J R, Hünig T, Schimpl A, Schubert U. The human immunodeficiency virus type-1 (HIV-1) Vpu protein interferes with an early step in the biosynthesis of major histocompatibility complex (MHC) class I molecules. J Exp Med. 1997;185:1295–1305. doi: 10.1084/jem.185.7.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klimkait T, Strebel K, Hoggan M D, Martin M A, Orenstein J M. The human immunodeficiency virus type 1-specific protein Vpu is required for efficient virus maturation and release. J Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Köning R, Fleury S, Germain R N. The structural basis of CD4-MHC class II interactions: co-receptor contributions to T cell receptor antigen recognition and oligomerization-dependent signal transduction. Curr Top Microbiol Immunol. 1996;205:19–46. doi: 10.1007/978-3-642-79798-9_2. [DOI] [PubMed] [Google Scholar]
- 31.Lamb R A, Pinto L H. Do Vpu and Vpr of human immunodeficiency virus type 1 and NB of influenza B virus have ion channel activities in the viral life cycles? Virology. 1997;229:1–11. doi: 10.1006/viro.1997.8451. [DOI] [PubMed] [Google Scholar]
- 32.Le A, Steiner J L, Ferrell G A, Shaker J C, Sifers R N. Association between calnexin and a secretion-incompetent variant of human a1-antitrypsin. J Biol Chem. 1994;269:7514–7519. [PubMed] [Google Scholar]
- 33.Lee D H, Goldberg A L. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J Biol Chem. 1996;271:272800–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- 34.Lenburg M E, Landau N R. Vpu-induced degradation of CD4: requirement of specific amino acid residues in the cytoplasmic domain of CD4. J Virol. 1993;67:7238–7245. doi: 10.1128/jvi.67.12.7238-7245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maldarelli F, Chen M Y, Willey R L, Strebel K. Human immunodeficiency virus type 1 Vpu protein is an oligomeric type 1 integral membrane protein. J Virol. 1993;67:5056–5061. doi: 10.1128/jvi.67.8.5056-5061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCracken A A, Brodsky J F. Assembly of ER-associated protein degradation in vitro: dependence of cytosol, calnexin, and ATP. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nandi D, Monaco J J. The genetics of proteasomes and antigen processing. Annu Rev Genet. 1995;29:729–754. doi: 10.1146/annurev.ge.29.120195.003501. [DOI] [PubMed] [Google Scholar]
- 38.Orlowski M, Cardozo C, Michaud C. Evidence for the presence of five distinct proteolytic components in the pituitary multicatalytic proteinase complex. Properties of two components cleaving bonds on the carboxyl side of branched chain and small neutral amino acids. Biochemistry. 1993;23:1563–1572. doi: 10.1021/bi00057a022. [DOI] [PubMed] [Google Scholar]
- 39.Palmer A, Rivett A J, Thomson S, Hendil K B, Butcher G W, Fuertes G, Knecht E. Subpopulations of proteasomes in rat liver nuclei, microsomes and cytosol. Biochem J. 1996;316:401–407. doi: 10.1042/bj3160401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips D M, Bourinbaiar A S. Mechanism of HIV spread from lymphocytes to epithelia. Virology. 1992;86:261–273. doi: 10.1016/0042-6822(92)90080-9. [DOI] [PubMed] [Google Scholar]
- 41.Pind S, Riordan J R, Williams D B. Participation of the endoplasmic reticulum chaperone calnexin (p88, IP90) in the biogenesis of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1994;269:12784–12788. [PubMed] [Google Scholar]
- 42.Qu D, Teckman J H, Omura S, Perlmutter D H. Degradation of a mutant secretory protein, alpha1-antitrypsin Z, in the endoplasmic reticulum requires proteasome activity. J Biol Chem. 1996;271:22791–22795. doi: 10.1074/jbc.271.37.22791. [DOI] [PubMed] [Google Scholar]
- 43.Sadasivan B, Lehner P J, Ortmann B, Spies T, Cresswell P. Roles of calreticulin and novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996;5:103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 44.Schneider C, Newman R A, Sutherland D R, Asser U, Greaves M F. A one-step purification of membrane proteins using a high efficiency immunomatrix. J Biol Chem. 1982;257:10766–10769. [PubMed] [Google Scholar]
- 45.Schubert U, Bour S, Ferrer-Montiel A F, Montal M, Maldarelli F, Strebel K. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J Virol. 1996;70:809–819. doi: 10.1128/jvi.70.2.809-819.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schubert U, Clouse K A, Strebel K. Augmentation of virus secretion by the HIV-1 Vpu protein is cell type independent and occurs in cultured human primary macrophages and lymphocytes. J Virol. 1995;69:7699–7711. doi: 10.1128/jvi.69.12.7699-7711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schubert U, Ferrer-Montiel A F, Oblatt-Montal M, Henklein P, Strebel K, Montal M. Identification of an ion channel activity of the Vpu transmembrane domain and its plausible involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996;398:12–18. doi: 10.1016/s0014-5793(96)01146-5. [DOI] [PubMed] [Google Scholar]
- 48.Schubert U, Henklein P, Boldyreff B, Wingender E, Strebel K, Porstmann T. The human immunodeficiency virus type 1 encoded Vpu protein is phosphorylated by casein kinase-2 (CK-2) at positions Ser52 and Ser56 within a predicted α-helix-turn-α-helix-motif. J Mol Biol. 1994;236:16–25. doi: 10.1006/jmbi.1994.1114. [DOI] [PubMed] [Google Scholar]
- 49.Schubert U, Schneider T, Henklein P, Hoffmann K, Berthold E, Hauser H, Pauli G, Porstmann T. Human-immunodeficiency-virus-type-1-encoded Vpu protein is phosphorylated by casein kinase II. Eur J Biochem. 1992;204:875–883. doi: 10.1111/j.1432-1033.1992.tb16707.x. [DOI] [PubMed] [Google Scholar]
- 50.Schubert U, Strebel K. Differential activities of the human immunodeficiency virus type 1-encoded Vpu protein are regulated by phosphorylation and occur in different cellular compartments. J Virol. 1994;68:2260–2271. doi: 10.1128/jvi.68.4.2260-2271.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strebel K, Klimkait T, Martin M A. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science. 1988;241:1221–1223. doi: 10.1126/science.3261888. [DOI] [PubMed] [Google Scholar]
- 52.Strebel K, Klimkait T, Maldarelli F, Martin M A. Molecular and biochemical analyses of human immunodeficiency virus type 1 Vpu protein. J Virol. 1989;63:3784–3791. doi: 10.1128/jvi.63.9.3784-3791.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tifft C J, Proia R L, Camerini-Otero R D. The folding and cell surface expression of CD4 requires glycosylation. J Biol Chem. 1992;267:3266–3273. [PubMed] [Google Scholar]
- 54.Townsend A, Bastin J, Keith G, Brownlee G, Andrew M, Coupar B, Boyle D, Chan S, Smith G. Defective presentation to class I-restricted cytotoxic T lymphocytes in vaccinia-infected cells is overcome by enhanced degradation of antigen. J Exp Med. 1988;168:211–1224. doi: 10.1084/jem.168.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trono D. HIV accessory proteins: leading roles for the supporting cast. Cell. 1994;82:189–192. doi: 10.1016/0092-8674(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 56.Tsubuki S, Saito Y, Tomioka M, Hisashi I, Kawashima S. Differential inhibition of calpain and proteasome activities by peptidyl aldehydes of di-leucine and tri-leucine. J Biochem. 1996;11:572–576. doi: 10.1093/oxfordjournals.jbchem.a021280. [DOI] [PubMed] [Google Scholar]
- 57.Varshavsky A. The N-end rule. Cell. 1992;69:725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- 58.Vinitsky, A., L. C. Antón, H. Link-Snyder, M. Orlowski, J. R. Bennink, and J. W. Yewdell. The generation of MHC class I associated peptides is only partially inhibited by proteasome inhibitors: involvement of non-proteasomal proteases in antigen processing? J. Immunol., in press. [PubMed]
- 59.Vinitsky A, Michaud C, Powers J C, Orlowski M. Inhibition of the chymotrypsin-like activity of the pituitary multicatalytic proteinase complex. Biochemistry. 1992;31:9421–9428. doi: 10.1021/bi00154a014. [DOI] [PubMed] [Google Scholar]
- 60.Ward C L, Omura S, Kopito R R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 61.Wiertz E J H J, Jones T R, Sun L, Bogyo M, Geuze H J, Ploegh H L. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum of the cytosol. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 62.Wiertz E J H J, Tortorella D, Bogyo M, Yu J, Mothes W, Jones T R, Rapoport T A, Ploegh H L. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteosome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 63.Willey R L, Maldarelli F, Martin M A, Strebel K. Human immunodeficiency virus type 1 Vpu protein regulates the formation of intracellular gp160-CD4 complexes. J Virol. 1992;66:226–234. doi: 10.1128/jvi.66.1.226-234.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Willey R L, Maldarelli F, Martin M A, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J Virol. 1992;66:7193–7200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wray V, Federau T, Henklein P, Klabunde S, Kunert O, Schomburg D, Schubert U. The solution structure of the hydrophilic region of the HIV-1 encoded virus protein U (Vpu) by CD and NMR spectroscopy. Int J Peptide Protein Res. 1995;45:35–43. doi: 10.1111/j.1399-3011.1995.tb01565.x. [DOI] [PubMed] [Google Scholar]
- 66.Zacksenhaus E, Sheinin R. Molecular cloning, primary structure and expression of the human X linked A1S9 gene cDNA which complements the ts A1S9 mouse L cell defect in DNA replication. EMBO J. 1990;9:2923–2929. doi: 10.1002/j.1460-2075.1990.tb07483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]