NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-β (transforming growth factor-β) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-β type I receptor (original) (raw)
Abstract
Inhibitory Smad, Smad7, is a potent inhibitor of TGF-β (transforming growth factor-β) superfamily signalling. By binding to activated type I receptors, it prevents the activation of R-Smads (receptor-regulated Smads). To identify new components of the Smad pathway, we performed yeast two-hybrid screening using Smad7 as bait, and identified NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) as a direct binding partner of Smad7. NEDD4-2 is structurally similar to Smurfs (Smad ubiquitin regulatory factors) 1 and 2, which were identified previously as E3 ubiquitin ligases for R-Smads and TGF-β superfamily receptors. NEDD4-2 functions like Smurfs 1 and 2 in that it associates with TGF-β type I receptor via Smad7, and induces its ubiquitin-dependent degradation. Moreover, NEDD4-2 bound to TGF-β-specific R-Smads, Smads 2 and 3, in a ligand-dependent manner, and induced degradation of Smad2, but not Smad3. However, in contrast with Smurf2, NEDD4-2 failed to induce ubiquitination of SnoN (Ski-related novel protein N), although NEDD4-2 bound to SnoN via Smad2 more strongly than Smurf2. We showed further that overexpressed NEDD4-2 prevents transcriptional activity induced by TGF-β and BMP, whereas silencing of the NEDD4-2 gene by siRNA (small interfering RNA) resulted in enhancement of the responsiveness to TGF-β superfamily cytokines. These data suggest that NEDD4-2 is a member of the Smurf-like C2-WW-HECT (WW is Trp-Trp and HECT is homologous to the E6-accessory protein) type E3 ubiquitin ligases, which negatively regulate TGF-β superfamily signalling through similar, but not identical, mechanisms to those used by Smurfs.
Keywords: bone morphogenetic protein (BMP), homologous to the E6-accessory protein C-terminus (HECT), transforming growth factor-β (TGF-β), neural precursor cell expressed, developmentally down-regulated 4-2 (NEDD4-2), Smad, Ski-related novel protein N (SnoN)
Abbreviations: BMP, bone morphogenetic protein; BMPR-IB, BMP type IB receptor; Co-Smad, common-partner Smad; I-Smad, inhibitory Smad; HECT, homologous to the E6-accessory protein C-terminus; NEDD4-2, neural precursor cell expressed, developmentally down-regulated 4-2; RING, really interesting new gene; ROC1, regulator of Cullins 1; R-Smad, receptor-regulated Smad; SCF, Skp1/Cullin1/F-box protein; siRNA, small interfering RNA; Smurf, Smad ubiquitin regulatory factor; SnoN, Ski-related novel protein N; TβR-I, transforming growth factor-β type I receptor; TGF-β, transforming growth factor-β
INTRODUCTION
Members of the TGF-β (transforming growth factor-β) superfamily cytokines, including TGF-β, activin, nodal, BMPs (bone morphogenetic proteins) and anti-Müllerian hormone, are multifunctional proteins that regulate cellular growth, differentiation, apoptosis and morphogenesis [1]. TGF-β superfamily cytokines bind to type I and type II serine/threonine kinase receptors, and transduce intracellular signals through Smad proteins [2–5]. R-Smads (receptor-regulated Smads) and Co-Smads (common-partner Smads) positively regulate signalling by the TGF-β superfamily. Upon phosphorylation by the type I receptors, R-Smads form heteromeric complexes with Co-Smad, Smad4, and translocate into the nucleus. In the nucleus, Smad complexes bind to transcriptional factor(s), as well as transcriptional co-activators and co-repressors, and regulate transcription of target genes. Among the R-Smads, Smad2 and Smad3 act in TGF-β, activin and nodal pathways, whereas Smad1, Smad5 and Smad8 act in BMP and anti-Müllerian hormone pathways. I-Smads (inhibitory Smads), including Smad6 and Smad7, bind to type I receptors and prevent phosphorylation of R-Smads, resulting in the inhibition of TGF-β superfamily signalling [6,7].
Ubiquitin-dependent protein degradation plays key roles in various biological processes, including signal transduction, cell-cycle progression and transcriptional regulation [8,9]. An E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases induce ubiquitination of proteins. E3 ligases play a crucial role in recognition of target proteins and in the subsequent protein degradation by 26 S proteasomes [10]. Of the E3 ubiquitin ligases, the RING (really interesting new gene)-type and HECT (homologous to the E6-accessory protein C-terminus)-type ligases are among the best characterized.
Ubiquitin–proteasome pathways play a pivotal role in the regulation of TGF-β superfamily signalling. A RING-type E3 ubiquitin ligase ROC1–SCF (regulator of Cullins 1–Skp1/Cullin1/F-box protein) complex was shown to degrade activated Smad3 and Smad4 [11,12]. Smurf (Smad ubiquitin regulatory factor) 1 was originally identified as a HECT-type E3 ubiquitin ligase for BMP-specific R-Smads, Smads 1 and 5 [13]. Smurf2, a Smurf1-related E3 ubiquitin ligase, interacts with Smad1, as well as Smad2, and induces their ubiquitin-mediated degradation [14,15]. In addition, Smurfs 1 and 2 interact with nuclear Smad7 and induce translocation of Smad7 to the cytoplasm in a CRM-1 (chromosome region maintenance 1)-dependent fashion [16–18]. The Smurf1–Smad7 complex is then targeted to the cell membrane through the N-terminal C2 domain in Smurf1, and associates with TβR-I (TGF-β type I receptor) [19]. After binding to TβR-I, Smurfs 1 and 2 induce ubiquitin-mediated degradation of TβR-I. In addition, Jab1/CSN5, which is a component of the COP9 (constitutively photomorphogenic 9) signalosome complex, has been reported to associate with Smad7, and induce nuclear export and degradation of Smad7 [20]. In contrast, some ubiquitin ligases have been reported to degrade negative regulators of the TGF-β/BMP pathway, and enhance signalling by members of the TGF-β superfamily. Smurf2 binds to Smad co-repressor SnoN (Ski-related novel protein N) via Smad2, and degrades SnoN, followed by enhancement of the TGF-β signal under certain conditions [21]. Recently, we showed that a RING-type E3 ubiquitin ligase, Arkadia, binds to and degrades Smad7, leading to enhancement of TGF-β/BMP signalling [22].
NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) and a structurally related protein, NEDD4, have been shown to interact with the epithelial sodium channel, and to down-regulate its activity via the ubiquitin–proteasome pathway [23–31]. In the present study, we identified NEDD4-2 as a novel binding partner of Smad7, and examined the function of NEDD4-2 in TGF-β superfamily signalling. We showed that NEDD4-2 interacts with TβR-I via Smad7 and induces receptor degradation. Moreover, NEDD4-2 interacts with Smad2 and enhances its ubiquitin-mediated degradation. TGF-β- and BMP-induced transcriptional activities are inhibited by overexpression of NEDD4-2 and are enhanced by knock down of the NEDD4-2 gene. Functional differences between NEDD4-2 and Smurfs will be discussed.
MATERIALS AND METHODS
Yeast two-hybrid screening
To construct a bait plasmid, full-length mouse Smad7 cDNA was inserted in-frame into pGBKT7 GAL4 DNA-binding vector. This construct was transformed into the yeast strain AH109 and was used to screen a pre-transformed HeLa cDNA library in the pGAD vector (Clontech) on SD−L−W−A (synthetic defined medium, deficient in leucine, tryptophan and adenine). Positive colonies were picked after 6–13 days and re-selected on SD−L−W−A−H (synthetic defined medium deficient in leucine, tryptophan, histidine and adenine). Library plasmids were rescued from the yeast and sequenced.
DNA construction and transfection
The original constructions of constitutively active forms of TGF-β type I and BMP type IB receptors [TβR-I(TD) and BMPR-IB(QD) respectively], Smad1, Smad2, Smad3, Smad4, Smad5, Smad8 and NEDD4 were described previously [19,32]. The open reading frame of NEDD4-2 was generated by a PCR-based approach from a cDNA clone of KIAA0439 (Kazusa DNA Research Institute, Chiba, Japan) as a template, and subcloned into _Eco_RI/_Xho_I-digested FLAG–pcDNA3 [32]. The catalytically inactive form of NEDD4-2 [NEDD4-2(CA)], in which Cys894 was replaced with alanine, was generated by a PCR-based approach. The cDNA for Smurf2 was kindly provided by Dr X.-H. Feng (Departments of Surgery and Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX, U.S.A.) [14]. COS7 cells, HEK-293 cells, HEK-293T cells and HepG2 cells were transiently transfected using FuGENE6 (Roche Applied Science) or LIPOFECTAMINE 2000 reagent (Invitrogen) as described previously [22].
Immunofluorescence labelling
Immunohistochemical staining of FLAG–NEDD4-2, FLAG–NEDD4, FLAG–Smurf2 or FLAG–Smad7 in transfected cells was performed using mouse anti-FLAG antibody, followed by incubation with FITC-labelled goat anti-mouse IgG as described previously [17]. For double-staining of Smad7 and E3 ubiquitin ligases, immunohistochemical staining of FLAG–Smad7 and 6Myc–E3 ligases was performed using mouse anti-FLAG or rabbit anti-Myc antibody, followed by incubation with FITC-labelled goat anti-mouse IgG or RITC (rhodamine isothiocyanate)-labelled goat anti-rabbit IgG respectively. Cell nuclei were stained with propidium iodide. Intracellular localization was determined by confocal laser scanning microscopy.
Immunoprecipitation and Immunoblotting
Cells were lysed with Nonidet P-40 lysis buffer (20 mM Tris/HCl, pH 7.5, 150 mM NaCl and 1% Nonidet P-40). Immunoprecipitation and immunoblotting were performed as described previously [33]. For inhibition of proteasomal degradation, cells were incubated with 10 μM lactacystin (Calbiochem) for 6 h just before cell lysis, unless otherwise indicated.
Affinity cross-linking and immunoprecipitation
Recombinant TGF-β1 (R&D Systems) was iodinated using the chloramine T method as described previously [34]. The immunoprecipitation of the cross-linked complex and analysis by SDS/PAGE were performed as described previously [33].
Pulse–chase analysis
Transfected COS7 cells were labelled for 10 min at 37 °C with 50 μCi of [35S]methionine and [35S]cysteine (Amersham Biosciences)/ml in methionine- and cysteine-free DMEM (Dulbecco's modified Eagle's medium), and chased in DMEM supplemented with 0.2% (v/v) foetal bovine serum, and unlabelled methionine and cysteine for the time periods indicated, as described previously [17]. Cells were then lysed, and the protein extracts were subjected to immunoprecipitation, followed by SDS/PAGE. The gels were fixed, dried, and then examined using a Fuji BAS 2500 Bio-Imaging Analyzer (Fuji Photo Film).
Luciferase assay
HepG2 cells, HEK-293 cells and HeLa cells were transiently transfected with an appropriate combination of reporter constructs, expression plasmids and pcDNA3. At 24 h after transfection, cell lysates were prepared, and luciferase activity was measured by the Dual-Luciferase Reporter System (Promega) as described previously [7]. Values were normalized to Renilla luciferase activity.
RNA interference and oligonucleotides
siRNAs (small interfering RNAs) were introduced into cells as described previously [22]. The following 21-mer oligonucleotide pairs were used for RNA interference: NEDD4-2 siRNA from nucleotides 727–747 (GenBank® accession number AB007899), 5′-AAGUGGACAAUUUAGGCCGAA-3′ and 5′-UUCGGCCUAAAUUGUCCACUU-3′; and a control siRNA from Euglena gracilis chloroplast DNA between s16 S and 16 S rRNA (GenBank® accession number X05005), 5′-AAGCGCGCAAAGUAGGAUUCG-3′ and 5′-CGAAUCCUACUUUGCGCGCUU-3′.
Real-time PCR
Quantitative real-time PCR analysis was performed using ABI PRISM 7000 Sequence Detection System (Applied Biosystems). The cDNA templates from human tissues were purchased from BD Biosciences (Human MTC Panel I/II), and those from human carcinoma cell lines were described previously [35]. The primer sequences used were as follows: human NEDD4-2, 5′-TCCAATGGTCCTCAGCTGTTTA-3′ (forward) and 5′-ATTTTCCACGGCCATGAGA-3′ (reverse); human Smurf2, 5′-GGCAGAACCAATTGAAAGACCA-3′ (forward) and 5′-GTTTCTGAACAAGGTCTCGCTT-3′ (reverse), and human GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-GAAGGTGAAGGTCGGAGTC-3′ (forward) and 5′-GAAGATGGTGATGGGATTTC-3′ (reverse).
RESULTS
NEDD4-2 interacts with Smads 2, 3, 6 and 7
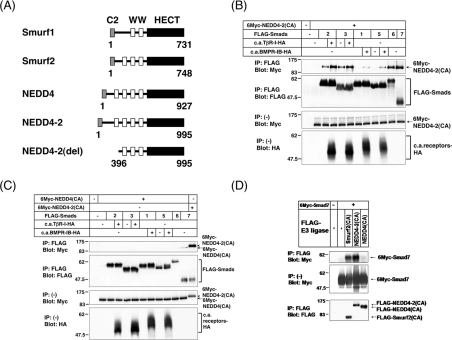
To identify proteins that play a crucial role in TGF-β superfamily signalling, we performed a yeast two-hybrid screening of a HeLa cDNA library using full-length Smad7 as bait. In total, we isolated 209 positive clones from 2.9×107 transformants. Among the positive clones, we have identified four NEDD4-2-encoding clones which lack the N-terminal 395 amino acids [NEDD4-2(del)] (Figure 1A). NEDD4-2 has four WW (Trp–Trp) domains that interact with the PY (Pro–Tyr) motif.
Figure 1. Interaction of NEDD4-2 with Smads.
(A) Schematic representation of Smurf1, Smurf2, NEDD4 (KIAA0093; GenBank® accession number D42055), NEDD4-2 (KIAA0439; GenBank® accession number AB007899) and NEDD4-2(del), which lacks the N-terminal 395 amino acids of NEDD4-2. (B and C) Interaction of NEDD4-2(CA) (B) or NEDD4(CA) (C) with various Smads in transfected cells. COS7 cells were transfected with the indicated plasmids, and lysates from cells were subjected to immunoprecipitation (IP) with anti-FLAG antibody (M2; Sigma), followed by immunoblotting (Blot) using anti-Myc antibody (9E10). The top panels show the interaction, and the bottom three panels the expression, of each protein as indicated. c.a.TβR-I and c.a.BMPR-IB denote constitutively active TGF-β type I receptor and constitutively active BMP type IB receptor respectively. Experiments were repeated three times (B and C), with essentially the same results. (D) Comparison of the binding of Smad7 to NEDD4-2(CA), Smurf2(CA) and NEDD4(CA). COS7 cells were transfected with 6Myc-tagged Smad7 and FLAG-tagged E3 ligases, and lysates from cells were subjected to immunoprecipitation (IP) with anti-FLAG antibody, followed by anti-Myc immunoblotting (Blot). The top panel shows the interaction, and the bottom two panels show the expression of each protein, as indicated. Experiments were repeated three times with essentially the same results.
Because most Smads, with the exception of Smads 4 and 8, have a PY motif in their linker regions, we examined the interaction of NEDD4-2 with other Smads in transfected COS7 cells. The mutant NEDD4-2(CA), in which Cys894 was replaced by alanine, resulting in lack of E3 ubiquitin ligase activity, was used for this study. NEDD4-2(CA) effectively interacted with Smad6 and Smad7 (Figure 1B). Moreover, NEDD4-2(CA) strongly associated with Smads 2 and 3 in the presence of TβR-I(TD). However, NEDD4-2(CA) associated with Smads 1 and 5 less effectively than with Smads 2 and 3, even in the presence of BMPR-IB(QD). NEDD4-2(CA) did not associate with Smad4 or Smad8, since Smad4 and Smad8 do not have the PY motif (results not shown).
We next analysed the interaction of NEDD4 with Smads. A NEDD4 ligase-inactive mutant, NEDD4(CA), failed to interact with any of the Smads examined (Figure 1C). When we compared the binding of NEDD4-2 to Smad7 with that of Smurf2 or NEDD4, NEDD4-2(CA), but not NEDD4(CA), was found to associate with Smad7 as strongly as Smurf2(CA) (Figure 1D).
NEDD4-2 induces translocation of Smad7 from the nucleus to the cytoplasm
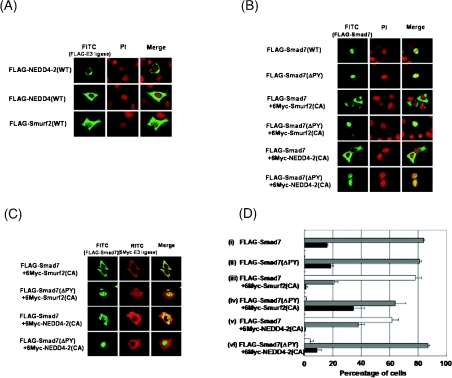
We next examined the effect of NEDD4-2 on the subcellular localization of Smad7 using immunohistochemical staining. When transfected alone into HepG2 cells, Smurf2 was mainly detected in the cytoplasm, but also gave weak staining in the nucleus. NEDD4 and NEDD4-2 localized predominantly in the cytoplasm (Figure 2A). In contrast, Smad7 and Smad7(ΔPY), which lacks the PY motif and fails to bind to Smurf2 or NEDD4-2 (results not shown), mainly localized in the nucleus of HepG2 cells (Figures 2B, rows 1 and 2, and 2D, i and ii). However, Smad7 was found to be located mainly in the cytoplasm in the presence of NEDD4-2(CA), like Smurf2(CA) (Figures 2B, rows 3 and 5, and 2D, iii and v) [16]. Consistent with this result was the observation that Smurf2(CA) and NEDD4-2(CA) co-localized with Smad7 in the cytoplasm (Figure 2C, rows 1 and 3). Moreover, Smurf2(CA) and NEDD4-2(CA) did not affect the subcellular localization of Smad7(ΔPY) (Figures 2B, rows 4 and 6, 2C, rows 2 and 4, and 2D, iv and vi), since Smurf2(CA) and NEDD4-2(CA) failed to interact with Smad7(ΔPY). Similar results were obtained in COS7 and HEK-293T cells (results not shown). These results suggest that NEDD4-2 induces translocation of Smad7 from the nucleus to the cytoplasm.
Figure 2. NEDD4-2 recruits Smad7 from the nucleus to the cytoplasm.
(A) Subcellular localization of wild-type (WT) Smurf2, NEDD4-2 and NEDD4 was investigated in transfected cells. HepG2 cells were transiently transfected with FLAG-tagged Smurf2, NEDD4-2 and NEDD4. Cells were fixed and stained as described in the Materials and methods section. Staining for FLAG-tagged proteins (FITC, green) and nuclear staining by propidium iodide (PI, red) was performed. (B–D) NEDD4-2(CA) recruits Smad7 from nucleus to cytoplasm. Subcellular localization of Smad7 in the presence or absence of NEDD4-2(CA) or Smurf2(CA) was analysed. In (B), HepG2 cells were transfected with FLAG–Smad7 or FLAG–Smad7(ΔPY) alone (top two panels), or together with 6Myc-Smurf2(CA) or 6Myc-NEDD4-2(CA) (bottom four panels). Staining for Smad7(WT) or Smad7(ΔPY) (FITC, green), and nuclear staining by propidium iodide (PI, red) was performed. In (C), HepG2 cells were transiently transfected with various plasmids, as indicated. Anti-FLAG staining for Smad7 or Smad7(ΔPY) (FITC, green), and anti-Myc staining for Smurf2(CA) or NEDD4-2(CA) [RITC (rhodamine isothiocyanate), red] was performed. (D) The distribution of Smad7 or Smad7(ΔPY) in transfected cells was scored as nucleus (black bar), nucleus and cytoplasm (grey bar), or cytoplasm (white bar). More than 300 cells were counted per experiment. Experiments were repeated three times with essentially the same results.
NEDD4-2 binds to TβR-I and induces its ubiquitin-mediated degradation
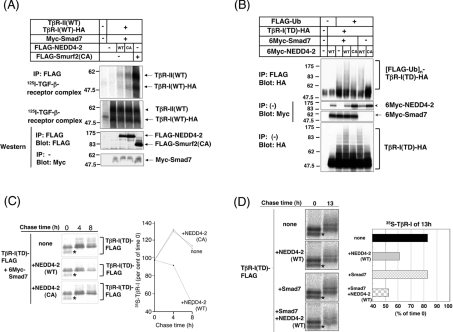
Previous studies have shown that Smurfs 1 and 2 interact with TβR-I via Smad7, and induce ubiquitin-mediated degradation of the receptor [16,17]. To investigate whether NEDD4-2 also binds to and degrades TβR-I, we first performed an affinity cross-linking assay in transfected COS7 cells. [125I]TGF-β cross-linked receptor complexes were not co-precipitated with NEDD4-2(CA) in the absence of Smad7 (results not shown). However, in the presence of Smad7, they were co-precipitated with NEDD4-2(CA), and weakly with NEDD4-2(WT) (Figure 3A). The binding of NEDD4-2(CA) to TβR-I was less efficient than that of Smurf2(CA). Next, we tested ubiquitination of TβR-I by the Smad7–NEDD4-2 complex. Ubiquitination of TβR-I(TD) was induced by NEDD4-2(WT), but not by NEDD4-2(CA), and this was enhanced by the presence of Smad7 (Figure 3B). In Figure 3(C), we show analysis of the degradation of TβR-I(TD) by the Smad7–NEDD4-2 complex using pulse–chase analysis. FLAG-tagged TβR-I(TD) proteins were observed as two differentially migrating bands. Because membrane receptors are post-translationally modified by the addition of N-linked oligosaccharides, the slowly migrating bands most likely represent a mature form of TβR-I(TD), whereas the rapidly migrating bands (asterisks in Figures 3C and 3D) represent its immature form. In the presence of Smad7, NEDD4-2(WT), but not NEDD4-2(CA), induced degradation of TβR-I(TD) (Figure 3C). We also examined the effect of Smad7 on turnover of TβR-I(TD) by NEDD4-2(WT). Smad7 alone did not accelerate degradation of TβR-I(TD); however, Smad7 enhanced degradation of TβR-I(TD) by NEDD4-2 (Figure 3D). These data suggest that NEDD4-2 interacts with TβR-I via Smad7, and induces ubiquitination and degradation of the receptor.
Figure 3. NEDD4-2 interacts with TβR-I, and induces its ubiquitination and degradation.
(A) Association of NEDD4-2 with TβR-II–TβR-I complex via Smad7. COS7 cells were transfected with the plasmids indicated. Cells were affinity-labelled with [125I]TGF-β1 and cross-linked, and immunoprecipitation (IP) with anti-FLAG antibody was performed using cell lysates. Immune complexes were subjected to SDS/PAGE, and co-precipitated receptor complexes (top panel) and cell-surface receptors (second panel) were analysed using a Fuji BAS 2500 Bio-imaging Analyzer (Fuji Photo Film). The bottom two panels show the expression of each protein as indicated by immunoblotting. Experiments were repeated twice with essentially the same results. (B) NEDD4-2 induces ubiquitination of constitutively active TβR-I, TβR-I(TD). HEK-293T cells were transfected with the plasmids indicated, and treated with 10 μM lactacystin for 6 h before cell lysis. Lysates from cells were subjected to immunoprecipitation (IP) with anti-FLAG antibody, followed by haemagglutinin (HA) immunoblotting (Blot). The top panel shows ubiquitination of the receptor, and the bottom three panels show the expression of each protein, as indicated. Experiments were repeated twice with essentially the same results. (C) NEDD4-2(WT), but not NEDD4-2(CA), induces degradation of TβR-I(TD). COS7 cells were transfected with TβR-I(TD)–FLAG and 6Myc–Smad7, with or without 6Myc–NEDD4-2(WT) or 6Myc–NEDD4-2(CA). Cells were pulse-labelled for 10 min with [35S]methionine and [35S]cysteine, and then chased for the indicated periods in medium containing unlabelled methionine and cysteine. Immunoprecipitation with anti-FLAG antibody was performed using total cell lysates. Immune complexes were subjected to SDS/PAGE and analysis using a Fuji BAS 2500 Bio-imaging Analyzer. Asterisks indicate immature forms of TβR-I(TD)-FLAG. 35S-labelled TβR-I(TD)–FLAG is quantified and plotted at each time point as the percentage of amount present at zero time. Experiments were repeated three times with essentially the same results. (D) Smad7 enhances degradation of TβR-I(TD) by NEDD4-2. COS7 cells were transfected with 6Myc–NEDD4-2(WT), 6Myc–Smad7, or both. Pulse–chase analysis was performed as in (C). Asterisks indicate immature forms of TβR-I(TD)–FLAG. Experiments were repeated twice with essentially the same results.
NEDD4-2 induces ubiquitination and degradation of Smad2
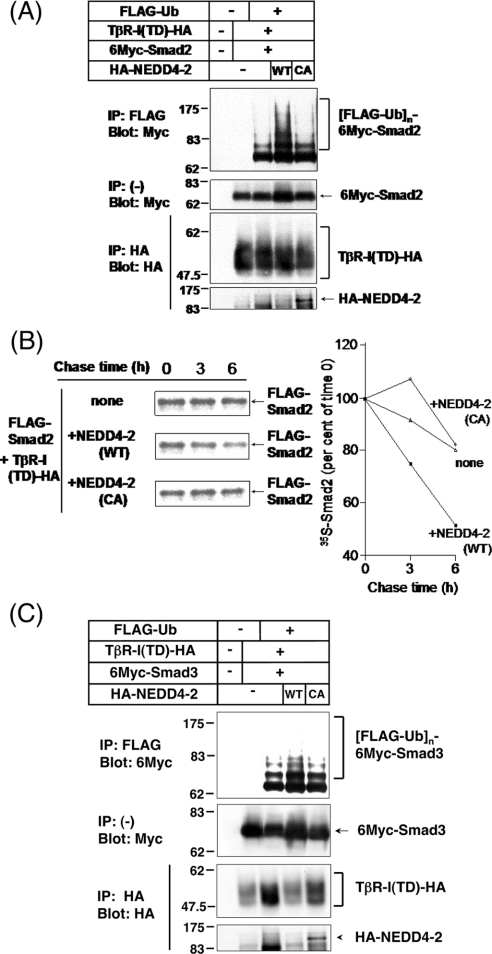
Since NEDD4-2 associates not only with I-Smads, but also with TGF-β-activated Smads 2 and 3 (Figure 1B), and since Smurf2 acts as an E3 ligase for Smad2, we next examined the effect of NEDD4-2 on the degradation of Smads 2 and 3. We first examined the effect of NEDD4-2 on ubiquitination of Smad2 in transfected HEK-293T cells. In the presence of TβR-I(TD), NEDD4-2(WT), but not NEDD4-2(CA), strongly induced ubiquitination of Smad2 (Figure 4A). We next analysed degradation of Smad2 by NEDD4-2 using a pulse–chase assay. NEDD4-2, but not NEDD4-2(CA), induced degradation of Smad2 in the presence of TβR-I(TD) (Figure 4B). Approx. 50% of the Smad2 protein was degraded after 6 h. These results suggest that NEDD4-2 associates with Smad2 in a TGF-β signal-dependent fashion, and induces ubiquitination and degradation of Smad2.
Figure 4. NEDD4-2 induces ubiquitin-mediated degradation of Smad2, but only inefficient ubiquitination of Smad3.
(A) NEDD4-2 induces ubiquitination of Smad2. HEK-293T cells were transfected with the plasmids indicated and treated with 10 μM lactacystin for 6 h before cell lysis. Lysates from cells were subjected to immunoprecipitation (IP) with anti-FLAG antibody followed by Myc-immunoblotting (Blot). The top panel shows ubiquitination of Smad2, and the bottom three panels show the expression of each protein, as indicated. Experiments were repeated three times with essentially the same results. Ub, ubiqutin. (B) NEDD4-2 induces degradation of Smad2. COS7 cells were transfected with FLAG–Smad2 and TβR-I(TD)–HA (where HA is haemagglutinin), with or without 6Myc–NEDD4-2(WT) or 6Myc–NEDD4-2(CA). Cells were pulse-labelled for 10 min with [35S]methionine and [35S]cysteine, and then chased for the indicated periods in medium containing unlabelled methionine and cysteine. Immunoprecipitation with anti-FLAG antibody was performed using total cell lysates. Immune complexes were subjected to SDS/PAGE and analysis using a Fuji BAS 2500 Bio-imaging Analyzer. 35S-labelled FLAG–Smad2 is quantified and plotted at each time point as the percentage of amount present at zero time. Experiments were repeated three times with essentially the same results. (C) NEDD4-2 only slightly induces ubiquitination of Smad3. HEK-293T cells were transfected with the plasmids indicated and treated with 10 μM lactacystin for 6 h before cell lysis. Lysates from cells were subjected to immunoprecipitation (IP) with anti-FLAG antibody followed by Myc-immunoblotting (Blot). Ub, ubiquitin. The top panel shows ubiquitination of Smad3, and the bottom three panels show the expression of each protein as indicated. Experiments were repeated twice with essentially the same results.
We also tested the effect of NEDD4-2 on ubiquitination of Smad3. When Smad3 was expressed together with ubiquitin and TβR-I(TD) in HEK-293T cells, ubiquitinated Smad3 was detected even in the absence of NEDD4-2, but NEDD4-2 only slightly enhanced ubiquitination of Smad3 (Figure 4C). These results suggested that NEDD4-2 acts as an E3 ligase for Smad2, but not as an efficient ligase for Smad3, although NEDD4-2 interacts with both Smad2 and Smad3.
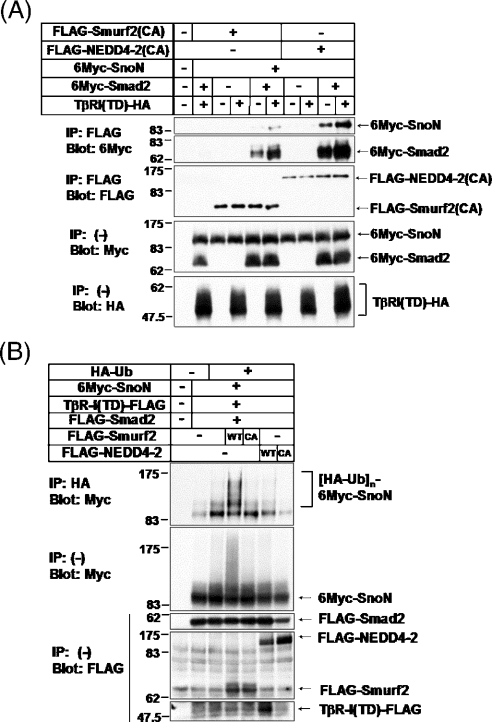
NEDD4-2 fails to induce ubiquitination of SnoN
Previous study has shown that Smurf2 associates with SnoN via Smad2 and induces ubiquitin-mediated degradation of SnoN [21]. We therefore investigated whether NEDD4-2 acts as an E3 ligase for SnoN. We first tested interaction of NEDD4-2 with SnoN via Smad2. NEDD4-2(CA) bound to SnoN in the presence of Smad2, and the interaction was enhanced in the presence of TβR-I(TD) (Figure 5A). It is important to note that NEDD4-2(CA) interacted with SnoN much more strongly than Smurf2(CA) did. We next examined whether NEDD4-2 induces ubiquitination of SnoN. Smurf2 strongly induced ubiquitination of SnoN, whereas NEDD4-2 failed to do so (Figure 5B). These results suggest that, in contrast with Smurf2, NEDD4-2 does not affect the degradation of SnoN, whereas the interaction of NEDD4-2 with SnoN is stronger than that of Smurf2; however, the possibility that the use of various cell lines might contribute to this apparent discrepancy cannot be excluded.
Figure 5. NEDD4-2 interacts with SnoN via Smad2, but fails to induce ubiquitination of SnoN.
(A) Binding of NEDD4-2(CA) to SnoN is stronger than that of Smurf2(CA). COS7 cells were transfected with the plasmids indicated. Cell lysates were subjected to immunoprecipitation (IP) with anti-FLAG antibody, followed by anti-Myc immunoblotting (Blot). The top two panels show the interaction, and the bottom three panels show the expression, of each protein, as indicated. Experiments were repeated twice with essentially the same results. (B) NEDD4-2 fails to induce ubiquitination of SnoN. HEK-293T cells were transfected with indicated plasmids. Cell lysates were subjected to immunoprecipitation (IP) with anti-HA (haemagglutinin) 12CA5 followed by anti-Myc immunoblotting (Blot). Ub, ubiquitin. The top panel shows ubiquitination of SnoN, and the bottom four panels show the expression of each protein as indicated. Experiments were repeated twice with essentially the same results.
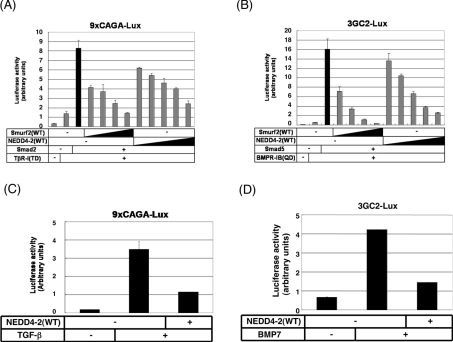
NEDD4-2 inhibits transcriptional activity induced by both TGF-β and BMP
To examine the function of NEDD4-2 in TGF-β superfamily signalling, we performed reporter assays using a TGF-β-responsive reporter construct, 9×CAGA-lux, and a BMP-responsive reporter construct, 3GC2-lux, in transfected HepG2 cells. As shown in Figure 4, NEDD4-2 causes ubiquitin-dependent degradation of Smad2, but not that of Smad3. We thus focused the effect of NEDD4-2 on Smad2-dependent transcription. NEDD4-2(WT) inhibited transcriptional activity induced by TβR-I(TD) and Smad2 in a dose-dependent manner (Figure 6A). Moreover, it inhibited transcriptional activity induced by BMPR-IB(QD) and Smad5 (Figure 6B) as well. NEDD4-2(WT) also repressed transcriptional activity induced by TGF-β and BMP7 (Figures 6C and 6D) in 293 cells. Similar results were obtained using HepG2 cells and HeLa cells.
Figure 6. NEDD4-2 inhibits transcriptional activity induced by both TGF-β and BMP.
(A and B) NEDD4-2 inhibits transcriptional activity induced by TβR-I(TD) (A) or BMPR-IB(QD) (B) in a dose-dependent manner. HepG2 cells were co-transfected with a 9×CAGA-lux construct (A) or a 3GC2-lux construct (B), and the plasmids indicated. Smurf2 cDNA was transfected using amounts of 0.05, 0.1, 0.2 and 0.4 μg. NEDD4-2 was transfected using amounts of 0.075, 0.15, 0.3, 0.6 and 1.2 μg. The expression level of Smurf2 was 3-fold higher than NEDD4-2 (results not shown). Experiments were repeated three times with essentially the same results. (C and D) NEDD4-2 inhibits transcriptional activity induced by TGF-β (C) or BMP7 (D). HEK-293 cells were transfected with a 9×CAGA-lux construct (C) or a 3GC2-lux construct with FLAG–Smad5 (D) and the plasmids indicated. Cells were treated with TGF-β (1 ng/ml) (C) or BMP7 (500 ng/ml) (D) for 24 h, and luciferase activities were measured. Experiments were repeated twice with essentially the same results.
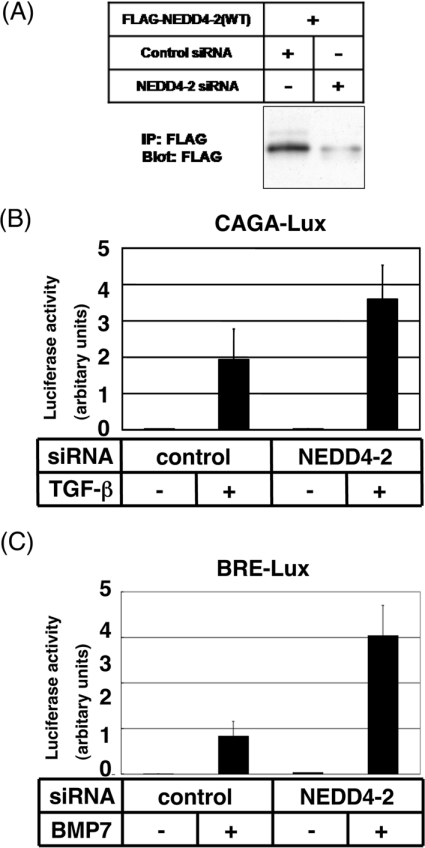
We next knocked-down the expression of endogenous NEDD4-2 protein by siRNAs. NEDD4-2 siRNA decreased expression levels of NEDD4-2 protein in transfected HEK-293 cells (Figure 7A). Interestingly, silencing of the NEDD4-2 gene resulted in enhancement of the transcriptional activity induced by TGF-β (Figure 7B) and BMP7 (Figure 7C) in HepG2 cells. These results suggest that endogenous NEDD4-2 in mammalian cells plays a role in repression of signalling by TGF-β superfamily cytokines.
Figure 7. Decreased expression level of NEDD4-2 results in enhancement of TGF-β and BMP signalling.
(A) Effects of the control and NEDD4-2 siRNAs on the expression of NEDD4-2 proteins were determined by immunoblotting (Blot) in transfected HEK-293 cells. IP, immunoprecipitation. (B and C) Enhancement of transcriptional activity induced by TGF-β (B) or BMP7 (C) in the presence of siRNA for NEDD4-2. HepG2 cells were transfected with the control or NEDD4-2 siRNAs, and a TGF-β-responsive 9×CAGA-lux (B) construct or a BRE-lux (BMP-responsive element-lux) construct (C) as indicated. Cells were treated with TGF-β (1 ng/ml) (B) or BMP7 (500 ng/ml) (C) for 24 h, and luciferase activities were measured. Experiments were repeated twice with essentially the same results.
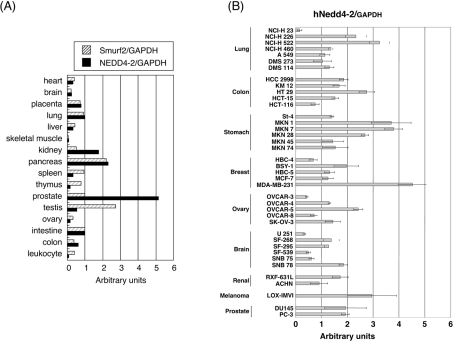
Expression of NEDD4-2 mRNA in human tissues and human carcinoma cell lines
We determined the expression levels of NEDD4-2 mRNA in various human tissues, and compared them with those of Smurf2 (Figure 8A). Among the tissues analysed, the expression of NEDD4-2 and Smurf2 showed similar levels in eight cases out of 16. On the other hand, high expression of NEDD4-2 was observed in kidney and pancreas, and the highest expression in prostate. In contrast, high expression of Smurf2 mRNA was found in pancreas and testis. These results suggest that NEDD4-2 and Smurf2 are expressed in distinct patterns in some human tissues.
Figure 8. Expression of NEDD4-2 mRNAs in human tissues and human cancer cell lines.
(A and B) Quantitative real-time PCR analyses were performed using various human tissues (A) and human cancer cell lines (B). Expression levels of mRNA for NEDD4-2 and/or Smurf2 were examined, and normalized to the amounts of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA, and results are given in arbitrary units.
We also determined the expression levels of NEDD4-2 mRNA in human carcinoma cell lines (Figure 8B). NEDD4-2 expression was up-regulated in several human carcinoma cell lines including lung cancer cell lines, NCI-H 226 and NCI-H 522, colon cancer cell line, HT29, gastric cancer cell lines, MKN-1, MKN-7 and MKN-28, breast cancer cell line, MDA-MB-231, ovary cancer cell line, OVCAR-5, and melanoma cell line, LOX-IMVI. Among these cancer cell lines, HT29, MKN-1 and MKN-28 have been reported to acquire TGF-β-resistance [36–38].
DISCUSSION
Recently, several reports have demonstrated that TGF-β superfamily signalling is regulated by HECT type E3 ubiquitin ligases [13–17]. Smurf1 induces ubiquitination and degradation of Smads 1 and 5, and of TβR-I and BMPR-Is through interacting I-Smads [13,17,39]. Smurf2 induces ubiquitin-mediated degradation of Smad1 and TβR-I; moreover, it has also been shown to induce degradation of Smad2 and SnoN [14–16,21].
In the present study, using yeast two-hybrid screening, we have identified NEDD4-2 as a new Smad7-binding partner that regulates TGF-β superfamily signalling. Like Smurfs, NEDD4-2 interacts with TβR-I through Smad7 and induces ubiquitin-dependent degradation of the receptor. In addition, NEDD4-2 interacts with Smad2 and induces its ubiquitin-meditated degradation. In contrast with Smurf2, however, NEDD4-2 failed to induce ubiquitination of transcriptional co-repressor SnoN, although it efficiently bound to SnoN via Smad2. Consistent with these results, NEDD4-2 prevented transcriptional activity of TGF-β signalling.
Like Smurfs, NEDD4-2 contains a C2 domain at the N-terminus, WW domains in the middle and a HECT domain at the C-terminus. NEDD4-2 interacts not only with Smad7, but also with other Smads, including Smads 2 and 3, since these Smad proteins have the PY motif that binds to the WW domain. Notably, although NEDD4-2 is most similar to NEDD4 of the mammalian HECT type E3 ligases (60% identity) and both proteins have been reported to down-regulate the activity of epithelial sodium channel, NEDD4-2, but not NEDD4, affects TGF-β superfamily signalling. Moreover, although NEDD4-2 binds to both Smad2 and Smad3, which are structurally similar to each other (92% identity), NEDD4-2 induces ubiquitin-dependent degradation of Smad2, but does not efficiently induce that of Smad3. We have reported previously that ROC1–SCFFbw1a is the ubiquitin E3 ligase for Smad3 in TGF-β signalling [11]. Thus different types of E3 ubiquitin ligases, e.g. HECT-type E3 ligases and RING-type E3 ligases, might play distinct roles in TGF-β signalling.
Although the function of NEDD4-2 resembles that of Smurf2, there are some differences between them. In contrast with the efficient binding of Smurf2 to BMP-specific R-Smads, NEDD4-2 only weakly interacts with Smads 1 and 5, and does not affect their turnover (results not shown). However, NEDD4-2 represses BMP signalling, possibly through degradation of BMP type I receptors via I-Smads (Figure 6B and D).
SnoN is a transcriptional co-repressor that interacts with Smad2/3 and Smad4, and represses TGF-β signalling through recruitment of HDAC (histone deacetylase) to Smads [40–42]. Bonni et al. [21] have reported that Smurf2 associates with SnoN via Smad2, and induces ubiquitin-mediated degradation of SnoN, but not that of Smad2. Thus Smurf2 is thought to enhance TGF-β signals under certain conditions. However, we showed that NEDD4-2 failed to induce ubiquitin-mediated degradation of SnoN (Figure 5B), although NEDD4-2 bound strongly to SnoN via Smad2. These data suggest that NEDD4-2 functions through similar, but not identical, mechanisms to Smurf2.
In conclusion, we identified NEDD4-2 as a new member of the Smurf-like C2-WW-HECT-type E3 ubiquitin ligases that negatively regulates TGF-β superfamily signalling. Like Smurf2, NEDD4-2 induces ubiquitin-dependent degradation of Smad2 and TβR-I, whereas, in contrast with Smurf2, NEDD4-2 fails to degrade SnoN. Overexpressed NEDD4-2 suppresses transcriptional activities induced by the TGF-β superfamily members, whereas silencing of the NEDD4-2 gene by siRNA for NEDD4-2 results in enhancement of signalling by TGF-β and BMP. Notably, expression profiles of NEDD4-2 and Smurf2 are distinct in some human tissues. Moreover, NEDD4-2 is overexpressed in several carcinoma cell lines, including TGF-β-resistant HT-29 colon cancer cell line and MKN-1/-28 gastric cancer cell lines [36–38]. Overexpression of NEDD4-2 might result in resistance to growth inhibition by TGF-β. It will be important to determine in the future how the degradation of R-Smads and TβR-I by NEDD4-2 is regulated under physiological and pathological conditions.
Acknowledgments
We thank X.-H. Feng for Smurf2 cDNA and N. Kusuhara for KIAA0439 cDNA. We are grateful to Yuri Inada and Aki Hanyu for technical help. This study was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sport, Science and Technology of Japan. This work was also supported by Nippon Boehringer Ingelheim (VRIA) and the Viral Hepatitis Research Foundation of Japan.
References
- 1.Roberts A. B., Sporn M. B. The transforming growth factor-βs. In: Sporn M. B., Roberts A. B., editors. Peptide Growth Factors and Their Receptors, Part I. Heidelberg: Springer-Verlag; 1990. pp. 419–472. [Google Scholar]
- 2.Heldin C.-H., Miyazono K., ten Dijke P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 3.Massagué J. TGF-β signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 4.Derynck R., Zhang Y., Feng X.-H. Smads: transcriptional activators of TGF-β responses. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 5.Attisano L., Wrana J. L. Smads as transcriptional co-modulators. Curr. Opin. Cell Biol. 2000;12:235–243. doi: 10.1016/s0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- 6.Imamura T., Takase M., Nishihara A., Oeda E., Hanai J., Kawabata M., Miyazono K. Smad6 inhibits signalling by the TGF-β superfamily. Nature (London) 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 7.Hanyu A., Ishidou Y., Ebisawa T., Shimanuki T., Imamura T., Miyazono K. The N domain of Smad7 is essential for specific inhibition of transforming growth factor-β signaling. J. Cell Biol. 2001;155:1017–1028. doi: 10.1083/jcb.200106023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 9.Jesenberger V., Jentsch S. Deadly encounter: ubiquitin meets apoptosis. Nat. Rev. Mol. Cell Biol. 2002;3:112–121. doi: 10.1038/nrm731. [DOI] [PubMed] [Google Scholar]
- 10.Laney J. D., Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 11.Fukuchi M., Imamura T., Chiba T., Ebisawa T., Kawabata M., Tanaka K., Miyazono K. Ligand-dependent degradation of Smad3 by a ubiquitin ligase complex of ROC1 and associated proteins. Mol. Biol. Cell. 2001;12:1431–1443. doi: 10.1091/mbc.12.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan M., Tang Y., Tytler E. M., Lu C., Jin B., Vickers S. M., Yang L., Shi X., Cao X. Smad4 protein stability is regulated by ubiquitin ligase SCF β-TrCP1. J. Biol. Chem. 2004;279:14484–14484. doi: 10.1074/jbc.C400005200. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H., Kavsak P., Abodollah S., Wrana J. L., Thomsen G. H. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature (London) 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 14.Lin X., Liang M., Feng X.-H. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-β signaling. J. Biol. Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Chang C., Gehling D. J., Hemmati-Brivanlou A., Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF-β receptor for degradation. Mol. Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 17.Ebisawa T., Fukuchi M., Murakami G., Chiba T., Tanaka K., Imamura T., Miyazono K. Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 18.Tajima Y., Goto K., Yoshida M., Shinomiya K., Sekimoto T., Yoneda Y., Miyazono K., Imamura T. Chromosomal region maintenance 1 (CRM1)-dependent nuclear export of Smad ubiquitin regulatory factor 1 (Smurf1) is essential for negative regulation of transforming growth factor-β signaling by Smad7. J. Biol. Chem. 2003;278:10716–10721. doi: 10.1074/jbc.M212663200. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki C., Murakami G., Fukuchi M., Shimanuki T., Shikauchi Y., Imamura T., Miyazono K. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J. Biol. Chem. 2002;277:39919–39925. doi: 10.1074/jbc.M201901200. [DOI] [PubMed] [Google Scholar]
- 20.Kim B. C., Lee H. J., Park S. H., Lee S. R., Karpova T. S., McNally J. G., Felici A., Lee D. K., Kim S. J. Jab1/CSN5, a component of the COP9 signalosome, regulates transforming growth factor β signaling by binding to Smad7 and promoting its degradation. Mol. Cell. Biol. 2004;24:2251–2262. doi: 10.1128/MCB.24.6.2251-2262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonni S., Wang H. R., Causing C. G., Kavsak P., Stroschein S. L., Luo K., Wrana J. L. TGF-β induces assembly of a Smad2–Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat. Cell Biol. 2001;3:587–595. doi: 10.1038/35078562. [DOI] [PubMed] [Google Scholar]
- 22.Koinuma D., Shinozaki M., Komuro A., Goto K., Saitoh M., Hanyu A., Ebina M., Nukiwa T., Miyazawa K., Imamura T., Miyazono K. Arkadia amplifies TGF-β superfamily signalling through degradation of Smad7. EMBO J. 2003;22:6458–6470. doi: 10.1093/emboj/cdg632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinudom A., Harvey B. J., Komwatana P., Young J. A., Kumar S., Cook D. I. Nedd4 mediates control of an epithelial Na+ channel in salivary duct cells by cytosolic Na+ Proc. Natl. Acad. Sci. U.S.A. 1998;95:7169–7173. doi: 10.1073/pnas.95.12.7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goulet C. C., Volk K. A., Adams C. M., Prince L. S., Stokes J. B., Snyder P. M. Inhibition of the epithelial Na+ channel by interaction of Nedd4 with a PY motif deleted in Liddle's syndrome. J. Biol. Chem. 1998;273:30012–30017. doi: 10.1074/jbc.273.45.30012. [DOI] [PubMed] [Google Scholar]
- 25.Abriel H., Kamynina E., Horisberger J.-D., Staub O. Regulation of the cardiac voltage-gated Na+ channel (H1) by the ubiquitin-protein ligase Nedd4. FEBS Lett. 2000;466:377–380. doi: 10.1016/s0014-5793(00)01098-x. [DOI] [PubMed] [Google Scholar]
- 26.Harvey K. F., Dinudom A., Komwatana P., Jolliffe C. N., Day M. L., Parasivam G., Cook D. I., Kumar S. All three WW domains of murine Nedd4 are involved in the regulation of epithelial sodium channels by intracellular Na+ J. Biol. Chem. 1999;274:12525–12530. doi: 10.1074/jbc.274.18.12525. [DOI] [PubMed] [Google Scholar]
- 27.Harvey K. F., Dinudom A., Cook D. I., Kumar S. The Nedd4-like protein KIAA0439 is a potential regulator of the epithelial sodium channel. J. Biol. Chem. 2001;276:8597–8601. doi: 10.1074/jbc.C000906200. [DOI] [PubMed] [Google Scholar]
- 28.Farr T. J., Coddington-Lawson S. J., Snyder P. M., McDonald F. J. Human Nedd4 interacts with the human epithelial Na+ channel: WW3 but not WW1 binds to Na+-channel subunits. Biochem. J. 2000;345:503–509. [PMC free article] [PubMed] [Google Scholar]
- 29.Kamynina E., Debonneville C., Bens M., Vandewalle A., Staub O. A novel mouse Nedd4 protein suppresses the activity of the epithelial Na+ channel. FASEB J. 2001;15:204–214. doi: 10.1096/fj.00-0191com. [DOI] [PubMed] [Google Scholar]
- 30.Kamynina E., Tauxe C., Staub O. Distinct characteristics of two human Nedd4 proteins with respect to epithelial Na+ channel regulation. Am. J. Physiol. Renal Physiol. 2001;281:F469–F477. doi: 10.1152/ajprenal.2001.281.3.F469. [DOI] [PubMed] [Google Scholar]
- 31.Snyder P. M., Olson D. R., McDonald F. J., Bucher D. B. Multiple WW domains, but not the C2 domain, are required for inhibition of the epithelial Na+ channel by human Nedd4. J. Biol. Chem. 2001;276:28321–28326. doi: 10.1074/jbc.M011487200. [DOI] [PubMed] [Google Scholar]
- 32.Kawabata M., Inoue H., Hanyu A., Imamura T., Miyazono K. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. 1998;17:4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebisawa T., Tada K., Kitajima I., Tojo K., Sampath T. K., Kawabata M., Miyazono K., Imamura T. Characterization of bone morphogenetic protein-6 signaling pathways in osteoblast differentiation. J. Cell Sci. 1999;112:3519–3527. doi: 10.1242/jcs.112.20.3519. [DOI] [PubMed] [Google Scholar]
- 34.Frolik C. A., Wakefield L. M., Smith D. M., Sporn M. B. Characterization of a membrane receptor for transforming growth factor-β in normal rat kidney fibroblasts. J. Biol. Chem. 1984;259:10995–11000. [PubMed] [Google Scholar]
- 35.Dan S., Tsunoda T., Kitahara O., Yanagawa R., Zembutsu H., Katagiri T., Yamazaki K., Nakamura Y., Yamori T. An integrated database of chemosensitivity to 55 anticancer drugs and gene expression profiles of 39 human cancer cell lines. Cancer Res. 2002;62:1139–1147. [PubMed] [Google Scholar]
- 36.Li C. Y., Suardet L., Little J. B. Potential role of WAF1/Cip1/p21 as a mediator of TGF-β cytoinhibitory effect. J. Biol. Chem. 1995;270:4971–4974. doi: 10.1074/jbc.270.10.4971. [DOI] [PubMed] [Google Scholar]
- 37.Ito M., Yasui W., Nakayama H., Yokozaki H., Ito H., Tahara E. Reduced levels of transforming growth factor-β type I receptor in human gastric carcinomas. Jpn. J. Cancer Res. 1992;83:86–92. doi: 10.1111/j.1349-7006.1992.tb02356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito M., Yasui W., Kyo E., Yokozaki H., Nakayama H., Ito H., Tahara E. Growth inhibition of transforming growth factor β on human gastric carcinoma cells: receptor and postreceptor signaling. Cancer Res. 1992;52:295–300. [PubMed] [Google Scholar]
- 39.Murakami G., Watabe T., Takaoka K., Miyazono K., Imamura T. Inhibition of BMP signaling by Smurf1 and inhibitory Smads. Mol. Biol. Cell. 2003;14:2809–2817. doi: 10.1091/mbc.E02-07-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nomura T., Khan M. M., Kaul S. C., Dong H.-D., Wadhwa R., Colmenares C., Kohno I., Ishii S. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 1999;13:412–423. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akiyoshi S., Inoue H., Hanai J., Kusanagi K., Nemoto N., Miyazono K., Kawabata M. c-Ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with Smads. J. Biol. Chem. 1999;274:35269–35277. doi: 10.1074/jbc.274.49.35269. [DOI] [PubMed] [Google Scholar]
- 42.Stroschein S. L., Wang W., Zhou S., Zhou Q., Luo K. Negative feedback regulation of TGF-β signaling by the SnoN oncoprotein. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]