A Third-Generation Lentivirus Vector with a Conditional Packaging System (original) (raw)
Abstract
Vectors derived from human immunodeficiency virus (HIV) are highly efficient vehicles for in vivo gene delivery. However, their biosafety is of major concern. Here we exploit the complexity of the HIV genome to provide lentivirus vectors with novel biosafety features. In addition to the structural genes, HIV contains two regulatory genes, tat and rev, that are essential for HIV replication, and four accessory genes that encode critical virulence factors. We previously reported that the HIV type 1 accessory open reading frames are dispensable for efficient gene transduction by a lentivirus vector. We now demonstrate that the requirement for the tat gene can be offset by placing constitutive promoters upstream of the vector transcript. Vectors generated from constructs containing such a chimeric long terminal repeat (LTR) transduced neurons in vivo at very high efficiency, whether or not they were produced in the presence of Tat. When the rev gene was also deleted from the packaging construct, expression of gag and pol was strictly dependent on Rev complementation in trans. By the combined use of a separate nonoverlapping Rev expression plasmid and a 5′ LTR chimeric transfer construct, we achieved optimal yields of vector of high transducing efficiency (up to 107 transducing units [TU]/ml and 104 TU/ng of p24). This third-generation lentivirus vector uses only a fractional set of HIV genes: gag, pol, and rev. Moreover, the HIV-derived constructs, and any recombinant between them, are contingent on upstream elements and trans complementation for expression and thus are nonfunctional outside of the vector producer cells. This split-genome, conditional packaging system is based on existing viral sequences and acts as a built-in device against the generation of productive recombinants. While the actual biosafety of the vector will ultimately be proven in vivo, the improved design presented here should facilitate testing of lentivirus vectors.
Lentiviruses have attracted the attention of gene therapy investigators (45) for their ability to integrate into nondividing cells (8, 15, 16, 25, 26). We previously developed replication-defective vectors from the lentivirus human immunodeficiency virus (HIV) and showed that they transduce target cells independent of mitosis (32). The vectors proved highly efficient for in vivo gene delivery and achieved stable long-term expression of the transgene in several target tissues, such as the brain (5, 33), the retina (31), and the liver and muscle of adult rats (21). A major concern, however, is the biosafety of vectors derived from a highly pathogenic human virus.
The complexity of the lentivirus genome may be exploited to build novel biosafety features in the design of a retrovirus vector. In addition to the structural gag, pol, and env genes common to all retroviruses, HIV contains two regulatory genes, tat and rev, essential for viral replication, and four accessory genes, vif, vpr, vpu, and nef, that are not crucial for viral growth in vitro but are critical for in vivo replication and pathogenesis (27).
The Tat and Rev proteins regulate the levels of HIV gene expression at transcriptional and posttranscriptional levels, respectively. Due to the weak basal transcriptional activity of the HIV long terminal repeat (LTR), expression of the provirus initially results in small amounts of multiply spliced transcripts coding for the Tat, Rev, and Nef proteins. Tat increases dramatically HIV transcription by binding to a stem-loop structure (transactivation response element [TAR]) in the nascent RNA, thereby recruiting a cyclin-kinase complex that stimulates transcriptional elongation by the polymerase II complex (46). Once Rev reaches a threshold concentration, it promotes the cytoplasmic accumulation of unspliced and singly spliced viral transcripts, leading to the production of the late viral proteins. Rev accomplishes this effect by serving as a connector between an RNA motif (the Rev-responsive element [RRE]), found in the envelope coding region of the HIV transcript, and components of the cell nuclear export machinery. Only in the presence of Tat and Rev are the HIV structural genes expressed and new viral particles produced (27).
In a first generation of HIV-derived vectors (32), viral particles were generated by expressing the HIV type 1 (HIV-1) core proteins, enzymes, and accessory factors from heterologous transcriptional signals and the envelope of another virus, most often the G protein of the vesicular stomatitis virus (VSV G) (9) from a separate plasmid. In a second version of the system, the HIV-derived packaging component was reduced to the gag, pol, tat, and rev genes of HIV-1 (51). In either case, the vector itself carried the HIV-derived _cis_-acting sequences necessary for transcription, encapsidation, reverse transcription, and integration (2, 4, 22, 24, 29, 30, 32, 35). It thus encompassed, from the 5′ to 3′ end, the HIV 5′ LTR, the leader sequence and the 5′ splice donor site, approximately 360 bp of the gag gene (with the gag reading frame closed by a synthetic stop codon), 700 bp of the env gene containing the RRE and a splice acceptor site, an internal promoter (typically the immediate-early enhancer/promoter of human cytomegalovirus [CMV] or that of the phosphoglycerokinase gene [PGK]), the transgene, and the HIV 3′ LTR. Vector particles are produced by cotransfection of the three constructs in 293T cells (32). In this design, significant levels of transcription from the vector LTR and of accumulation of unspliced genomic RNA occur only in the presence of Tat and Rev.
Here, we demonstrate that the _trans_-acting function of Tat becomes dispensable if part of the upstream LTR in the transfer vector construct is replaced by constitutively active promoter sequences. Furthermore, we show that the expression of rev in trans allows the production of high-titer HIV-derived vector stocks from a packaging construct which contains only gag and pol. This design makes the expression of the packaging functions conditional on complementation available only in producer cells. The resulting gene delivery system, which conserves only three of the nine genes of HIV-1 and relies on four separate transcriptional units for the production of transducing particles, offers significant advantages for its predicted biosafety.
MATERIALS AND METHODS
Transfer vector constructs.
pHR′CMV-LacZ and pHR′CMV-Luciferase have been described elsewhere (32). pHR2 is a lentivirus transfer vector in which the polylinker and downstream nef sequences up to the _Kpn_I site of pHR′ have been replaced with a _Cla_I/_Spe_I/_Sna_BI/_Sma_I/_Bam_HI/_Sac_II/_Eco_RI polylinker. pHR2 was generated by replacing the 3.7-kb _Cla_I-_Sac_I fragment of pHR′CMVlacZ with a 607-bp _Cla_I-_Sac_I fragment generated by PCR using pHR′CMVlacZ as the template with oligonucleotide primers 5′-CCATCGATGGACTAGTCCTACGTA TCCCCGGGGACGGGATCCGCGGAATTCCGTTTAAGACCAATGAC-3′ and 5′-TTATAATGTCAAGGCCTCTC-3′, followed by digestion with _Cla_I and _Sac_I.
pHR2PGK-NGFR, pHR2CMV-NGFR, and pHR2MFG-NGFR are lentivirus transfer vectors in which the truncated low-affinity nerve growth factor receptor (NGFR) (6) transgenes under the control of the murine PGK, human CMV, and Moloney leukemia virus (MLV) promoters, respectively, have been inserted into the polylinker of pHR2. The pHR2PGK-NGFR transgene encodes no intron sequences, the pHR2CMV-NGFR vector includes the intron from plasmid pMD (34), and the pHR2MFG-NGFR vector contains the MLV intron from MFG-S (34).
pRRL, pRLL, pCCL, and pCLL are lentivirus transfer vectors containing chimeric Rous sarcoma virus (RSV)-HIV or CMV-HIV 5′ LTRs and vector backbones in which the simian virus 40 polyadenylation and (enhancerless) origin of replication sequences have been included downstream of the HIV 3′ LTR, replacing most of the human sequence remaining from the HIV integration site. In pRRL, the enhancer and promoter (nucleotides −233 to −1 relative to the transcriptional start site; GenBank accession no. J02342) from the U3 region of RSV are joined to the R region of the HIV-1 LTR. In pRLL, the RSV enhancer (nucleotides −233 to −50) sequences are joined to the promoter region (from position −78 relative to the transcriptional start site) of HIV-1. In pCCL, the enhancer and promoter (nucleotides −673 to −1 relative to the transcriptional start site; GenBank accession no. K03104) of CMV were joined to the R region of HIV-1. In pCLL, the CMV enhancer (nucleotides −673 to −220) was joined to the promoter region (position −78) of HIV-1. Exact sequences and details of construction are available on request.
pHR2hPGK-GFP, pCCLhPGK-GFP, pCLLhPGK-GFP, pRRLhPGK-GFP, and pRLLhPGK.GFP are lentivirus transfer vectors containing the enhanced green fluorescent protein (eGFP) (750-bp _Bam_HI-_Not_I fragment from pEGFP-1; Clontech) coding region, under the control of the human PGK promoter (nucleotides 5 to 516; GenBank accession no. M11958), inserted into the polylinker region of each parental vector. pRRLGFP was obtained by deletion of the _Xho_I-_Bam_HI fragment containing the PGK promoter from pRRLhPGK-GFP.
pRRLhPGK.GFP.SIN-18 is a vector in which 3′ LTR sequences from −418 to −18 relative to the U3/R border have been deleted from pRLLhPGK.GFP (52).
Packaging constructs.
The _tat_-defective packaging construct pCMVΔR8.93 was obtained by swapping an _Eco_RI-_Sac_I fragment from plasmid R7/pneo(−) (12) with the corresponding fragment of pCMVΔR8.91, a previously described plasmid expressing Gag, Pol, Tat, and Rev (51). This fragment has a deletion affecting the initiation codon of the tat gene and a frameshift created by the insertion of an _Mlu_I linker into the _Bsu_36I site as described previously. pCMVΔR8.74 is a derivative of pCMVΔR8.91 in which a 133-bp _Sac_II fragment, containing a splice donor site, has been deleted from the CMV-derived region upstream of the HIV sequences to optimize expression.
pMDLg/p is a CMV-driven expression plasmid that contains only the gag and pol coding sequences from HIV-1. First, p_kat_2Lg/p was constructed by ligating a 4.2-kb _Cla_I-_Eco_RI fragment from pCMVΔR8.74 with a 3.3-kb _Eco_RI-_Hin_dIII fragment from p_kat_2 (14) and a 0.9-kb _Hin_dIII-_Nco_I fragment from p_kat_2 along with an _Nco_I-_Cla_I linker consisting of synthetic oligonucleotides 5′-CATGGGTGCGAGAGCGTCAGTATTAAGCGGGGGAGAATTAGAT-3′ and 5′-CG ATCTAATTCTCCCCCGCTTAATACTGACGCTCTCGCACC-3′. Next, pMDLg/p was constructed by inserting the 4.25-kb _Eco_RI fragment from p_kat_2Lg/p into the _Eco_RI site of pMD-2. pMD-2 is a derivative of pMD.G (34) in which the pXF3 plasmid backbone of pMD.G has been replaced with a minimal pUC plasmid backbone and the 1.6-kb VSV G-encoding _Eco_RI fragment has been removed.
pMDLg/pRRE differs from pMDLg/p by the addition of a 374-bp RRE-containing sequence from HIV-1 (HXB2) immediately downstream of the pol coding sequences. To generate pMDLg/pRRE, the 374-bp _Not_I-_Hin_dIII RRE-containing fragment from pHR3 was ligated into the 9.3-kb _Not_I-_Bgl_II fragment of pVL1393 (Invitrogen) along with a _Hin_dIII-_Bgl_II oligonucleotide linker consisting of synthetic oligonucleotides 5′-AGCTTCCGCGGA-3′ and 5′-GATCTCCGCGGA-3′ to generate pVL1393RRE (pHR3 was derived from pHR2 by the removal of HIV env coding sequences upstream of the RRE sequences in pHR2). A _Not_I site remains at the junction between the gag and RRE sequences. pMDLg/pRRE was then constructed by ligating the 380-bp _Eco_RI-_Sst_II fragment from pV1393RRE with the 3.15-kb _Sst_II-_Nde_I fragment from pMD-2FIX (pMD-2FIX is a human factor IX-containing variant of pMD-2 which has an _Sst_II site at the 3′ end of the factor IX insert), the 2.25-kb _Nde_I-_Avr_II fragment from pMDLg/p, and the 3.09-kb _Avr_II-_Eco_RI fragment from pkat1Lg/p (14).
pRSV-Rev and pTK-Rev (generous gifts of T. Hope, Salk Institute) are rev cDNA-expressing plasmids in which the joined second and third exons of HIV-1 rev are under the transcriptional control of the RSV U3 and herpes simplex virus type 1 thymidine kinase (TK) promoters, respectively. Both expression plasmids utilize polyadenylation signal sequences from the HIV LTR in a pUC118 plasmid backbone.
Vector production and assays.
Vectors were produced by transient transfection into 293T cells as previously described (33), with the following modifications. A total of 5 × 106 293T cells were seeded in 10-cm-diameter dishes 24 h prior to transfection in Iscove modified Dulbecco culture medium (JRH Biosciences) with 10% fetal bovine serum, penicillin (100 IU/ml), and streptomycin (100 μg/ml) in a 5% CO2 incubator, and the culture medium was changed 2 h prior to transfection. A total of 20 μg of plasmid DNA was used for the transfection of one dish: 3.5 μg of the envelope plasmid pMD.G, 6.5 μg of packaging plasmid, and 10 μg of transfer vector plasmid. The precipitate was formed by adding the plasmids to a final volume of 450 μl of 0.1× TE (1× TE is 10 mM Tris [pH 8.0] plus 1 mM EDTA) and 50 μl of 2.5 M CaCl2, mixing well, then adding dropwise 500 μl of 2× HEPES-buffered saline (281 mM NaCl, 100 mM HEPES, 1.5 mM Na2HPO4 [pH 7.12]) while vortexing and immediately adding the precipitate to the cultures. The medium (10 ml) was replaced after 14 to 16 h; the conditioned medium was collected after another 24 h, cleared by low-speed centrifugation, and filtered through 0.22-μm-pore-size cellulose acetate filters. For in vitro experiments, serial dilutions of freshly harvested conditioned medium were used to infect 105 cells in a six-well plate in the presence of Polybrene (8 μg/ml). Viral p24 antigen concentration was determined by immunocapture (Alliance; DuPont-NEN). Vector batches were tested for the absence of replication-competent virus by monitoring p24 antigen expression in the culture medium of transduced SupT1 lymphocytes for 3 weeks. In all cases tested, p24 was undetectable (detection limit, 3 pg/ml) once the input antigen had been eliminated from the culture. Transducing activity was expressed in transducing units (TU).
Northern blot analysis.
Total RNA was isolated from 1 × 107 to 2 × 107 cells harvested at confluence by using RNAsol B as suggested by the manufacturer; 10 to 20 μg of RNA was loaded per well on 1% agarose gels, using NorthernMax (Ambion, Austin, Tex.) reagents as described by the manufacturer. Transfer was to Zetabind membranes (Cuno Inc., Meridien, Conn.) by either capillary transfer or pressure blotting (Stratagene). 32P-labeled probes were made by random priming.
Intracerebral injection of vectors.
Twelve Fischer 344 male rats weighing approximately 220 g were obtained from Harlan Sprague-Dawley (Indianapolis, Ind.). The rats were housed with access to ad libitum food and water on a 12-h light/dark cycle and were maintained and treated in accordance with published National Institutes of Health guidelines. All surgical procedures were performed with the rats under isoflurane gas anesthesia, using aseptic procedures. After a rat was anesthetized in a sleep box, it was placed in a small animal stereotaxic device (Kopf Instruments, Tujunga, Calif.) using the earbars, which do not break the tympanic membrane. The rats were randomly divided into one control and four treatment groups. After the rats were placed in the stereotaxic frame, 2 μl of lentivirus vector concentrated by ultracentrifugation at 50,000 × g for 140 min at 20°C (33) in phosphate-buffered saline (PBS) was injected consecutively into the striatum in both hemispheres over 4 min at a rate of 0.5 μl/min (coordinates, AP 0.0, LAT ±3.0, DV −5.5, −4.5, −3.5 with the incisor bar set at 3.3 mm below the intra-aural line [36]), using a continuous-infusion system as described previously in detail (28). During the injection, the needle was slowly raised 1 mm in the dorsal direction every 40 s (3-mm total withdrawal). One minute after cessation of the injection, the needle was retracted an additional 1 mm and then left in place for an additional 4 min before being slowly withdrawn from the brain.
Histology.
One month after vector injection, each animal was deeply anesthetized with intraperitoneal pentobarbital and perfused through the aorta with sterile PBS, followed by ice-cold 4% paraformaldehyde perfusion. The brains were removed from the skulls, postfixed in 4% paraformaldehyde by immersion for 24 h, and then transferred into a 30% sucrose–PBS solution for 3 to 4 days, until the brains sank to the bottom of their containers. The brains were then frozen on dry ice, and 40-μm-thick coronal sections were cut on a sliding microtome. Sections were collected in series in microtiter well plates that contained a glycerin-based antifreeze solution, and they were kept at −30°C until further processing. Immunocytochemistry was performed according to the general procedure described previously (44). After several PBS rinses and an incubation in 3% hydrogen peroxide, the sections were placed in a 3% normal goat serum. The sections were then incubated in the primary anti-GFP antibody (1:1,000; Clontech, Palo Alto, Calif.) in 1% normal goat serum–0.1% Triton X-100 overnight at room temperature. After rinsing, the sections were incubated in the biotinylated rabbit anti-goat secondary antibody (Vector, Burlingame, Calif.) for 3 h. After rinsing, the sections were incubated with horseradish peroxidase-streptavidin and then reacted by using a purple chromagen kit (VIP; Vector), mounted, dried, dehydrated, and coverslipped.
RESULTS
Tat is required to produce a vector of efficient transducing activity.
To investigate the role of Tat in the production of transducing particles, expression from lentivirus vectors was first examined by Northern analysis (Fig. 1). The patterns of RNAs induced by transfer vectors in which the transgene was driven by an internal PGK, CMV, or retrovirus MFG promoter were studied in both producer and target cells. In transfected 293T cells, expression occurred mainly from the internal promoter. When a packaging construct expressing both Tat and Rev was cotransfected, a dramatic enhancement of transcription from the LTR was observed, with an accumulation of unspliced vector RNA. In cells transduced with the vectors, that is, in the absence of Tat and Rev, transcription from the LTR was almost completely suppressed, the residual transcripts underwent splicing, and the internal promoter was responsible for most of the expression.
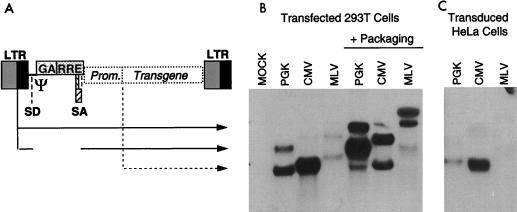
FIG. 1.
Northern analysis of the RNA expression from lentivirus vectors. Three pHR2 vectors carrying an expression cassette for the same transgene (truncated low-affinity NGFR) and driven by three different promoters (PGK, CMV, and retroviral MFG) were analyzed in producer and transduced cells. Total RNA was extracted and analyzed by Northern blotting with a probe specific for the transgene sequence. (A) Schematic of the vector construct depicts the species of RNA driven by the internal promoter (Prom.; broken arrow, shorter transcript) and the viral LTR (solid arrows, longer transcripts; the two species differ for the splicing of the viral intron). The splice donor and acceptor sites (SD and SA), the packaging sequence (Ψ), the truncated gag sequence (GA), and the RRE are indicated. (B) The vector constructs were transfected in 293T cells without or with the packaging construct. (C) Vector particles produced by the 293T transfectants were used to transduce HeLa cells. In the absence of the viral transactivators, supplied by the core packaging construct only in the producer cells, vector expression occurs mainly from the internal promoter. Note the dramatic enhancement of the upstream transcription and the accumulation of unspliced RNA (carrying the Ψ sequence) in the presence of the packaging construct. In the transduced cells, the LTR is silenced. Note that the three expression cassettes differ in the size of the promoters and 5′ untranslated sequence. In each case, the smallest RNA species represents transcripts initiated from the internal promoter, while the intermediate-size and larger species correspond to spliced and unspliced LTR-driven RNAs, respectively.
A packaging plasmid carrying two mutations in tat (pCMVΔR8.93) was then constructed. The first mutation is a deletion of the T in the ATG initiation codon of the tat gene; the second is an insertion of a _Mlu_I linker producing a translation stop codon after residue 46 of the Tat protein. These changes confer a _tat_-defective phenotype to HIV-1 (12). After transfection of the control or _tat_-defective packaging constructs into 293T cells, comparable yields of vector particles were recovered in the culture medium, as assayed by using the Gag p24 antigen (see Table 3). Such Tat independence was expected from the replacement of the HIV LTR by the constitutive CMV promoter in the packaging construct. However, the particles produced in the absence of Tat had a dramatically reduced transducing activity (Table 1): 5 to 15% of that of particles produced by the control Tat-positive packaging construct.
TABLE 3.
GFP transduction into HeLa cells by lentivirus vectors made by transfer constructs with a wild-type or 5′ chimeric LTR and packaging constructs with or without a functional tat genea
| Transfer construct | tat gene in packaging construct | Endpoint titer (TU/ml) | p24 antigen (ng/ml) | Transduction efficiency (TU/ng of p24) |
|---|---|---|---|---|
| pHR2 | + | 4.1 × 106 | 297 | 13,805 |
| pHR2 | − | 2.4 × 105 | 545 | 440 |
| pRRL | + | 1.3 × 107 | 546 | 23,810 |
| pRRL | − | 4.9 × 106 | 344 | 14,244 |
TABLE 1.
Transducing activities of lentivirus vectors made with and without a functional tat gene in the packaging constructa
| Transfer vector | Target cells | Mean transducing activity (TU/ng of p24) ± SEb | |
|---|---|---|---|
| With tat in packaging construct | Without tat in packaging construct | ||
| pHR′CMV-LacZ | 293T | 1,056 ± 54 | 152 ± 26 |
| pHR2PGK-eGFP | HeLa | 5,666 | 384 |
| pHR′CMV-Luciferase | HeLa | 3,000 ± 152 | 152 ± 26 |
| HeLa-tat | 3,777 ± 348 | 486 ± 59 | |
| pHR′Luciferasec | HeLa | 46 ± 1 | 0.3 ± 0.003 |
| HeLa-tat | 3,296 ± 276 | 174 ± 75 |
We also tested whether the Tat-defective phenotype could be rescued by complementation in target cells (Table 1). HeLa-tat cells, a cell line expressing Tat from the HIV-1 LTR (13), were transduced by vectors produced with or without Tat. The expression of Tat in target cells did not compensate for the loss in transduction efficiency of vector produced without Tat.
As expected from the Northern analysis, functional inactivation of the tat gene resulted in a lower abundance of vector RNA in producer cells. This was indicated by the decrease in luciferase activity in cells producing a luciferase vector without an internal promoter. In this case, transgene expression directly reflects the abundance of transcripts originating from the LTR. 293T cells producing luciferase vectors without Tat had only 5% of the luciferase content of cells producing the same vector with Tat ([1.0 ± 0.2] × 109 relative light units [RLU]/dish without Tat; [20.2 ± 0.7] × 109 RLU/dish with Tat). This ratio corresponded very closely to that observed in cells transduced by either type of vector in the course of the same experiment (Table 1), suggesting that the abundance of vector RNA in producer cells is a rate-limiting factor in the transduction by lentivirus vectors.
One could thus conclude that Tat is required in producer cells to activate transcription from the HIV LTR and to generate vector particles with a high transducing activity.
The tat requirement is offset by placing a constitutive promoter upstream of the transfer vector.
If the only function of Tat is trans activation of vector transcription from the LTR, the _tat_-defective phenotype should be rescued by placing a strong constitutive promoter upstream of the vector transcript. Three transcriptional domains have been identified in the HIV promoter in the U3 region of the LTR: the core or basal domain, the enhancer, and the modulatory domain (27). Transcription starts at the U3/R boundary, the first nucleotide of R being numbered 1. The core promoter contains binding sites for the TATA-binding protein (−28 to −24) and SP-1 (three binding sites between −78 to −45). The enhancer contains two binding sites for NF-κB which overlap with a binding site for NFATc (−104 to −81). The modulatory domain contain binding sites for several cellular factors, including AP-1 (−350 to −293), NFAT-1 (−256 to −218), USF-1 (−166 to −161), Ets-1 (−149 to −141), and LEF (−136 to −125). A panel of 5′ chimeric transfer constructs carrying substitutions of either all or part of the U3 region of the 5′ LTR was generated. All substitutions were made to preserve the transcription initiation site of HIV. Partial substitutions joined new enhancer sequences to the core promoter of the HIV LTR (−78 to 1), while full substitutions replaced also the promoter. pRLL and pRRL vectors carried the enhancer and the enhancer/promoter, respectively, of RSV; pCLL and pCCL vectors carried the enhancer and the enhancer/promoter of human CMV.
Control pHR2 and 5′ chimeric transfer constructs carrying a PGK-eGFP expression cassette were tested by transfection of 293T cells with control or _tat_-defective packaging constructs, and the expression of the eGFP transgene was analyzed by fluorescence-activated cell sorting (FACS). The RRL chimeric construct yielded a higher level of eGFP expression than the pHR2 vector, reflecting the constitutive transcriptional activity of the new sequence (Fig. 2A). Interestingly, the chimeric vector also displayed upregulation by Tat, as shown by the increased eGFP expression of cells cotransfected with the control packaging construct. Tat upregulation was proven to be a direct effect by transfecting a pRRL-eGFP vector lacking an internal promoter with control or _tat_-defective packaging constructs and analyzing GFP expression by FACS (Fig. 2B). Comparable results were obtained with the other chimeric LTR vectors (not illustrated). Vector particles were then collected from the transfected producer cells and assayed for transduction of eGFP into HeLa cells and human primary lymphocytes (peripheral blood lymphocytes [PBL]). As shown in Table 2, all vectors had efficient transducing activity, as assessed by endpoint titration on HeLa cells or maximal transduction frequency of PBL. The vector produced by the pRRL chimera was as efficient as that produced by the pHR2 construct and was selected to test transduction independent of Tat. As shown in Table 3, the pRRL construct yielded a vector of only slightly reduced transducing activity (60%) when the packaging construct was tat defective. The residual effect of Tat on transduction was in agreement with the ability of Tat to upregulate transcription from the chimeric LTR.
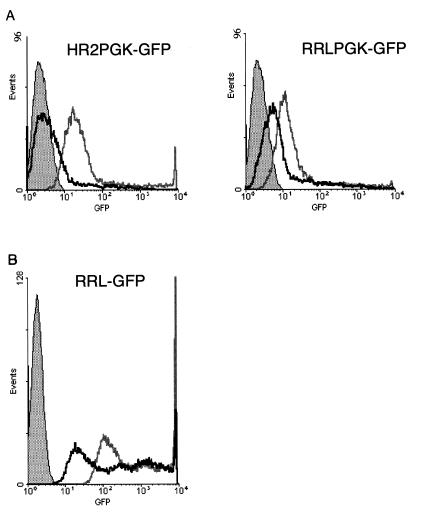
FIG. 2.
Transcriptional activities of wild-type and 5′ chimeric vector constructs in the absence and presence of Tat. (A) Control pHR2 and the 5′ chimeric pRRL transfer construct carrying a PGK-eGFP expression cassette were transfected into 293T cells with a packaging construct having a functional (pCMVΔR8.91; grey line) or inactive (pCMVΔR8.93; black line) tat gene. GFP expression was analyzed by FACS. The filled area represents nontransfected cells. In the absence of Tat, the chimeric construct yielded a level of GFP expression higher than that achieved by the pHR2 construct. Both constructs were further upregulated by Tat. (B) A pRRL construct carrying the eGFP gene without an internal promoter was transfected with a packaging construct carrying a functional (grey line, open area) or inactive (black line, open area) tat gene. Direct upregulation of the chimeric promoter by Tat was observed. The filled area represents nontransfected cells.
TABLE 2.
GFP transduction by lentivirus vectors made by transfer constructs with a wild-type or 5′ chimeric LTR
| Transfer construct | Endpoint titer on HeLa cells (TU/ml)a | Transduction efficiency on human lymphocytes (% positive cells)b |
|---|---|---|
| pHR2 | 2.3 × 107 | 30 |
| pCCL | 4.6 × 106 | 14 |
| pCLL | 7.9 × 106 | 18 |
| pRRL | 1.8 × 107 | 29 |
| pRLL | 8.9 × 106 | 18 |
The use of the chimeric LTR construct allowed removal of Tat from the packaging system with a minimal loss in the transduction efficiency of the vector in vitro. To test vector performance in the more challenging setting of in vivo delivery into brain neurons, high-titer vector stocks were generated from the pHR2 and pRRL constructs with and without Tat. The four stocks of eGFP vector were matched for particle content by p24 antigen and injected bilaterally in the neostriata of groups of three adult rats. The animals were sacrificed after 1 month, and serial sections of the brain were analyzed for eGFP fluorescence (not shown) and immunostained by antibodies against eGFP (Fig. 3). The results obtained in vivo matched the in vitro data. Vector produced by the pHR2 construct only achieved significant transduction of the neurons when packaged in the presence of Tat. Vector produced by the pRRL chimera was as efficient when made with or without Tat. The transduction extended throughout most of the striatum and reached a very high density of positive cells in the sections closest to the injection site. No signs of pathology were detectable in the injected tissue, except for a small linear scar marking the needle track, by hematoxylin and eosin staining of the sections (data not shown).
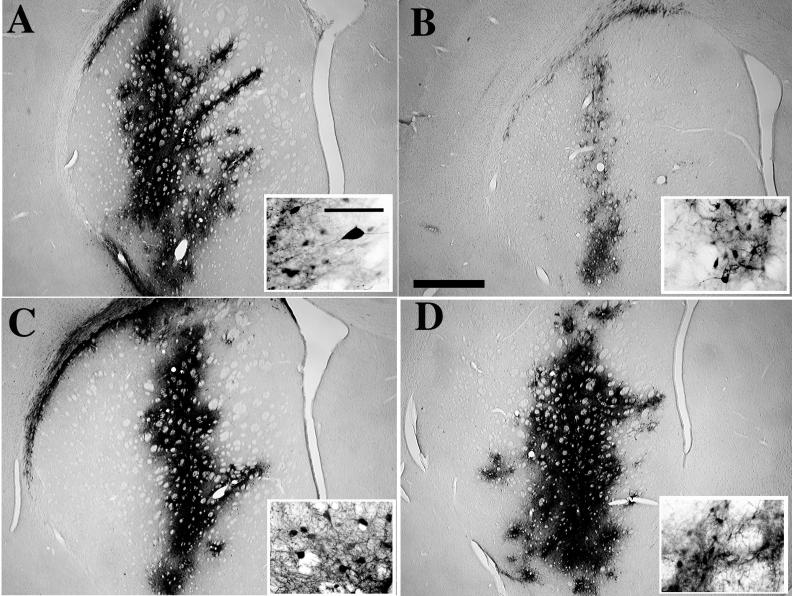
FIG. 3.
In vivo transduction of eGFP into brain cells by lentivirus vectors produced with and without Tat. Vectors carrying a PGK-eGFP expression cassette were produced by the pHR2 (A and B) or the 5′ chimeric pRRL (C and D) transfer construct and a packaging construct with (pCMVΔR8.91; A and C) or without (pCMVΔR8.93; B and D) a functional tat gene, concentrated by ultracentrifugation, and normalized for particle content prior to injection into the corpora striata of adult rats. One month after injection, brain sections were stained for immunoreactivity to the GFP protein. While both types of vectors transduced neurons very efficiently when made with Tat, only the vector made by the chimeric transfer construct worked as well when produced without Tat. Representative sections close to the injection site are shown for one of six striata injected per each type of vector. The bar in panel B represents 1 mm; that in the inset in panel A represents 100 μm.
These results provide evidence that Tat is dispensable for efficient transduction by a lentivirus vector.
A new split-genome conditional packaging system.
The possibility of deleting the tat gene prompted us to explore a new design of the packaging component of the HIV vector system, in which two separate nonoverlapping expression plasmids, one for the gag and pol genes and the other for the rev gene, were used. The gag and pol reading frames were expressed within the context of the MD cassette, which employs the CMV promoter and intervening sequence and the human β-globin poly(A) site (34). All HIV sequences upstream of the gag initiation codon were removed, and the leader was modified for optimal fit to the Kozak consensus for translation. This construct, however, expressed almost no p24 antigen when transfected alone in 293T cells. This observation is in agreement with the previously reported presence of _cis_-repressive or inhibitory sequences in the gag and pol genes (40, 41). The HIV RRE was then inserted downstream of the pol gene, and the resulting plasmid was cotransfected with a rev expression vector (Table 4). High levels of p24 antigen production were observed in this case, the highest yields being obtained when rev was driven by an RSV promoter. When the gag-pol and the rev constructs were cotransfected with the pRRL chimeric transfer vector and the VSV G-expressing plasmid, high-titer vector was obtained in the culture medium. Both the yield of particles and their transducing efficiency were similar to those obtained with previous versions of the system. Northern analysis of producer cells confirmed that unspliced vector genomic RNA accumulated only in the presence of Rev (data not shown). Thus, both the expression of the gag and pol genes and the accumulation of packageable vector transcripts are dependent on trans complementation by a separate Rev expression construct. Such a conditional packaging system provides an important safety feature unavailable to oncoretrovirus vectors.
TABLE 4.
GFP transduction into HeLa cells by lentivirus vectors made by linked or split packaging constructs and a pRRL transfer constructa
| Packaging construct | Separate rev plasmidb | p24 antigen (ng/ml) | Endpoint titer (TU/ml) | Transduction efficiency (TU/ng of p24) |
|---|---|---|---|---|
| pCMVΔR8.74 | 364 | 1.07 × 107 | 29,436 | |
| pMDLg/pRRE | <0.1 | ND | NA | |
| pMDLg/pRRE | TK-Rev, 5 μg | 29 | 6.9 × 105 | 23,793 |
| pMDLg/pRRE | TK-Rev, 12 μg | 94 | 2.02 × 106 | 21,489 |
| pMDLg/pRRE | RSV-Rev, 2.5 μg | 774 | 1.0 × 107 | 13,495 |
| pMDLg/pRRE | RSV-Rev, 5 μg | 776 | 7.6 × 106 | 9,761 |
| pMDLg/pRRE | RSV-Rev, 12 μg | 565 | 4.8 × 106 | 8,495 |
DISCUSSION
The predicted biosafety of a viral vector depends in part on how much segregation of the _cis_- and _trans_-acting functions of the viral genome is achieved by the vector design and is maintained during vector production. A vector particle is assembled by viral proteins expressed in the producer cell from a construct(s) stripped of the _cis_-acting sequences required for the transfer of the viral genome to target cells (packaging construct). These _cis_-acting sequences are instead linked to the transgene in the transfer vector. As the vector particle packages only the genetic information contained in this latter construct, the infection process is limited to a single round without spreading. Through recombination, it is possible that sequences encoding viral proteins rejoin the _cis_-acting elements of the transfer vector. If the resulting recombinant expresses all required functions, it is able to replicate (i.e., it is a replication-competent retrovirus [RCR]) and presents a risk to the recipient. The formation of heterozygous vector particles containing RNAs from both the packaging and transfer vectors, followed by homologous recombination during reverse transcription, is the mechanism most often incriminated in the emergence of RCR during the production of retroviral vectors. The likelihood of this type of recombination is dependent on residual _cis_-acting sequences in the packaging plasmid, allowing some level of encapsidation, and on the extent of homology between packaging and vector constructs (10).
A first strategy to improve the biosafety of a vector is to use nonoverlapping split-genome packaging constructs that require multiple recombination events with the transfer vector for RCR generation. Earlier studies described several approaches to generate replication-defective HIV vectors (7, 35, 38, 42). However, these vectors could be produced only to low infectious titers, were restricted to CD4-positive cellular targets, and carried the risk of generating wild-type HIV by recombination of the components. A major advance was achieved when an improved vector design was combined with the use of the envelope of another virus (32, 33, 39). The lentivirus vector that we describe here is packaged by three nonoverlapping expression constructs, two expressing HIV proteins and the other expressing the envelope of a different virus. Moreover, all HIV sequences known to be required for encapsidation and reverse transcription (2, 22, 24, 27, 29, 30, 35) are absent from these constructs, with the exception of the portion of the gag gene that contributes to the stem-loop structure of the HIV-1 packaging motif (29).
A second strategy to improve vector biosafety took advantage of the complexity of the lentivirus genome. The minimal set of HIV-1 genes required to generate an efficient vector was identified, and all other HIV reading frames were eliminated from the system. As the products of the removed genes are important for the completion of the virus life cycle and for pathogenesis, no recombinant can acquire the pathogenetic features of the parental virus. We previously demonstrated that all four accessory genes of HIV could be deleted from the packaging construct without compromising gene transduction (51). In this work, we went further by deleting another factor crucial for HIV replication, the tat gene. Its product is one of the most powerful transcriptional activators known and plays a pivotal role in the exceedingly high replication rates that characterize HIV-induced disease (18, 19, 47).
It was found that Tat was required in producer cells to generate vector of efficient transducing activity but that this requirement was offset by inducing constitutive high-level expression of vector RNA. Due to the low basal transcription from the HIV LTR, Tat was necessary to increase the abundance of vector transcripts and allow their efficient encapsidation by the vector particles. When made in the absence of Tat, vector particles had 10- to 20-fold-reduced transducing activity. However, when strong constitutive promoters replaced the HIV sequence in the 5′ LTR of the transfer construct, vectors made without Tat exhibited a less than twofold reduction in transducing activity. As Tat strongly upregulated transcription from the chimeric LTR, the transducing activity of the output particles must reach saturation. The abundance of vector RNA in producer cells thus appears to be a rate-limiting factor for transduction until it reaches a threshold. Conceivably, an upper limit is set by the total output of particles available to encapsidate vector RNA. As the total particle output varied with the types of vector and internal promoter used, this may explain the quantitative differences obtained in response to tat deletion.
Successful deletion of the tat gene was unexpected in view of a reported additional role for Tat in reverse transcription (17, 20). While the reasons for this discrepancy are not obvious, it should be noted that the transduction pathway of the lentivirus vector mimics only in part the infection pathway of HIV. The vector is pseudotyped by the envelope of an unrelated virus and contains only the core proteins of HIV, without any accessory gene product. The VSV envelope targets the vector to the endocytic pathway, and it has been shown that redirection of HIV-1 from its normal route of entry by fusion at the plasma membrane significantly changes the biology of the infection. For example, Nef and cyclophilin A are required for the optimal infectivity of wild-type HIV-1 but not of a (VSV G) HIV pseudotype (1). It is also possible that the kinetics of reverse transcription are more critical for the establishment of viral infection than for gene transduction, given the differences in size and sequence between the virus and vector genome.
Tat-independent transduction by an HIV-based vector was recently reported by Kim et al. for in vitro cellular targets (23). In the vector designed by these authors, however, Tat and Rev were expressed from the transfer vector and thus were also present in target cells. A CMV-HIV hybrid LTR was used; this construct yielded vector titers approximately 30% of that obtained with an intact LTR. When the tat gene was inactivated, the titer did not change. Srinivasakumar et al. (43) previously reported a rather low (5- to 10-fold) dependence on Tat of an HIV-based vector produced by cells stably expressing the HIV structural proteins. In this case, titers of 5 × 103 TU/ml with Tat and 7 × 102 TU/ml without Tat were obtained on HeLa-CD4 cells. Although these titers are much lower than those reported here, the vector particles carried the HIV envelope, an indication that Tat is not absolutely required for transduction by vector particles which in that case mirror more closely the wild-type virus. It remained possible, however, that a dependence on Tat may be revealed in more challenging gene deliveries into the body tissues that are the actual targets of gene therapy. This could have been due to a stricter Tat requirement for optimal transduction efficiency or for the production of high-titer vector stocks or to differences in cell-type-specific factors. Our results now establish that Tat is fully dispensable for lentivirus vector transduction even when high titers are achieved and, most importantly, for gene delivery in vivo into terminally differentiated neurons of an adult rat brain.
The Northern analysis of producer and target cells shows that the Tat dependence of LTR-driven expression restricts the production of vector genomic RNA to producer cells. This applies as well to vectors made by the 5′ chimeric constructs, as the U3 sequences of both LTRs of the resulting provirus are derived from the vector 3′ LTR. However, the functional replacement of the tat gene in the packaging construct by promoter sequences upstream of the transfer construct makes the generation of a transcriptionally active recombinant much more unlikely. This will be even more significant in stable producer cell lines that avoid the risk of plasmid recombination during cotransfection.
We also exploited the Rev dependence of gag-pol expression and of the accumulation of unspliced, packageable transcripts. Yu et al. (50) previously showed that the dependence on Rev can be used to make expression of HIV genes inducible. We describe a core packaging system split in two separate nonoverlapping expression constructs, one for the gag and pol reading frames optimized for Rev-dependent expression and the other for the rev cDNA. This third-generation packaging system matches the performance of its predecessors in terms of both yield and transducing efficiency. However, it increases significantly the predicted biosafety of the vector. It has been suggested that the Rev-RRE axis could be replaced by the use of constitutive RNA transport elements of other viruses, although at the price of decreased efficiency (11, 23, 43). We would suggest that maintaining the Rev dependence of the system allows for an additional level of biosafety through the splitting of the HIV-derived components of the packaging system.
The conditional packaging system described here can be combined with a self-inactivating vector construct carrying a major deletion in the 3′ LTR (52). This vector design (Fig. 4) offers significant biosafety features. The contribution of HIV is reduced to a fraction of _cis_-acting sequences in the vector, leaving out in particular most of the LTR, and to only three genes, gag, pol, and rev, in the packaging constructs, compared with the nine genes necessary for the in vivo replication and pathogenesis of wild-type HIV-1 (3, 18, 27, 49). The actual biosafety of a vector must be proven in vivo. However, given the serious limitations of the available animal models of HIV-induced disease, the biosafety of HIV-derived vectors will ultimately be proven only in human hosts. Therefore, the vector design must ensure the highest predictable biosafety for clinical testing to be acceptable.
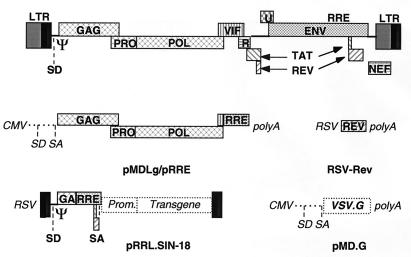
FIG. 4.
Schematic drawing of the HIV provirus and the four constructs used to make a lentivirus vector of the third generation. The viral LTRs, the reading frames of the viral genes, the major 5′ splice donor site (SD), the packaging sequence (Ψ), and the RRE are boxed and indicated in bold type. The conditional packaging construct, pMDLg/pRRE, expresses the gag and pol genes from the CMV promoter and intervening sequences and polyadenylation site of the human β-globin gene. As the transcripts of the gag and pol genes contain _cis_-repressive sequences, they are expressed only if Rev promotes their nuclear export by binding to the RRE. All tat and rev exons have been deleted, and the viral sequences upstream of the gag gene have been replaced. A nonoverlapping construct, RSV-Rev, expresses the rev cDNA. The transfer construct, pRRL.SIN-18, contains HIV-1 _cis_-acting sequences and an expression cassette for the transgene. It is the only portion transferred to the target cells and does not contain wild-type copies of the HIV LTR. The 5′ LTR is chimeric, with the enhancer/promoter of RSV replacing the U3 region (RRL) to rescue the transcriptional dependence on Tat. The 3′ LTR has an almost complete deletion of the U3 region, which includes the TATA box (from nucleotides −418 to −18 relative to the U3/R border). As the latter is the template used to generate both copies of the LTR in the integrated provirus, transduction of this vector results in transcriptional inactivation of both LTRs; thus, it is a self-inactivating vector (SIN-18). The fourth construct, pMD.G, encodes a heterologous envelope to pseudotype the vector, here shown coding for VSV G. Only the relevant parts of the constructs are shown.
It is noteworthy that the fraction of the HIV-1 genome that is left in the vector is probably smaller than could be achieved with any of the nonprimate lentiviruses, the genomic complexity of which is lower than that of HIV-1 (37). Also, the risks associated with the introduction in humans of a recombinant arising from a nonprimate lentivirus, even in a form that in its cognate animal species appears to be attenuated, are very difficult to assess, as illustrated by the ongoing debate on xenotransplantation (48). In contrast, the almost two decades spent studying a virus that has now spread in tens of millions of people worldwide have revealed a considerable amount of information on the pathogenic features of HIV-1, in particular on the dependence of virulence on a crucial set of viral genes. Based on these data, we would like to suggest that the HIV-based vectors described here are good candidates for the clinical trial of lentivirus vectors in human gene therapy.
ACKNOWLEDGMENTS
We are indebted to Tom Hope for providing the Rev expression plasmids, to Melinda Van Roey and Heidi Oline for help with the animal experiments, and to Jennifer Davis and Mitch Finer for suggestions and critical reading of the manuscript.
This work was partly supported by a grant and by a fellowship from the Swiss National Science Foundation to D.T. and R.Z., respectively.
REFERENCES
- 1.Aiken C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol. 1997;71:5871–5877. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldrovandi G M, Zack J A. Replication and pathogenicity of human immunodeficiency virus type 1 accessory gene mutants in SCID-hu mice. J Virol. 1996;70:1505–1507. doi: 10.1128/jvi.70.3.1505-1511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz R D, Hammarskjöld M L, Helga-Maria C, Rekosh D, Goff S P. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology. 1995;212:718–723. doi: 10.1006/viro.1995.1530. [DOI] [PubMed] [Google Scholar]
- 5.Blömer U, Naldini L, Kafri T, Trono D, Verma I M, Gage F H. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bordignon C, Bonini C, Verzeletti S, Nobili N, Maggioni D, Traversari C, Giavazzi R, Servida P, Zappone E, Benazzi E, Porta F, Ferrari G, Mavilio F, Rossini S, Blaese R M, Candotti F. Transfer of the HSV-tk gene into donor peripheral blood lymphocytes for in vivo modulation of donor anti-tumor immunity after allogeneic bone marrow transplantation. Hum Gene Ther. 1995;6:813–819. doi: 10.1089/hum.1995.6.6-813. [DOI] [PubMed] [Google Scholar]
- 7.Buchschacher G L J, Panganiban A T. Human immunodeficiency virus vectors for inducible expression of foreign genes. J Virol. 1992;66:2731–2739. doi: 10.1128/jvi.66.5.2731-2739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J-K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and non-mammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffin J M. Retroviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1767–1846. [Google Scholar]
- 11.Corbeau P, Kraus G, Wong-Staal F. Transduction of human macrophages using a stable HIV-1/HIV-2-derived gene delivery system. Gene Ther. 1998;5:99–104. doi: 10.1038/sj.gt.3300563. [DOI] [PubMed] [Google Scholar]
- 12.Feinberg M B, Baltimore D, Frankel A L. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci USA. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felber B K, Drysdale C M, Pavlakis G N. Feedback regulation of human immunodeficiency virus type 1 expression by the Rev protein. J Virol. 1990;64:3734–3741. doi: 10.1128/jvi.64.8.3734-3741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finer M H, Dull T J, Qin L, Farson D, Roberts M R. kat: a high efficiency retroviral transduction system for primary human T lymphocytes. Blood. 1994;83:43–50. [PubMed] [Google Scholar]
- 15.Gallay P, Chin D, Hope T J, Trono D. HIV-1 infection of nondividing cells mediated through the recognition of integrase by the import/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- 17.Harrich D, Ulich C, Garcia-Martinez L F, Gaynor R B. Tat is required for efficient reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haynes B F, Pantaleo G, Fauci A S. Toward an understanding of the correlates of protective immunity to HIV infection. Science. 1996;271:324–328. doi: 10.1126/science.271.5247.324. [DOI] [PubMed] [Google Scholar]
- 19.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 20.Huang L M, Joshi A, Willey R, Orenstein J, Jeang K T. Human immunodeficiency viruses regulated by alternative trans-activators: genetic evidence for a novel non-transcriptional function of Tat in virion infectivity. EMBO J. 1994;13:2886–2896. doi: 10.1002/j.1460-2075.1994.tb06583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kafri T, Blömer U, Peterson D A, Gage F H, Verma I M. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat Genet. 1997;17:314–317. doi: 10.1038/ng1197-314. [DOI] [PubMed] [Google Scholar]
- 22.Kaye J F, Richardson J H, Lever A M L. cis-acting sequences involved in human immunodeficiency virus type 1 RNA packaging. J Virol. 1995;69:6588–6592. doi: 10.1128/jvi.69.10.6588-6592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim V N, Mitrophanous K, Kingsman S M, Kingsman A J. Minimal requirement for a lentivirus vector based on human immunodeficiency virus type 1. J Virol. 1998;72:811–816. doi: 10.1128/jvi.72.1.811-816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis P F, Hensel M, Emerman M. Human immunodeficiency virus infection of cell arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis P F, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luciw P A. Human immunodeficiency viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1881–1975. [Google Scholar]
- 28.Mandel, R. J., K. G. Rendahl, K. S. Spratt, R. O. Snyder, L. K. Cohen, and S. E. Leff. Characterization of intrastriatal recombinant adeno-associated virus mediated gene transfer of human tyrosine hydroxylase and human GTP-cyclohydroxylase I in a rat model of Parkinson’s disease. J. Neurosci., in press. [DOI] [PMC free article] [PubMed]
- 29.McBride M S, Panganiban A. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBride M S, Schwartz M D, Panganiban A. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyoshi H, Takahashi M, Gage F H, Verma I M. Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc Natl Acad Sci USA. 1997;94:10319–10323. doi: 10.1073/pnas.94.19.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 33.Naldini L, Blömer U, Gage F H, Trono D, Verma I M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ory D S, Neugeboren B A, Mulligan R C. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parolin C, Dorfman T, Palu G, Gottlinger H, Sodroski J. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J Virol. 1994;68:3888–3895. doi: 10.1128/jvi.68.6.3888-3895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paxinos G, Watson C. The rat brain in stereotaxic coordinates. San Diego, Calif: Academic Press; 1987. [Google Scholar]
- 37.Poeschla E, Wong-Staal F, Looney D J. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat Med. 1998;4:354–357. doi: 10.1038/nm0398-354. [DOI] [PubMed] [Google Scholar]
- 38.Poznansky M, Lever A, Bergeron L, Haseltine W, Sodroski J. Gene transfer into human lymphocytes by a defective human immunodeficiency virus type 1 vector. J Virol. 1991;65:532–536. doi: 10.1128/jvi.65.1.532-536.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiser J, Harmison G, Kluepfel-Stahl S, Brady R O, Karlsson S, Schubert M. Transduction of nondividing cells pseudotyped defective high-titer HIV type 1 particles. Proc Natl Acad Sci USA. 1996;93:15266–15271. doi: 10.1073/pnas.93.26.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider R, Campbell M, Nasioulas G, Felber B K, Pavlakis G N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber B K, Pavlakis G N. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimada T, Fujii H, Mitsuya A, Nienhuis W. Targeted and highly efficient gene transfer into CD4+ cells by a recombinant human immunodeficiency virus retroviral vector. J Clin Investig. 1991;88:1043–1047. doi: 10.1172/JCI115365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srinivasakumar N, Chazal N, Helga-Maria C, Prasad S, Hammarskjold M, Rekosh D. The effect of viral regulatory protein expression on gene delivery by human immunodeficiency virus type 1 vectors produced in stable packaging cell lines. J Virol. 1997;71:5841–5848. doi: 10.1128/jvi.71.8.5841-5848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sternberger L A, Hardy P H, Cuculis J J, Meyer H G. The unlabelled antibody-enzyme method of immunohistochemistry. Preparation and properties of soluble antigen-antibody complex (horseradish peroxidase-antihorseradish peroxidase) and its use in the identification of spirochetes. J Histochem Cytochem. 1970;18:315–333. doi: 10.1177/18.5.315. [DOI] [PubMed] [Google Scholar]
- 45.Verma I M, Somia N. Gene therapy promises, problems and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 46.Wei P, Garber M E, Fang S-M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 47.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 48.Weiss R A. Transgenic pigs and virus adaptation. Nature. 1998;391:327–328. doi: 10.1038/34772. [DOI] [PubMed] [Google Scholar]
- 49.Wyand M S, Manson K H, Lackner A A, Desrosiers R C. Resistance of neonatal monkeys to live attenuated vaccine strains of simian immunodeficiency virus. Nat Med. 1997;3:32–36. doi: 10.1038/nm0197-32. [DOI] [PubMed] [Google Scholar]
- 50.Yu H, Rabson A B, Kaul M, Ron Y, Dougherty J P. Inducible human immunodeficiency virus type 1 packaging cell lines. J Virol. 1996;70:4530–4537. doi: 10.1128/jvi.70.7.4530-4537.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zufferey R, Nagy D, Mandel R J, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 52.Zufferey, R., T. Dull, R. J. Mandel, A. Bukovsky, D. Quiroz, L. Naldini, and D. Trono. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol., in press. [DOI] [PMC free article] [PubMed]