Regulation of actin cytoskeleton by mDab1 through N-WASP and ubiquitination of mDab1 (original) (raw)
Abstract
Migration of cells is critical to development of the central nervous system. Reelin, which was identified from the reeler mutant mice having a defect in the multilamellar structure of the brain, is thought to be a key signalling molecule that functions as a cue for determination of cell position. mDab1 (mouse Disabled homologue 1) functions downstream of Reelin. However, the mechanism by which mDab1 regulates cell migration during brain development is unknown. In the present paper, we show that mDab1 associates with N-WASP (neuronal Wiskott–Aldrich syndrome protein) in vitro and in brains of embryonic mice. mDab1 activates N-WASP directly, and induces actin polymerization through the Arp2/3 (actin-related protein 2/3) complex. mDab1 induces formation of filopodia when it is overexpressed in COS-7 cells. This filopodium formation is dependent on N-WASP, because expression of an N-WASP mutant that cannot induce Arp2/3-complex-mediated actin polymerization suppressed filopodium formation. The PTB (phosphotyrosine-binding) domain of mDab1 binds to N-WASP via the NRFY (Asn-Arg-Phe-Tyr) sequence close to the CRIB (Cdc42/Rac-interactive binding) motif of N-WASP and activates N-WASP in vitro. When mDab1 is phosphorylated by Fyn kinase in COS-7 cells, mDab1 is ubiquitinated in a Cbl-dependent manner, and mDab1 does not induce filopodium in the presence of activated Fyn. These findings suggest that mDab1 regulates the actin cytoskeleton through N-WASP, which is negatively regulated by phosphorylation-mediated ubiquitination of mDab1.
Keywords: actin-related protein 2/3 complex (Arp2/3 complex), Cbl, Fyn, mouse Disabled homologue 1 (mDab1), neuronal Wiskott–Aldrich syndrome protein (N-WASP), ubiquitination
Abbreviations: aa, amino acids; Arp2/3, actin-related protein 2/3; CA, Cys381→Ala mutant; Cdc42, cell-division control 42; GE, Gly306→Glu mutant; GST, glutathione S-transferase; KM, Lys299→Met mutant; mDab1, mouse Disabled homologue 1; PTB, phosphotyrosine-binding; TKB, tyrosine-kinase-binding; VCA, verprolin/cofilin/acidic; WASP, Wiskott–Aldrich syndrome protein; N-WASP, neuronal WASP; WH1, WASP homology 1; WT, wild-type; YF, Tyr531→Phe mutant
INTRODUCTION
Cell migration plays an essential role in brain development. Reeler mice were first identified due to their irregular gait [1], and the causative mutation is in a large secretory protein, Reelin [2,3]. Reeler mice show defects in the laminar structure of the brain, indicating that Reelin is an important molecule in brain development [1]. Reeler mice have an ‘inside-out’ laminar structure, in which cells normally located in the outer layer of the brain are instead located in the inner layer. Drosophila disabled (dab) is the gene responsible for axon guidance and fasciculation. Mouse Disabled homologue 1 (mDab1) is the homologue of Drosophila Disabled, and was identified as a mutation that causes a phenotype similar to that of reeler mice [4–8]. Mutations in the mDab1 gene, scrambler and yotari, yielded phenotypes indistinguishable from that of reeler, suggesting that both mDab1 and Reelin play essential roles in the cell migration or guidance necessary for laminar formation of brain. Upon Reelin stimulation, mDab1 is phosphorylated at several tyrosine residues [8]. This phosphorylation is thought to be important, because expression of an mDab1 mutant that lacks tyrosine residues results in a phenotype similar to that of mDab1-deficient mice [9]. Recently, Fyn and Src kinases were found to phosphorylate mDab1 in response to Reelin. Upon Reelin-mediated phosphorylation, phosphorylated mDab1 binds Lis1 (lissencephaly 1), which is involved in brain development [10]. Interestingly, mDab1 levels increase in the absence of Fyn or Src, suggesting that phosphorylation of mDab1 by Fyn or Src regulates mDab1 levels [11,12]. Protein levels in cells are often regulated by the protein-degradation system through ubiquitination. Phosphorylated mDab1 is a substrate for ubiquitination-dependent degradation [13]. Therefore mDab1 is thought to be down-regulated by Reelin–Fyn/Src-mediated phosphorylation of mDab1 during brain development.
mDab1 has a PTB (phosphotyrosine-binding) domain at its N-terminus. This PTB domain does not bind with phosphorylated peptide; instead, it binds phosphoinositides and unphosphorylated peptide sequences containing the NPXY (Asn-Pro-Xaa-Tyr) motif, providing an interface for lipid–protein and protein–protein interactions [14–16]. Through this domain, mDab1 binds to the lipoprotein receptor, which also binds to Reelin.
In cell migration, the rearrangement of the actin cytoskeleton plays an essential role. At the leading edge of migrating cells, including growth cones at the tips of axons and dendrites, dynamic rearrangement of actin leads to the formation of filopodia and lamellipodia. In these processes, WASP (Wiskott–Aldrich syndrome protein) family proteins, including N-WASP (neuronal WASP), activate the Arp2/3 (actin-related protein 2/3) complex, which is the nucleation core for actin polymerization [17,18]. Surprisingly, when mDab1 is expressed in non-neuronal cultured cells, it causes N-WASP-dependent extension of filopodia. This finding suggests that mDab1 directly regulates actin cytoskeleton rearrangements necessary for cell migration during laminar formation.
EXPERIMENTAL
Cell culture and expression vectors
COS-7 cells were cultured as described previously [19]. Transfection was performed with LIPOFECTAMINE 2000 (Invitrogen) according to the manufacturer's instructions. mDab1 (mDab1-555; a gift from Dr Kazunori Nakajima of Keio University, Tokyo, Japan), PTB domain [aa (amino acids) 1–251] or ΔPTB (aa 239–555) was tagged with the N-terminal FLAG or myc and then subcloned into the pEF-BOS expression vector [20]. The transfected cells were starved overnight before immunostaining as described previously [19]. The PTB domain (aa 1–251) of GST (glutathione S-transferase)-fusion mDab1 (0.1 mg/ml) was injected into COS-7 cells, and was observed as described previously [21]. GST alone was injected as negative control. Cdc42 (cell-division control 42) and N-WASP expression vectors were generated as described previously [22,23].
Proteins
Various mutant N-WASPs were expressed in Sf9 insect cells with a bac-to-bac baculovirus expression system (Gibco BRL) with either a GST tag or a His6 tag. Recombinant-virus-infected Sf9 cells were lysed, clarified and purified with glutathione–Sepharose 4B (Amersham Biosciences) or Ni-NTA (Ni2+-nitrilotriacetate)–agarose (Qiagen) as described previously [23]. GST-fusion VCA (verprolin/cofilin/acidic) and PTB domain or WT (wild-type) mDab1 were purified as described previously [19,24]. The GST moiety was removed for some experiments by thrombin digestion [19]. Arp2/3 complex was purified as described previously [25]. Actin was purified from rabbit skeletal muscle, and monomeric actin (G-actin) was isolated by gel filtration on Superdex 200 (Amersham Biosciences) in G buffer (2 mM Tris/HCl, pH 8.0, 0.2 mM CaCl2, 0.5 mM dithiothreitol and 0.2 mM ATP).
Antibodies and immunoprecipitation analysis
Anti-N-WASP polyclonal antibody was produced as described previously [21]. Anti-Fyn polyclonal antibody (sc-16) and antimyc monoclonal antibody (9E10) were from Santa Cruz Biotechnology. Anti-FLAG (M2) monoclonal antibody was from Sigma. Anti-mDab1 (AB5840) rabbit polyclonal antibody was from Chemicon. Non-immune mouse or rabbit IgG (sc-2025 or sc-2027) was from Santa Cruz Biotech.
COS-7 cells were lysed in lysis buffer containing 40 mM Tris/HCl (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 0.5% (v/v) Triton X-100 and 1×Complete™ protease inhibitor cocktail (Roche). Then FLAG-tagged mDab1 was immunoprecipitated with anti-FLAG antibody (×1000 dilution) or non-immune IgG (×100 dilution). In immunoprecipitations using mouse brain, pregnant Jcl:ICR mice were anaesthetized deeply and the embryos were extracted at embryonic day 14. Approx. nine brains were lysed in 1 ml of the lysis buffer. After sonication (level 4 using a Taitec VP 5S ultrasonic homogenizer) and clarification by centrifugation (20000 g), the brain lysate was diluted 2-fold by lysis buffer. Approx. 400 μl of diluted brain lysate was mixed with anti-mDab1 antibody (×200 dilution) or non-immune IgG (×100 dilution). Then the antibodies were bound to Protein-G–agarose (Pierce). After washing with lysis buffer, the precipitates were subjected to Western blot analysis.
Pyrene–actin assay
The pyrene–actin assay was performed as described previously [26]. Final concentrations of Arp2/3 complex, G-actin and pyrene–actin were 60 nM, 2 μM and 0.2 μM respectively. The concentration of WT or mutant N-WASP was 100 nM. The PTB domain of mDab1 was used at 600 nM.
Ubiquitination assay
HEK-293T (human embryonic kidney) cells were transfected with these constructs, including constitutively active (Tyr531→Phe; YF) and dominant-negative (Lys299→Met; KM) Fyn in pME18S [27], HA (haemagglutinin)-tagged WT TKB (tyrosine-kinase-binding) domain mutant (Gly306→Glu; GE) and RING finger mutant (Cys381→Ala; CA) of Cbl in pcDNA3.1 (gifts from Dr Wallace Y. Langdon of the University of Western Australia, Crawley, WA, Australia) [28], myc-tagged mDab1 in pEF-BOS, and FLAG-tagged ubiquitin in pcDNA3.1. After transfection, approx. 107 cells were harvested in 1 ml of TNE buffer (50 mM Tris/HCl, pH 8, 100 mM NaCl, 1% Nonidet P40, 1 mM EDTA, 1 mM Na3VO4 and 5 μg/ml aprotinin). A 400 μl volume of this lysate was mixed with 2 μg of anti-myc antibody (9E10; Santa Cruz Biotech) for immunoprecipitation. After washing of immobilized antibody with TNE supplemented with 0.1% (w/v) SDS and 1% (w/v) sodium deoxycholate, the precipitates were analysed by Western blotting with anti-Fyn (FYN-3, SC-16; Santa Cruz Biotech) and anti-FLAG (M2; Sigma) antibodies.
RESULTS AND DISCUSSION
mDab1 is expressed predominantly in the central nervous system [5], and is thought to regulate cell migration. Cell migration signals often converge on ubiquitous actin regulatory molecules. Thus, to assess whether mDab1 influences cell morphology, we ectopically expressed WT mDab1 in COS-7 cells, and examined changes in the actin cytoskeleton. The cells with exogenous mDab1 expression had prominent microspikes in COS-7 cells (Figure 1). mDab1 expression also caused formation of microspikes in HeLa cells (results not shown). Therefore mDab1 can regulate the actin cytoskeleton in non-neuronal cells.
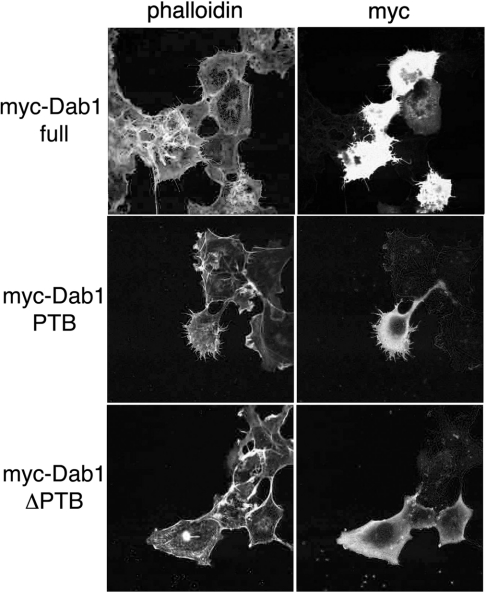
Figure 1. Induction of microspike formation by mDab1.
WT mDab1 or mutant (PTB domain alone or ΔPTB) mDab1 was expressed ectopically in COS-7 cells. Expression was detected with myc tag, and actin filaments were examined by phalloidin staining.
We next examined which domain of mDab1 is necessary for microspike formation. mDab1 has a PTB domain [5]. We expressed exogenous mDab1 without the PTB domain (ΔPTB) and the PTB domain alone. Expression of the PTB domain of mDab1 caused microspike formation, but expression of ΔPTB did not induce changes in actin cytoskeleton. Therefore the PTB domain is necessary for mDab1-mediated regulation of actin cytoskeleton.
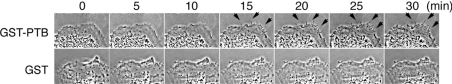
Microspikes are observed when cells extend protrusive structures such as filopodia or retract their membranes (retraction fibres). To distinguish between these two possibilities, we injected the PTB domain purified from Escherichia coli into COS-7 cells and observed changes in morphology by time-lapse microscopy. The cells injected with the PTB domain of mDab1 formed filopodia (Figure 2), indicating that microspikes observed in the ectopic expression experiments (Figure 1) are filopodia.
Figure 2. Induction of filopodium formation by mDab1.
COS-7 cells injected with GST–mDab1 PTB domain or GST alone were observed by time-lapse microscopy. Phase-contrast images of cells before injection (at zero time) to 30 min after injection are shown. Arrows indicate outgrowing filopodia.
Filopodium formation is dependent on reorganization of the actin cytoskeleton. The small GTPase Cdc42 is thought to be a key regulator in this process [29]. Cdc42 binds N-WASP, leading to a conformational change that activates N-WASP [21]. Activated N-WASP then activates the Arp2/3 complex, which induces actin polymerization for extension of filopodia [26].
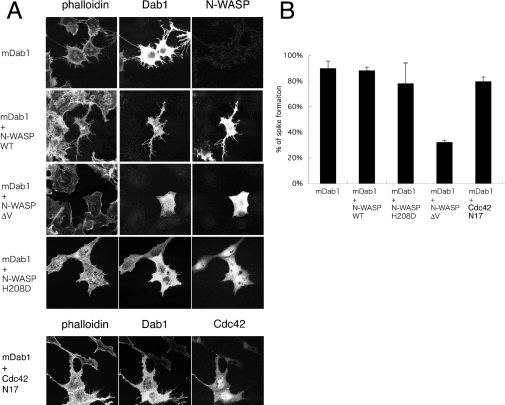
To examine whether mDab1-induced formation of filopodia is dependent on N-WASP, we co-expressed mDab1 with a dominant-negative N-WASP that contains a deletion in the verprolin homology (V) domain (ΔV) that inhibits Arp2/3-complex-dependent actin polymerization [26,30]. The cells with ΔV N-WASP and mDab1 formed fewer microspikes than cells expressing WT N-WASP and mDab1 or mDab1 alone (Figure 3). Therefore mDab1 appears to regulate the actin cytoskeleton through N-WASP.
Figure 3. N-WASP-dependent microspike formation by mDab1.
(A) Microspikes were visualized by phalloidin staining of actin filaments in cells transfected with vector expressing mDab1 or co-transfected with vectors expressing mDab1 and WT, ΔV or H208D N-WASP, or dominant-negative [Asn17 (N17) mutant] Cdc42. The transfected cells were detected by anti-N-WASP or anti-myc antibody. (B) The percentages of transfected cells with microspikes are presented. Results are means±S.D.
We next examined the relation between mDab1 and Cdc42 by expressing the H208D (His208→Asp) mutant of N-WASP, which suppresses Cdc42-induced expression of filopodia [21]. Cells expressing H208D N-WASP and mDab1 formed microspikes at frequencies similar to those of cells expressing WT N-WASP and mDab1 or mDab1 alone. However, the microspikes on cells expressing H208D N-WASP were shorter than those on cells with WT N-WASP (Figure 3). The mDab1-induced microspikes were not inhibited in the presence of dominant-negative Cdc42 Asn17 mutant (Figure 3). Therefore formation of microspikes induced by mDab1 is independent of Cdc42.
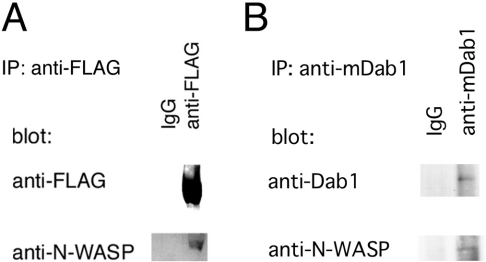
These data suggest that mDab1 activates N-WASP directly. To test this hypothesis, we examined the physical interaction between N-WASP and mDab1 by immunoprecipitation. In COS-7 cells, exogenously expressed mDab1 was immunoprecipitated with endogenous N-WASP (Figure 4A), indicating a direct interaction between N-WASP and mDab1. In lysates of E14 mouse brains, N-WASP was immunoprecipitated with mDab1, indicating that N-WASP and mDab1 interact during brain development (Figure 4B).
Figure 4. Association between N-WASP and mDab1.
(A) FLAG-tagged mDab1 was expressed in COS-7 cells and immunoprecipitated with anti-FLAG-antibody or non-immune antibody as a negative control. The precipitate was analysed by Western blotting with anti-FLAG or anti-N-WASP antibody. (B) Lysate of brains from E14 mice was subjected to immunoprecipitation (IP) with anti-mDab1 antibody (Chemicon) or non-immune antibody. Precipitate was analysed by Western blotting with anti-mDab1 or anti-N-WASP antibody.
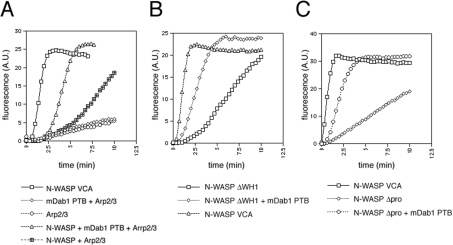
We then examined whether purified mDab1 could activate N-WASP and Arp2/3-complex-mediated actin polymerization in vitro by the pyrene–actin assay, in which an increase in fluorescence indicates actin filament formation. N-WASP alone is inactive in Arp2/3-complex-induced actin polymerization in vitro, because autoinhibition by the interaction between VCA and CRIB (Cdc42/Rac-interactive binding) region prevents the association of N-WASP with Arp2/3 complex [26] (Figure 5A). We used the PTB domain of mDab1 in this assay, because we were unable to purify WT mDab1, and the PTB domain alone is sufficient to induce formation of microspikes in cultured cells (Figure 2). As shown in Figure 5(A), rapid actin polymerization was observed in the presence of the PTB domain of mDab1, N-WASP and the Arp2/3 complex. Lack of the PTB domain, N-WASP or Arp2/3 complex diminished the rapid actin polymerization. Because the PTB domain alone had no effect on actin polymerization with Arp2/3 complex (Figure 5A), the PTB domain of mDab1 activated N-WASP. Activation of N-WASP by the PTB domain was enhanced by the addition of Cdc42, indicating co-operativity of mDab1 and Cdc42 (results not shown). Activation of N-WASP is independent of the WH1 (WASP homology 1) domain and proline-rich region (the domain structure is shown in Figure 6A), because deletion of these regions did not affect N-WASP activation by the mDab1 PTB domain. Thus the mDab1 PTB domain appears to disrupt the autoinhibition of N-WASP, possibly by interfering with the association between the CRIB and VCA domains (Figures 5B and 5C). These data indicate that mDab1 activates N-WASP directly.
Figure 5. Activation of N-WASP by mDab1 in vitro.
Pyrene–actin assay to monitor Arp2/3-complex-mediated actin polymerization. Increase in fluorescence indicates actin polymerization. Actin polymerization induced by WT (A), ΔWH1 (B) and Δproline-rich (C) N-WASP in the presence or the absence of PTB domain of mDab1 is shown. A.U., absorbance units.
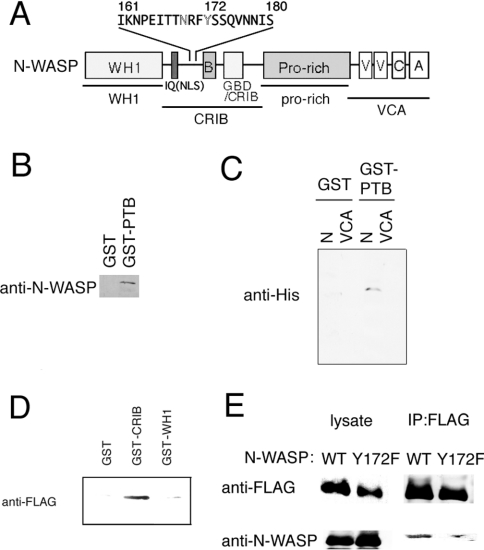
Figure 6. mDab1-binding site in N-WASP.
(A) Schematic representation of the domain structure of N-WASP. Domains of N-WASP used in pull-down assays are underlined. A putative mDab1-binding site (NRFY sequence) is highlighted. IQ (calmodulin-binding) motif is indicated. NLS, nuclear localization sequence. (B) and (C) Pull-down assay using GST or GST–mDab1 PTB domain immobilized on agarose beads. Purified WT N-WASP (B) or His6-tagged N-terminal region or VCA region of N-WASP (C) was mixed with GST or GST–mDab1 PTB domain immobilized on agarose beads. After washing of the beads, the bound proteins were analysed by Western blotting. (D) Pull-down assay with GST, GST–N-WASP WH1 or GST–N-WASP CRIB domain. FLAG-tagged mDab1 was expressed in COS-7 cells, and the COS-7 lysate was mixed with immobilized GST or GST-fusion N-WASP fragments. Bound proteins were detected by Western blotting with anti-FLAG antibody. (E) Association of mDab1 with Tyr172 of N-WASP. FLAG-tagged mDab1 and WT or Y172F mutant of N-WASP (without tags) were expressed in COS-7 cells and immunoprecipitated with anti-FLAG-antibody. The precipitate was analysed by Western blotting with anti-FLAG or anti-N-WASP antibody.
We next confirmed a direct interaction between the mDab1 PTB domain and N-WASP, and narrowed the region of N-WASP required for association with mDab1 (Figure 6). Pull-down assays revealed an association between the PTB domain of mDab1 and WT N-WASP (Figure 6B). As shown in Figure 6(C), the N-terminal region of N-WASP, including the WH1 domain and CRIB motif, is required for association with the PTB domain. The VCA domain (Figure 6C) and proline-rich region (results not shown), were not involved in binding with the mDab1 PTB domain.
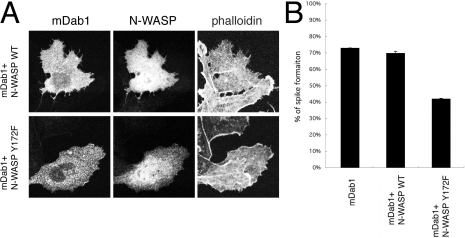
Because the N-WASP mutant lacking the WH1 domain (ΔWH1) was activated by the PTB domain (Figure 5), the region containing the CRIB motif is thought to be the PTB region (Figure 6A). As shown in Figure 6(D), the region containing the CRIB motif is necessary for mDab1 binding. The PTB domain of mDab1 is known to bind the non-phosphorylated NPXY motif. [14]. There is no NPXY sequence on N-WASP, but the sequence NRFY (Asn-Arg-Phe-Tyr) was found near the CRIB motif in N-WASP (Figure 6A). Proline can be substituted for the arginine residue in the NPXY motif [14]. Thus the NRFY sequence in N-WASP is likely to be the binding site. When we immunoprecipitated mDab1 expressed in COS-7 cells, WT N-WASP was co-precipitated with mDab1, whereas the Y172F (Tyr172→Phe) mutant of N-WASP, which is a mutant of the NRFY sequence, was not co-precipitated as effectively as WT N-WASP (Figure 6E). The residual N-WASP in immunoprecipitates from cells expressing Y172F N-WASP and mDab1 was presumably from endogenous N-WASP. mDab1-induced microspike formation was impaired in cells expressing the Y172F mutant of N-WASP, whereas it was not in cells expressing WT N-WASP (Figures 7A and 7B).
Figure 7. Effect of PTB domain binding mutant on mDab1-induced microspike formation.
(A) Phalloidin staining of actin filaments of cells transfected with myc-tagged mDab1 and WT or Y172F mutant N-WASP (without tags). The transfected cells were visualized with anti-N-WASP antibody or anti-myc antibody. (B) The percentages of transfected cells with microspikes are presented. Results are means±S.D.
These data indicate that mDab1 associates with the NRFY sequence in the CRIB motif of N-WASP. The region around CRIB is known to be responsible for the autoinhibitory interaction of N-WASP, suggesting that the association of mDab1 with this region disrupts the intramolecular interaction directly, activating Arp2/3-complex-mediated actin polymerization.
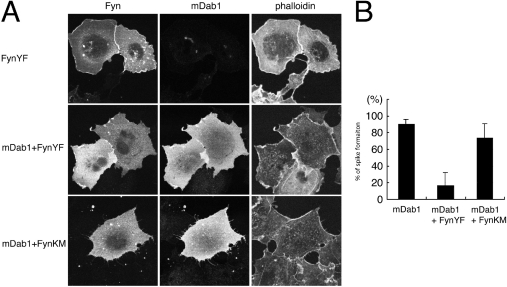
mDab1 is phosphorylated upon Reelin stimulation, presumably through Src family tyrosine kinases [8,11,12]. This phosphorylation could influence the cytoskeletal effect of mDab1. Therefore we co-expressed constitutively active (YF) or dominant-negative (KM) Fyn with mDab1 and examined the morphology of these cells. YF Fyn is known to phosphorylate mDab1 [5,13] (results not shown). In COS-7 cells, expression of YF Fyn alone caused membrane ruffling instead of microspike formation (Figure 8). Cells expressing YF Fyn and mDab1 showed fewer microspikes in comparison with cells expressing KM Fyn and mDab1 or mDab1 alone (Figure 8). Therefore phosphorylation of mDab1 appears to reduce the ability to induce microspike formation.
Figure 8. Effect of Fyn on mDab1-mediated microspike formation in COS-7 cells.
(A) Phalloidin staining of actin filaments of cells transfected with myc-tagged mDab1 and constitutively active (YF) or dominant-negative (KM) Fyn. Phalloidin staining of cells transfected with YF Fyn alone is also shown. The transfected cells were visualized with anti-Fyn or anti-myc antibody. (B) The percentages of transfected cells with microspikes are presented. Results are means±S.D.
Recently, the amounts of Fyn and Src kinase were shown to be critical for brain levels of mDab1 [11,12]. mDab1 levels were increased significantly in Fyn or Src knockout mice, and mDab1 levels decreased when levels of Fyn or Src increased during brain development. We then examined whether mDab1 is degraded through the ubiquitin–proteasome degradation pathway when it is tyrosine-phosphorylated.
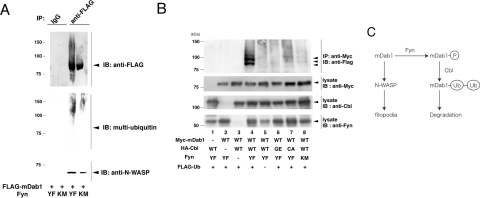
As shown in Figure 9(A), the level of ubiquitination of mDab1 was significantly higher in cells expressing YF Fyn in comparison with those expressing KM Fyn. However, mDab1 association with N-WASP is observed even in the presence of YF Fyn. Therefore changes in the mode of mDab1 binding after phosphorylation abolishes mDab1-induced microspike formation in COS-7 cells.
Figure 9. Ubiquitination of mDab1 by Cbl.
(A) Vector expressing FLAG-tagged WT mDab1 was co-transfected into COS-7 cells with vectors expressing YF or KM Fyn. mDab1 was immunoprecipitated with anti-FLAG antibody and subjected to Western blotting with anti-FLAG, anti-N-WASP or anti-multi-ubiquitin (FK2, MBL) antibody. (B) HEK-293T cells were transfected with vector expressing myc-tagged mDab1, FLAG-tagged ubiquitin, YF or KM mutant of Fyn, HA (haemagglutinin)-tagged WT, TKB domain mutant (GE mutant) or RING-finger-domain mutant (CA mutant) of Cbl. After transfection, cells were immunoprecipitated with anti-myc antibody and then subjected to Western blotting with the indicated antibodies. (C) Schematic diagram of the relationship between mDab1, Fyn or Src kinase, Cbl and N-WASP. IP, immunoprecipitation.
Recently, ubiquitination of mDab1 after phosphorylation was reported [13], although the mechanism of ubiquitination of mDab1 is still unclear. Many tyrosine-phosphorylated proteins are known to be ubiquitinated by the proto-oncogene Cbl. Cbl is abundant in brain [31], and contains a TKB domain that recognizes phosphorylated tyrosine residues as well as a RING finger essential for ubiquitin ligase activity [32]. We examined whether Cbl mediates ubiquitination of mDab1.
Ubiquitination of mDab1 was monitored by FLAG-tagged ubiquitin. Ubiquitination of mDab1 in the presence of YF Fyn was very weak when monitored by FLAG-tagged ubiquitin; however, addition of WT Cbl and the YF mutant of Fyn dramatically increased the ubiquitination of mDab1, whereas WT Cbl and KM Fyn did not (Figure 9B). In contrast, addition of a TKB domain mutant (GE mutant) or RING finger domain mutant (CA mutant) of Cbl and YF Fyn did not increase ubiquitination of mDab1 as effectively as WT Cbl. Thus Cbl appears to be a ubiquitin ligase responsible for ubiquitination of mDab1.
Because filopodia/microspike formation is essential for cell migration, phosphorylation of mDab1 leading to ubiquitination-triggered degradation and loss of microspike formation could reflect the mechanism underlying abnormal cell migration during brain development in reeler mice (Figure 9C).
Acknowledgments
We thank Dr Kazunori Nakajima (Keio University School of Medicine, Japan) for mDab1 cDNA. We thank Dr Wallace Y. Langdon (University of Western Australia, Crawley, WA, Australia) for Cbl constructs. This work was supported by grant-in-aids from the Ministry of Education, Culture, Sports, and Technology of Japan and from the Japan Science and Technology Corporation (JST).
References
- 1.Falconer D. S. Two mutations trembler and reeler, with neurological actions in the house mouse. J. Genet. 1951;50:192–201. doi: 10.1007/BF02996215. [DOI] [PubMed] [Google Scholar]
- 2.D'Arcangelo G., Miao G. G., Chen S. C., Soares H. D., Morgan J. I., Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature (London) 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- 3.Hirotsune S., Takahara T., Sasaki N., Hirose K., Yoshiki A., Ohashi T., Kusakabe M., Murakami Y., Muramatsu M., Watanabe S., et al. The reeler gene encodes a protein with an EGF-like motif expressed by pioneer neurons. Nat. Genet. 1995;10:77–83. doi: 10.1038/ng0595-77. [DOI] [PubMed] [Google Scholar]
- 4.Sheldon M., Rice D. S., D'Arcangelo G., Yoneshima H., Nakajima K., Mikoshiba K., Howell B. W., Cooper J. A., Goldowitz D., Curran T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature (London) 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- 5.Howell B. W., Gertler F. B., Cooper J. A. Mouse disabled (mDab1): a Src binding protein implicated in neuronal development. EMBO J. 1997;16:121–132. doi: 10.1093/emboj/16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howell B. W., Hawkes R., Soriano P., Cooper J. A. Neuronal position in the developing brain is regulated by mouse disabled-1. Nature (London) 1997;389:733–737. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- 7.Ware M. L., Fox J. W., Gonzalez J. L., Davis N. M., Lambert de Rouvroit C., Russo C. J., Chua S. C., Jr, Goffinet A. M., Walsh C. A. Aberrant splicing of a mouse disabled homolog, mdab1, in the scrambler mouse. Neuron. 1997;19:239–249. doi: 10.1016/s0896-6273(00)80936-8. [DOI] [PubMed] [Google Scholar]
- 8.Yoneshima H., Nagata E., Matsumoto M., Yamada M., Nakajima K., Miyata T., Ogawa M., Mikoshiba K. A novel neurological mutant mouse, yotari, which exhibits reeler-like phenotype but expresses CR-50 antigen/reelin. Neurosci. Res. 1997;29:217–223. doi: 10.1016/s0168-0102(97)00088-6. [DOI] [PubMed] [Google Scholar]
- 9.Howell B. W., Herrick T. M., Hildebrand J. D., Zhang Y., Cooper J. A. Dab1 tyrosine phosphorylation sites relay positional signals during mouse brain development. Curr. Biol. 2000;10:877–885. doi: 10.1016/s0960-9822(00)00608-4. [DOI] [PubMed] [Google Scholar]
- 10.Assadi A. H., Zhang G., Beffert U., McNeil R. S., Renfro A. L., Niu S., Quattrocchi C. C., Antalffy B. A., Sheldon M., Armstrong D. D., et al. Interaction of reelin signaling and Lis1 in brain development. Nat. Genet. 2003;35:270–276. doi: 10.1038/ng1257. [DOI] [PubMed] [Google Scholar]
- 11.Bock H. H., Herz J. Reelin activates SRC family tyrosine kinases in neurons. Curr. Biol. 2003;13:18–26. doi: 10.1016/s0960-9822(02)01403-3. [DOI] [PubMed] [Google Scholar]
- 12.Arnaud L., Ballif B. A., Forster E., Cooper J. A. Fyn tyrosine kinase is a critical regulator of disabled-1 during brain development. Curr. Biol. 2003;13:9–17. doi: 10.1016/s0960-9822(02)01397-0. [DOI] [PubMed] [Google Scholar]
- 13.Arnaud L., Ballif B. A., Cooper J. A. Regulation of protein tyrosine kinase signaling by substrate degradation during brain development. Mol. Cell. Biol. 2003;23:9293–9302. doi: 10.1128/MCB.23.24.9293-9302.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howell B. W., Lanier L. M., Frank R., Gertler F. B., Cooper J. A. The disabled 1 phosphotyrosine-binding domain binds to the internalization signals of transmembrane glycoproteins and to phospholipids. Mol. Cell. Biol. 1999;19:5179–5188. doi: 10.1128/mcb.19.7.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun M., Keshvara L., Park C. G., Zhang Y. M., Dickerson J. B., Zheng J., Rock C. O., Curran T., Park H. W. Crystal structures of the Dab homology domains in mouse disabled-1 and -2. J. Biol. Chem. 2003;278:36572–36581. doi: 10.1074/jbc.M304384200. [DOI] [PubMed] [Google Scholar]
- 16.Stolt P. C., Jeon H., Song H. K., Herz J., Eck M. J., Blacklow S. C. Origins of peptide selectivity and phosphoinositide binding revealed by structures of disabled-1 ptb domain complexes. Structure. 2003;11:569–579. doi: 10.1016/s0969-2126(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 17.Suetsugu S., Takenawa T. Regulation of cortical actin networks in cell migration. Int. Rev. Cytol. 2003;229:245–286. doi: 10.1016/s0074-7696(03)29006-9. [DOI] [PubMed] [Google Scholar]
- 18.Takenawa T., Miki H. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 2001;114:1801–1809. doi: 10.1242/jcs.114.10.1801. [DOI] [PubMed] [Google Scholar]
- 19.Suetsugu S., Miki H., Takenawa T. The essential role of profilin in the assembly of actin for microspike formation. EMBO J. 1998;17:6516–6526. doi: 10.1093/emboj/17.22.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizushima S., Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miki H., Sasaki T., Takai Y., Takenawa T. Induction of filopodium formation by WASP-related actin-depolymerizing protein N-WASP. Nature (London) 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 22.Suetsugu S., Hattori M., Miki H., Tezuka T., Yamamoto T., Mikoshiba K., Takenawa T. Sustained activation of N-WASP through phosphorylation is essential for neurite extension. Dev. Cell. 2002;3:645–658. doi: 10.1016/s1534-5807(02)00324-6. [DOI] [PubMed] [Google Scholar]
- 23.Suetsugu S., Miki H., Takenawa T. Identification of another actin-related protein (Arp) 2/3 complex binding site in neural Wiskott–Aldrich syndrome protein (N-WASP), that complements actin polymerization induced by the Arp2/3 complex activating (VCA) domain of N-WASP. J. Biol. Chem. 2001;276:33175–33180. doi: 10.1074/jbc.M102866200. [DOI] [PubMed] [Google Scholar]
- 24.Miki H., Miura K., Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 25.Egile C., Loisel T. P., Laurent V., Li R., Pantaloni D., Sansonetti P. J., Carlier M. F. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J. Cell. Biol. 1999;146:1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., Kirschner M. W. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi M., Kuramochi S., Fusaki N., Nada S., Kawamura-Tsuzuku J., Matsuda S., Semba K., Toyoshima K., Okada M., Yamamoto T. Functional and physical interaction of protein-tyrosine kinases Fyn and Csk in the T-cell signaling system. J. Biol. Chem. 1993;268:27413–27419. [PubMed] [Google Scholar]
- 28.Thien C. B., Walker F., Langdon W. Y. RING finger mutations that abolish c-Cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol. Cell. 2001;7:355–365. doi: 10.1016/s1097-2765(01)00183-6. [DOI] [PubMed] [Google Scholar]
- 29.Hall A. Rho GTPase and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 30.Miki H., Takenawa T. Direct binding of the verprolin-homology domain in N-WASP to actin is essential for cytoskeletal reorganization. Biochem. Biophys. Res. Commun. 1998;243:73–78. doi: 10.1006/bbrc.1997.8064. [DOI] [PubMed] [Google Scholar]
- 31.Nishio H., Otsuka M., Kinoshita S., Tokuoka T., Nakajima M., Noda Y., Fukuyama Y., Suzuki K. Phosphorylation of c-Cbl protooncogene product following ethanol administration in rat cerebellum: possible involvement of Fyn kinase. Brain Res. 2002;950:203–209. doi: 10.1016/s0006-8993(02)03038-x. [DOI] [PubMed] [Google Scholar]
- 32.Thien C. B., Langdon W. Y. Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]