Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation (original) (raw)
Abstract
X-chromosome inactivation (XCI) is highly dynamic during early mouse embryogenesis and strictly depends on the Xist noncoding RNA. The regulation of Xist and its antisense partner Tsix remains however poorly understood. We provide here the first evidence of transcriptional control of Xist expression. We show that RNA polymerase II (RNAPolII) preinitiation complex recruitment and H3 Lys 4 (H3-K4) methylation at the Xist promoter form the basis of the Xist expression profiles that drives both imprinted and random XCI. In embryonic stem (ES) cells, which are derived from the inner cell mass where imprinted XCI is reversed and both Xs are active, we show that Xist is repressed at the level of preinitiation complex (PIC) recruitment. We further demonstrate that Tsix, although highly transcribed in ES cells, is not itself responsible for the transcriptional down-regulation of Xist. Rather, Tsix induces efficient H3-K4 methylation over the entire Xist/Tsix unit. We suggest that chromatin remodeling of the Xist locus induced by biallelic Tsix transcription renders both Xist loci epigenetically equivalent and equally competent for transcription. In this model, Tsix, by resetting the epigenetic state of the Xist/Tsix locus, mediates the transition from imprinted to random XCI.
Keywords: X-chromosome inactivation, histone methylation, antisense transcription, preinitiation complex of transcription, noncoding RNAs
X-chromosome inactivation (XCI), the transcriptional silencing of one X chromosome in female mammals, is dynamically regulated during early development. In mice, X-inactivation is first imprinted in all the cells of the cleavage-stage embryo, with exclusive inactivation of the paternal X-chromosome (Huynh and Lee 2003; Mak et al. 2004; Okamoto et al. 2004). This initial form of imprinted X-inactivation is labile, however, and at the blastocyst stage, the paternal X is reactivated in cells of the inner cell mass (ICM) (Mak et al. 2004; Okamoto et al. 2004). This results in both X chromosomes being active during a short time window. Subsequently, random X-inactivation is initiated, and either the paternal or the maternal X can be chosen for inactivation. In contrast, X-inactivation remains imprinted in extra-embryonic tissues (Takagi and Sasaki 1975). The remarkable and initially unsuspected plasticity of XCI during early development implies the resetting, at the implantation stage, of the epigenotype of both Xs to confer on them an equal probability of inactivation.
The initiation of X-inactivation is controlled by the X-inactivation center (Xic), a complex X-linked locus responsible for the counting and choice mechanisms that ensures the inactivation of a single X in female cells and an absence of inactivation in male cells (for review, see Rougeulle and Avner 2003). The Xist gene, which lies within the Xic, produces an essential noncoding RNA that coats the X chromosome (Clemson et al. 1996) and triggers gene silencing in cis (Penny et al. 1996; Marahrens et al. 1997). Xist expression must be tightly regulated in order to ensure the dynamics of XCI, as only high levels of Xist RNA can induce X-inactivation. Indeed, in preimplantation embryos, Xist expression is imprinted and high Xist RNA levels are produced from the paternal X only (Kay et al. 1994). Drastic changes in Xist expression pattern take place in the ICM, where Xist becomes biallelically expressed. At this stage, Xist expression is strongly down-regulated, presumably to avoid the coating and inactivation of both X chromosomes. At the onset of random XCI, Xist is up-regulated specifically on the future inactive X (Borsani et al. 1991), irrespective of its parental origin. Two aspects of Xist regulation, the levels of Xist RNA and its chromosomal origin, appear therefore to be crucial for X-inactivation.
Very little is known about the transcriptional control of Xist expression, and current models for the up-regulation of Xist during random X-inactivation are centered on the stabilization of the transcript (Panning et al. 1997; Sheardown et al. 1997). Xist transcription is thought to initiate from two distinct promoters, P1 and P2 (Johnston et al. 1998), separated by 1.5 kb (P2 being downstream of P1). The functional significance of these two isoforms is related to the existence of a repeat element lying between both promoters that is essential for the silencing activity of Xist RNA (Wutz et al. 2002). Transcribing Xist from P1 rather than from P2 would be expected to radically change the silencing properties of the Xist RNA. Somewhat surprisingly, it was shown that transcripts initiating from P2, and therefore devoid of the repeat element, are in large excess compared to those initiating from P1. This has led to the suggestion that P2 is the major Xist promoter (Johnston et al. 1998). However, the activity of the P1 and/or P2 promoters may change in the course of imprinted and/or random X-inactivation, and the possibility that the regulation of Xist expression is mediated by transcriptional mechanisms remains open.
One of the most exciting characteristics of the Xist gene is its complete overlapping by a noncoding antisense transcription unit, Tsix (Debrand et al. 1999; Lee et al. 1999; Mise et al. 1999). Tsix has been shown to be a crucial _cis_-acting repressor of Xist up-regulation, and a tight correlation between antisense down-regulation and Xist RNA accumulation during random X-inactivation was established (for review, see Rougeulle and Avner 2004). Importantly, both deletion of the major Tsix promoter and truncation of the antisense transcription unit lead to a complete bias in random X-inactivation towards the mutated allele (Debrand et al. 1999; Lee and Lu 1999; Luikenhuis et al. 2001). This strongly suggests that Tsix is involved in the chromosomal origin of Xist expression during random X-inactivation. Interestingly, the phenotype of Tsix deletions cannot be reverted by inserting back a cDNA expressing a spliced variant of Tsix (Shibata and Lee 2004). This points to the possibility that the act of Tsix transcription itself is required for the function of Tsix. Whether Tsix modulates Xist transcription or acts at the level of Xist RNA stabilization or both remains an open question, however. It has also been proposed that Tsix is responsible for the paternal-restricted expression of Xist during imprinted X-inactivation, although the role of Tsix in the early, reversible imprinted Xist expression has yet to be clearly addressed (Lee 2000; Sado et al. 2001).
Histone modifications are involved in the epigenetic regulation of gene expression (Sims et al. 2003). By reconfiguring the chromatin template and establishing a local chromatin structure, such epigenetic modifications make promoter regions permissive or not for the subsequent assembly of the preinitiation complex (PIC), composed of RNA polymerase II (RNAPolII) and the general transcription factors, and/or for the activation of transcription (Lee and Young 2000). Interestingly, differential histone modifications are associated with the patterning of expression of several imprinted genes (Xin et al. 2001; Fournier et al. 2002; Lewis et al. 2004; Umlauf et al. 2004; Vu et al. 2004). In these cases, the paternal and maternal alleles are distinguished by selective enrichment of the promoter region by a set of epigenetic marks associated with either active (e.g., methylation of H3 Lys 4 [H3-K4]) or silent (e.g., H3-K9 and K27 methylation) chromatin. Although some data exist concerning the chromatin features of the Xist locus (Kimura et al. 2002; Rougeulle et al. 2003, 2004), a more systematic approach is clearly needed to assess the role of histone modifications in the regulation of Xist expression.
To gain insight into the epigenetic and transcriptional aspects of Xist regulation, we focused on the chromatin structure of the locus in relation to PIC recruitment at the Xist and Tsix promoters. Our detailed chromatin immunoprecipitation (ChIP) analysis exploited three cell types representative of different X-inactivation status. Trophoblast stem (TS) cells (Tanaka et al. 1998) carry an inactive X chromosome of paternal origin and can be used as a model system to study imprinted X-inactivation (Mak et al. 2002). In TS cells, Xist expression is restricted to the paternal allele, whereas Tsix is expressed from the maternal one (S. Pichard, D. Arnaud, C. Chureau, E. Heard, C. Ciaudo, C. Rougeulle, and P. Avner, in prep.). Mouse embryonic fibroblasts (MEFs) have already undergone random X-inactivation and can be used to analyze stable X-inactivation as established in somatic cells. In female MEFs, Xist is expressed only from the inactive X, while both Tsix alleles are silenced. Finally, embryonic stem (ES) cells, derived from the ICM of the blastocyst where imprinted X-inactivation is reversed and both Xs active, have been used extensively to study the initiation of random X-inactivation. In ES cells, both Xist and Tsix are biallelically expressed, although Tsix transcripts are in large excess compared to Xist transcripts (Shibata and Lee 2003).
Our analysis provides the first evidence of transcriptional control of Xist. We show that the strong monoallelic Xist expression required to achieve X-inactivation, whether imprinted or random, is driven by monoallelic recruitment of the transcriptional apparatus and H3-K4 dimethylation to the Xist P1 promoter. Conversely, in ES cells, the essential down-regulation of Xist expression appears in a large part to be mediated through the repression of PIC recruitment to the Xist P1 promoter. In contrast to current models, we demonstrate that the function of Tsix in ES cells is not to directly repress Xist transcription but rather to induce H3-K4 methylation over the entire Xist/Tsix locus. We propose that the role of such chromatin remodeling activity is to erase pre-established epigenetic marks associated with imprinted X-inactivation, in order to render both Xist homologs epigenetically equivalent and equally competent for random X-inactivation. This provides a new role for Tsix in the transition from imprinted to random X-inactivation.
Results
Monoallelic Xist expression is marked by allelic H3-K4 dimethylation but not by H3-K9 and K27 methylation
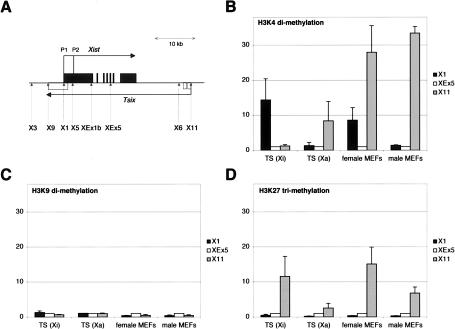
In female TS cells, Xist is expressed from the paternal, inactive X (Mak et al. 2002), whereas Tsix expression comes from the maternal, active X (S. Pichard, D. Arnaud, C. Chureau, E. Heard, C. Ciaudo, C. Rougeulle, and P. Avner, in prep.). To address whether methylation of H3-K4 and/or H3-K9 and H3-K27 over the promoters is involved in the allelic expression of both Xist and Tsix in imprinted X-inactivation, we used a female TS cell line carrying a paternal X chromosome of 129/Sv origin and a maternal X chromosome whose Xic region is derived from a Mus musculus musculus strain that carries the Pgk.1a allele. We exploited single nucleotide polymorphisms (SNPs) to distinguish the paternal (inactive) and maternal (active) X in the ChIP assay. Two allelic primers pairs were successfully designed for the Xist P1 promoter (position X1) and the Tsix promoter (position X11), respectively (Fig. 1A). As shown in Figure 1B, promoters of expressed Xist and Tsix alleles on the paternal and maternal X chromosomes respectively are enriched for H3-K4 dimethylation. However, and in contrast to other imprinted genes, the silent maternal Xist allele is not found enriched for either H3-K9 dimethylation or H3-K27 trimethylation, compared with the active one (Fig. 1C,D). The silent, paternal Tsix allele displays significant enrichment for H3-K27 trimethylation, but not for H3-K9 dimethylation.
Figure 1.
ChIP analysis of histone modifications within the Xist/Tsix region in female TS cells and in MEFs. (A) Schematic representation of the Xist/Tsix locus. Xist exons are represented by black boxes, and Tsix exons are represented by white boxes. Dotted arrows indicate the positions of the primer pairs used in the ChIP analysis. (B-D) Analysis of H3-K4 dimethylation (B), H3-K9 dimethylation (C), and H3-K27 trimethylation (D) in female TS cells and MEFs. In TS cells, two allelic primers (X1, black bars, and X11, gray bars) allowed us to distinguish the paternal allele (Xi) from the maternal (Xa). White bars (XEx5) correspond to a primer pair detecting both the paternal and maternal alleles. As both X chromosomes in female MEFs are of 129/Sv origin, they cannot be distinguished using allelic primers. All graphs were obtained after normalization with position XEx5 located in the body of Xist.
A similar pattern of histone modifications at the Xist promoter is detected in cells displaying random XCI, such as MEFs. Enrichment for H3-K4 dimethylation over the Xist promoter is specific to female cells compared with males (which do not express Xist as they have a single active X chromosome) (Fig. 1B). Similarly to TS cells, the silent Xist promoter in male MEFs is enriched neither for H3-K9 dinor for H3-K27 trimethylation (Fig. 1C,D). Promoter-restricted H3-K4 dimethylation therefore appears to be the epigenetic mark for monoallelic Xist expression in both imprinted and random X-inactivation. Importantly, neither H3-K9 dimethylation nor H3-K27 trimethylation is specifically associated with the silent Xist allele on the active X chromosome. It is formally possible, however, that other, still unidentified repressive modifications mark the silent Xist promoter.
Intriguingly, as previously reported by others (Kimura et al. 2002), the promoter region of Tsix is enriched for H3-K4 dimethylation in MEFs despite the fact that Tsix is not expressed in such cells (Fig. 1B). The function of such enrichment remains elusive but might be related to the choice process occurring at the onset of random X-inactivation, in which Tsix is known to be involved (Lee et al. 1999; for review, see Rougeulle and Avner 2004). Alternatively, the function of H3-K4 methylation at the Tsix promoter could be related to the counting process, i.e., the mechanism that ensures the inactivation of the appropriate number of X chromosomes in each cell. Recently, Morey et al. (2004) showed that a counting element lies within a 20-kb region that surrounds the anti-sense promoter. The presence of H3-K27 trimethylation at the Tsix promoter in MEFs (Fig. 1D) is likely to be related to the silencing of Tsix.
Monoallelic Xist expression in imprinted and random X-inactivation is mediated by allelic PIC recruitment to the Xist P1 promoter
To address whether the differential pattern of H3-K4 dimethylation at the expressed and silent Xist alleles reflects a transcriptional control of Xist expression, we performed a systematic analysis of RNAPolII and TFIIB binding to the Xist/Tsix region in TS cells and in MEFs.
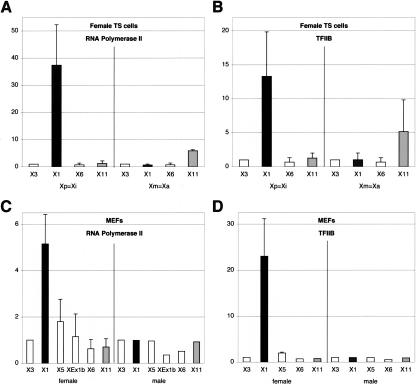
In TS cells, the distribution of RNAPolII and TFIIB parallels that of H3-K4 dimethylation, with a binding specific to the active Xist and Tsix promoters on the paternal and maternal chromosomes, respectively (Fig. 2A,B). This indicates that monoallelic expression of both Xist and Tsix in imprinted X-inactivation is mediated through monoallelic PIC recruitment to the corresponding promoters. Similarly, high levels of monoallelic Xist expression in random X-inactivation is driven by efficient recruitment of the transcriptional apparatus to a single allele, as shown in MEFs (Fig. 2C,D). The specific accumulation of general transcription factors at the Xist promoter region in female cells and its absence in males strongly suggest that recruitment occurs specifically on the inactive X. This further indicates that the lack of Xist expression in males results from the absence of transcription rather than from a post-transcriptional mechanism, in agreement with a previous report of DNA methylation at the Xist promoter on the active X (Norris et al. 1994). Similarly, there is no enrichment for basal transcription factors at the Tsix promoter in male and female MEFs.
Figure 2.
Binding of the transcription machinery to the Xist/Tsix region in female TS cells and in MEFs. (A,B) Binding of RNA polymerase II (A) and TFIIB (B) in female TS cells. As in Figure 1, the positions corresponding to the Xist P1 promoter (X1) and the Tsix promoter (X11) are in black and gray, respectively. Allelic primers for X1 and X11 allowed us to distinguish the maternal and paternal alleles. All the other positions (in white) detect both paternal and maternal alleles. Male TS cells exhibit a profile similar to that of the maternal X chromosome of female TS cells (data not shown). (C,D) Similar analysis in female and male MEFs. The graphs show the fold-enrichment value of each position compared with position X3.
Interestingly, in both TS cells (data not shown) and MEFs (Fig. 2C,D), the transcription machinery is mostly detected at the P1 promoter compared with P2. Binding of TFIIB appears 10-fold enriched over P1 compared with P2 in MEFs, and this is accompanied by a twofold enrichment of RNAPolII (Fig. 2C,D). The greater enrichment noted for TFIIB than for RNAPolII likely reflects the presence, at P2, of RNAPolII that has in fact initiated transcription at P1. In support of this interpretation, we detected, at a position located between P1 and P2, RNAPolII levels similar to that seen at P2 (data not shown). These data, which contrast with previous observations (Johnston et al. 1998), strongly suggest that P1 is the major Xist promoter, at least in cells expressing high levels of Xist.
We conclude that efficient monoallelic Xist expression in imprinted and random X-inactivation is regulated at the level of PIC recruitment to the P1 promoter, and associated with allelic distribution of H3-K4 dimethylation.
In undifferentiated ES cells, Xist down-regulation is mediated by the repression of PIC recruitment to the Xist promoter
Undifferentiated ES cells are unique in the expression pattern of the Xist/Tsix locus, with both genes being expressed from each individual X chromosome. While it has been shown that Tsix transcripts are in a 10- to 100-fold molar excess compared with Xist (Shibata and Lee 2003), it is unknown whether this difference in abundance of the RNAs is due to transcriptional or post-transcriptional mechanisms. To address this issue, we monitored the binding of the transcription machinery to the Xist/Tsix region in ES cells.
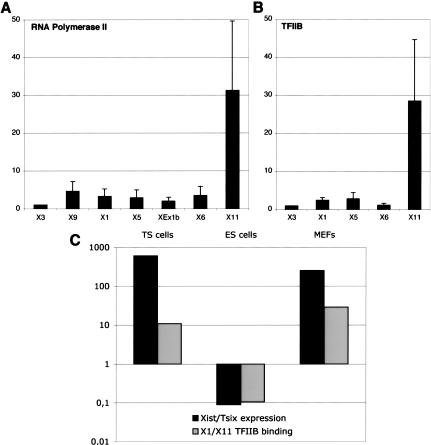
The distribution of RNAPolII reveals a strong bias in transcription towards Tsix (Fig. 3A), as only the Tsix promoter displays high levels of RNAPolII accumulation. All the positions tested within the overlapping Xist/Tsix region (positions X1, X5, XEx1b) show RNAPolII levels similar to that observed at positions transcribed only in Tsix orientation (X9, X6), indicating that in ES cells, RNAPolII specifically associated with the transcription of Xist cannot be detected by ChIP. Similarly, TFIIB binding shows an extreme skew towards the antisense promoter, although low but significant levels of TFIIB binding are detected at both Xist P1 and P2 promoters (Fig. 3B). These observations imply that the excess of Tsix transcripts compared with Xist transcripts is in large part supported by a differential distribution of RNAPolII within the region, which is in turn regulated by a differential efficiency of PIC assembly on the promoters.
Figure 3.
Xist down-regulation in undifferentiated ES cells is regulated by repression of PIC recruitment to the P1 Xist promoter. (A,B) Binding of RNA polymerase II (A) and TFIIB (B) in female ES cells (LF2). Identical results were obtained in male ES cells (data not shown). Graphs show the fold-enrichment value of each position compared with position X3. (C) Comparative analysis of the Xist/Tsix transcripts ratio (black bars) with the ratio of TFIIB binding to Xist P1 promoter versus Tsix promoter (gray bars) in female TS cells, ES cells, and MEFs. The RNA levels of Xist and Tsix were determined by real-time PCR using primers XcR1 for Xist and X6 for Tsix, and standardized against the mRNA levels of Arpo P0.
The extremely low levels of TFIIB binding detected at the Xist promoters further indicates that, in ES cells, reduced levels of Xist expression result from repression of PIC recruitment. The efficiency of PIC assembly at P1 is therefore dramatically different between cells in which X-inactivation has occurred (such as TS cells and MEFs) and cells in which X-inactivation has yet to occur (ES cells). Using quantitative RT-PCR, we determined the relative levels of expression of Xist and Tsix in female TS, ES cells, and MEFs (Fig. 3C, black bars). As expected, in TS and MEFs female cells, the ratio is skewed towards Xist. In contrast, ES cells show a skew towards Tsix, in agreement with previous observations (Shibata and Lee 2003). Calculation of the ratios of TFIIB binding to Xist P1 promoter versus Tsix promoter (Fig. 3C, gray bars) indicates that cells expressing predominantly Xist (after imprinted or random X-inactivation) show a bias of TFIIB binding towards the Xist P1 promoter. Conversely, cells expressing predominantly Tsix (a configuration specific to ES cells) show a bias of TFIIB binding towards the Tsix promoter. This comparative analysis is strongly supportive of the Xist/Tsix balance being regulated at the level of PIC recruitment to Xist and Tsix promoters.
Tsix is not responsible for the transcriptional repression of Xist
Given the distribution of the transcription machinery in ES cells and the known role of Tsix as a negative regulator of Xist accumulation, it is tempting to speculate that Tsix transcription across the Xist promoter is itself sufficient to repress high levels of PIC recruitment in ES cells. To address this issue, we used two previously characterized mutations that result in an absence of Tsix transcription (Luikenhuis et al. 2001; Morey et al. 2004). We exploited mutations on an XY background in order to overcome the need for polymorphisms, a potential limitation in the number of positions that can be analyzed. Male ES cells have been shown to be competent for Xist up-regulation and X-inactivation, as shown in male cells carrying Xic transgenes (Lee et al. 1996) or a 65-kb deletion 3′ to Xist (Morey et al. 2004). Identical results were obtained in mutant female ES cells lines (Supplementary Figure S1D,E).
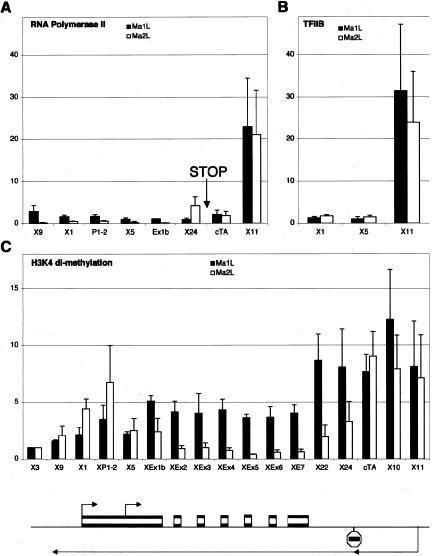
One mutant ES cell line, Ma2L, carries a splice acceptor site and a transcriptional stop signal located some 4 kb downstream of the Tsix promoter (Luikenhuis et al. 2001). The control cell line, Ma1L, contains a single loxP site at the equivalent position. Detailed profiling of RNAPolII in the Xist/Tsix region of the Ma2L ES cell line confirmed the efficiency of the introduced stop signal (Fig. 4A). The amount of RNAPolII at positions located upstream of the transcriptional stop (positions cTA and X11) is identical in both wild-type and mutant cells, showing that truncating the Tsix unit has no effect on the initiation of Tsix transcription. At position X24, which lies 700 bp downstream of the stop signal, the levels of RNAPolII were increased in the mutant, probably reflecting the accumulation of RNAPolII at the ectopic 3′ end of Tsix. The other positions tested, whether lying within or outside the Xist transcription unit, show a virtually complete loss of RNAPolII, confirming our previous conclusion that in ES cells the Xist/Tsix region transcribes almost exclusively Tsix.
Figure 4.
Transcriptional and chromatin analyses of _Tsix_-truncated undifferentiated male ES cells. Ma2L cells (white bars) carry a transcriptional stop signal 4 kb downstream of the Tsix promoter. Ma1L (black bars) is the corresponding control cell line. (A) ChIP analysis of RNA polymerase II distribution within the Xist/Tsix region in Ma1L and Ma2L cell lines. The position of the stop signal is indicated in the graph. (B) ChIP analysis of TFIIB binding to the Xist P1 and P2 promoters and the Tsix promoter in Ma1L and Ma2L. (C) Extensive ChIP analysis of H3-K4 dimethylation within the Xist/Tsix locus in Ma1L and Ma2L. The graphs show the fold-enrichment value of each position compared with position X3. The schematic map below the graph shows the position of the primers used and of the stop signal.
Strikingly, neither RNAPolII nor TFIIB (Fig. 4B) are enriched at the Xist promoters upon truncation of Tsix transcription, as would have been expected if Tsix were repressing Xist at the transcriptional level. This indicates that Tsix is not directly responsible for the repression of PIC recruitment at P1. To confirm this, we used a second cell line, CK35Δ65, that carries a 65-kb deletion 3′ to Xist encompassing the Tsix promoter (Morey et al. 2004). Similarly to the Tsix truncation, deletion of the antisense promoter does not result in increased levels of RNAPolII or TFIIB at P1/P2 Xist promoters (Supplementary Fig. S1B,C). Our results clearly indicate that although Tsix is highly transcribed in ES cells, neither Tsix transcription nor Tsix transcript is responsible for repressing Xist transcription, ruling out an often-cited model to mechanistically explain the effects of Tsix on the expression level of Xist. Importantly, this implies the existence of _Tsix_-independent mechanisms that repress Xist transcription specifically in ES cells. Our result furthermore provides an explanation for the failure of undifferentiated ES cells to induce strong Xist RNA accumulation and X-inactivation upon Tsix invalidation (Debrand et al. 1999; Lee and Lu 1999; Luikenhuis et al. 2001).
Tsix transcription induces complex modification of H3-K4 dimethylation within the Xist/Tsix locus
Given the low levels of PIC recruitment to the Xist P1 promoter in ES cells (Fig. 3A,B) and the association of H3-K4 dimethylation with the active Xist promoter in TS cells and in MEFs (Fig. 1B), we monitored the levels of H3-K4 dimethylation within the Xist/Tsix locus in ES cells, with the expectation of finding low levels of this epigenetic mark associated with the poorly active Xist promoter. The data presented in Figure 4C (black bars) were obtained with male ES cells, but a similar profile was observed in female ES cells (data not shown). Surprisingly, significant enrichment for H3-K4 dimethylation was detected at the Xist 5′ region. In contrast to MEFs and TS cells however, H3-K4 dimethylation is not restricted to the Xist promoter region but is associated with the entire Xist/Tsix unit (Fig. 4C, black bars). Interestingly, we found that H3-K4 dimethylation spreads from the 5′ extremity of Tsix (position X11) toward the 3′ end (position X9) in a gradient reminiscent of Tsix RNA levels described by others (Shibata and Lee 2003). We conclude that in ES cells, the Xist locus is in a primed state for transcription, characterized by a global enrichment for H3-K4 dimethylation.
We show here that in ES cells (1) the Xist/Tsix locus is mainly transcribed in the Tsix orientation (Fig. 3A), (2) H3-K4 dimethylation spreads over the entire Tsix transcription unit (Fig. 4C, black bars), and (3) deleting 65 kb 3′ to Xist (CK35Δ65) results in a specific and efficient loss of H3-K4 dimethylation within Xist (Morey et al. 2004). Altogether, these data point to a function for Tsix in the deposition of H3-K4 dimethylation within the Xist/Tsix region.
To confirm this, we analyzed the pattern of H3-K4 dimethylation in the Ma2L mutant ES cells that carry a truncated Tsix allele (white bars in Fig. 4C). Strikingly, blocking Tsix transcription leads to an almost complete loss of H3-K4 dimethylation at positions located downstream of the introduced transcriptional stop signal, whereas levels of dimethylated H3-K4 were found to be mostly unchanged at upstream positions (Fig. 4C). This result indicates that Tsix induces H3-K4 dimethylation within the region, probably through the recruitment of histone methyltransferase activity by either the RNAPolII transcribing Tsix or the Tsix RNA itself.
Intriguingly, Tsix appears to have a dual role in the regulation of H3-K4 dimethylation at the Xist promoter, since the absence of Tsix transcription (or transcript) induces elevated levels of dimethylation (and also trimethylation) (data not shown) over this region (positions X1 and XP1.2 in Fig. 4C). Interestingly, this increase concerns not only the Xist promoter itself but also an extended region that corresponds to the only spliced exon of Tsix known to overlap Xist RNA (Sado et al. 2001; Shibata and Lee 2003). We conclude that Tsix induces complex modification of H3-K4 dimethylation within Xist, demonstrating for the first time that Tsix remodels the chromatin structure of the Xist locus.
Discussion
Transcriptional regulation of Xist expression
We have reported here major variations in the PIC recruitment activity of the Xist P1 promoter that correlate with the X-inactivation status of the corresponding chromosome. The level of transcription basal machinery associated with the P1 promoter in cell types expressing high levels of Xist RNA, in which XCI has already occurred (female MEFs and TS cells) is much greater than the level detected in cells expressing low levels of Xist, in which XCI has yet to occur (ES cells). This indicates that Xist expression is regulated transcriptionally by the selective recruitment of the transcriptional apparatus to its promoter (Fig. 5). Since high levels of Xist expression were previously shown to be supported by the stabilization of the transcript, it is of clear interest to address whether or not these mechanisms act together. Because transcription and RNA processing are tightly coupled events (Proudfoot 2000), it is tempting to speculate that PIC assembly at the Xist promoter might also be linked to the recruitment of Xist RNA stabilization factors.
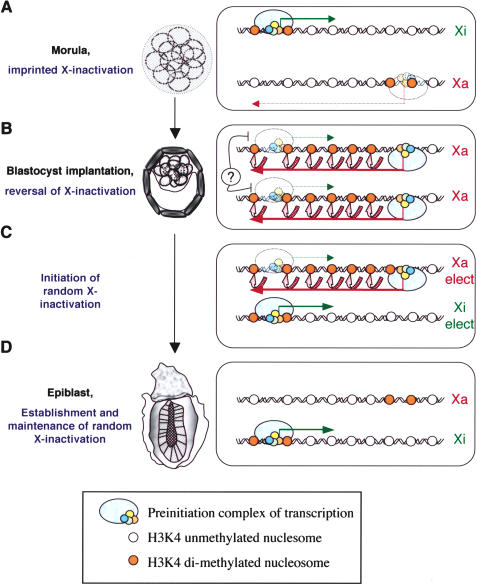
Figure 5.
A model of the regulation of the Xist/Tsix region during early mouse development. Developmental stages are shown on the left, and the status of the Xist/Tsix region in terms of transcription and chromatin at each stage is shown on the right. (A) In the preimplantation embryo, the segregation of PIC together with H3-K4 dimethylation over the paternal and maternal Xist and Tsix promoters ensures paternal Xist and maternal Tsix expression, respectively. Bold arrows represent high levels of expression, dotted arrows represent low levels of expression. (B) At the implantation stage, an unknown ICM-specific repressive mechanism of PIC recruitment at P1 Xist promoter avoids biallelic X-inactivation (represented by a question mark and a transparent PIC at the Xist promoter). In the meantime, biallelic Tsix activation erases the histone methylation pattern by inducing high levels of H3-K4 dimethylation across the region (pink arrows), rendering both Xist alleles epigenetically equivalent. (C) At the onset of random X-inactivation, monoallelic down-regulation of Tsix results in an increase of H3-K4 dimethylation over the corresponding Xist promoter region. The absence of ICM-specific Xist repression allows stable binding of the transcriptional apparatus, inducing high levels of monoallelic Xist expression and X-inactivation. (D) In the female epiblast, other epigenetic marks lock in the expression profile of Xist and the silencing of Tsix transcription.
Intriguingly, the strong variation in the binding of the transcription machinery that we detected at the Xist P1 promoter in the different cell types studied was not observed for P2. Our results indicate first that unlike P1, the activity of P2 is not developmentally regulated, and second, that P1 is the major Xist promoter, at least in cells expressing high levels of Xist. This latter observation contradicts earlier conclusions based on the RNA steady-state-level measurement of the two putative isoforms produced by these promoters (Johnston et al. 1998). This apparent paradox can be attributed to more efficient transcription from P2 than from P1, regulated either at the level of promoter clearance or at the elongation step. An alternative explanation is suggested by preliminary measurement by quantitative RT-PCR of Xist RNA at several positions both up- and downstream of P2. We observed a strong influence of the reverse transcription (RT) reaction temperature on the RT efficiency over the 5′ extremity of Xist RNA (data not shown). This suggests the existence of secondary structures of Xist transcripts that might have been responsible for artifactual quantification of molecules bearing this region. The Xist 5′ region is known to be rich in DNA repeats such as the so-called A repeat responsible for the silencing properties of Xist RNA.
The correlation in ES cells between low Xist RNA levels and the presence of limited quantities of RNAPolII and TFIIB at the Xist P1 promoter (note that other general transcription factors such as TFIIA, TFIIE, TFIIF, and TFIIH gave similar results) (Supplementary Fig. S2) suggests either the presence of a repressor or the lack of a specific activator in such cells. Importantly, the repression of P1 in ES cells might be part of the mechanism allowing the reactivation of the paternal X chromosome in the ICM. Our analysis of Tsix mutant cells reveals that Tsix is not directly responsible for the repression of PIC assembly at the Xist promoters, despite Tsix being highly transcribed in ES cells. This implies firstly that a _Tsix_-independent mechanism represses Xist transcription specifically in ES cells (Fig. 5B), and secondly that the elevation in Xist RNA levels resulting from Tsix invalidation in undifferentiated ES cells (Supplementary Fig. S1A) must be due to mechanisms other than PIC recruitment, affecting for example the stability of Xist RNA. Our results are in agreement with previous observations indicating that undifferentiated ES cells lacking Tsix do not accumulate high enough levels of Xist RNA to initiate X-inactivation (Debrand et al. 1999; Lee and Lu 1999; Luikenhuis et al. 2001; Sado et al. 2001). The molecular elucidation of the transcription factor network achieving the repression of PIC recruitment specifically in ES cells will provide invaluable information regarding X-inactivation.
The chromosomal origin of Xist expression is based on allelic H3-K4 dimethylation and PIC recruitment
H3-K4 methylation has emerged as an important mark in the epigenetic programming of the genome, as highlighted by the acquisition of H3-K4 dimethylation in reprogrammed somatic genomes (Kimura et al. 2004). We found that H3-K4 dimethylation also appears to be involved in the allelic programming of Xist expression, as it is the only mark tested to display a differential distribution that follows monoallelic Xist expression. The allelic pattern of H3-K4 dimethylation correlates with the binding of transcription basal machinery to the promoter. Thus, in both imprinted and random X-inactivation, monoallelic Xist expression is achieved by transcriptional mechanisms likely based on the epigenetic distinction of both alleles through H3-K4 dimethylation (Fig. 5A,D). However, whether allelic PIC recruitment is a cause or a consequence of allelic H3-K4 dimethylation remains to be determined. Interestingly, and in contrast to other examples of monoallelically expressed genes, neither H3-K9 dimethylation nor H3-K27 trimethylation is associated with the silent Xist allele, even in cells with imprinted XCI. The absence of such repressive marks might be important for the plasticity of Xist expression, which must shift from imprinted to random at the implantation stage of development, and it is in agreement with the role of Tsix in reprogramming H3-K4 methylation. However, we cannot exclude the possibility that other marks contribute to the epigenetic repression of the silent Xist promoter on the active X chromosome. Among them, DNA methylation is unlikely to be involved, since the Xist promoter region was shown to be hypomethylated in preimplantation embryos (MacDonald et al. 1998).
Tsix triggers H3-K4 dimethylation within Xist
In ES cells, the entire Tsix unit is enriched for H3-K4 methylation, and our results have clearly established that Tsix is responsible for such histone modification, providing the first demonstration of a function of Tsix in regulating chromatin conformation of the Xist locus. This is reminiscent of the situation in S. cerevisiae, where a link between H3-K4 methylation and transcription elongation was demonstrated (Bernstein et al. 2002; Krogan et al. 2003; Ng et al. 2003). Such transcription-dependent H3-K4 methylation is mediated by the Set1 histone methyltransferase (HMT) through its association with the elongating RNAPolII (Krogan et al. 2003; Ng et al. 2003). We suggest that the association of an as yet unknown HMT-containing complex with the Tsix transcribing polymerase is responsible for the methylation of the overall region in ES cells (Fig. 5B,C). Alternatively, it is formally possible that H3-K4 dimethylation is triggered by the Tsix transcript itself through interaction between the RNA and an HMT-containing complex, similarly to the proposed function of Xist RNA in directing H3-K9 dimethylation and H3-K27 trimethylation to the inactive X chromosome (Heard 2004). That no H3-K4 methylation can be detected within the body of Xist in TS cells, despite maternal transcription of Tsix, might be due either to the low transcriptional and thereby methylation activity of Tsix or to the absence of the HMT responsible for such a modification, or a key cofactor controlling its accessibility, in such cells.
Tsix is usually described as a negative regulator of Xist expression. This clearly contrasts with the role of Tsix in the deposition of a histone modification specific for active chromatin within Xist, which suggests that Tsix transcription might also be involved in the transcriptional potential of Xist in ES cells. We propose that, at the implantation stage of development, biallelic activation of Tsix is responsible for the erasure of the allelic H3-K4 methylation mark that characterizes imprinted Xist expression (Fig. 5). The enrichment of the Xist locus in H3-K4 dimethylation will result in both Xist homologs being epigenetically equivalent and equally competent for transcription and random X-inactivation. This provides a new function for Tsix in the reversion of imprinted Xist expression. The absence of a functional TSIX in human, which correlates with the absence of imprinted XCI in this species (for review, see Rougeulle and Avner 2004), is in agreement with our model.
Does Tsix down-regulation mark the future inactive X chromosome by restoring promoter-restricted H3-K4 dimethylation pattern on a single Xist allele?
The analysis of _Tsix_-truncated ES cell lines revealed that H3-K4 methylation of the Xist promoter region is not regulated solely by the transcription-dependent global methylation of the region, but that additional mechanisms are involved in which Tsix acts as a repressor. Because the region concerned corresponds to the only spliced exon of Tsix known to overlap Xist RNA (Sado et al. 2001; Shibata and Lee 2003), one might hypothesize that an RNA duplex between spliced transcripts of Tsix and Xist is responsible for the negative regulation of H3-K4 dimethylation within the Xist promoter region through targeting of the RNA interference (RNAi)-dependent heterochromatinization machinery. This notion is supported by the recent identification of the RNA-induced transcriptional silencing (RITS) complex that is believed to recruit components of the RNAi-mediated heterochromatinization pathway to genomic loci by interaction involving nascent-transcript and short interfering RNAs (siRNAs) produced by DICER (Verdel et al. 2004). Interestingly, DNA methylation, which can be induced by siRNAs, was shown to characterize to a certain extent the Xist promoter on both alleles in ES cells (Sado et al. 1996).
The down-regulation of Tsix during random XCI was shown to occur first on the future inactive X (Lee et al. 1999), and this was proposed to contribute to the choice process (Lee and Lu 1999). Our finding that disruption of Tsix transcription in ES cells results in increased levels of H3-K4 dimethylation over the Xist promoter region in cis provides novel insights into the mechanism of action of Tsix in the choice process of random X-inactivation: Monoallelic down-regulation of Tsix at the onset of random XCI will mark epigenetically the future active Xist promoter (Fig. 5C). The combined loss of the ICM-specific Xist repressive mechanism will result in allelic recruitment of the transcription machinery. The action of Tsix in the choice process of X-inactivation is therefore likely to be dual, with persistent Tsix transcription on the future active X repressing Xist RNA accumulation, while Tsix down-regulation epigenetically marks the future inactive X for monoallelic Xist expression.
In conclusion (Fig. 5), we demonstrate here that Xist transcription is a highly regulated event participating in both the expression level and the chromosomal origin of Xist RNA. Importantly, we show that Tsix has no effect on the transcriptional regulation of Xist. Rather, Tsix acts as a chromatin remodeler responsible for complex regulation of H3-K4 dimethylation within Xist in ES cells. We propose that the chromatin-related function of Tsix is crucial for the transition from imprinted to random Xist expression. Biallelic activation of Tsix at the implantation stage of mouse development erases epigenetic marks associated with imprinted XCI and renders both Xist loci epigenetically indistinguishable and competent for random X-inactivation. Finally, our results suggest that the known role of Tsix in random choice is in part mediated by the establishment, on the future inactive X, of a chromatin structure characteristic of monoallelic Xist expression.
Material and methods
Cell culture
MEFs were prepared from 13.5-d-old embryos and cultured in Dulbecco's modified Eagle medium (DMEM, GIBCO), 10% fetal calf serum (FCS, ES cell grade, GIBCO). ES cell lines were grown in DMEM, 15% FCS, and 1000 U/mL LIF (Chemicon). Female LF2 ES cells were cultured on gelatin-coated plates in the absence of feeder cells. Male Ma1L and Ma2L ES cells (a kind gift from R. Jaenisch, Whitehead Institute for Biomedical Research, Cambridge, MA) were cultured on Mitomycin C-treated male embryonic fibroblast feeder cells. Female TS cells (F3 cell line) were grown on male fibroblast feeder cells in RPMI 1640 medium (GIBCO), 20% FCS (Biowest), MEM sodium pyruvate 1 mM (GIBCO), 2-mercaptoethanol 100 μM (GIBCO), FGF4 25 ng/mL (RD systems), and Heparin 1 μg/mL (Labosi). Feeder cells from Ma1L, Ma2L, and F3 cell cultures were removed by adsorption before chromatin and RNA extractions.
ChIP
ChIP assays were carried out essentially as described (Heard et al. 2001), with the following modifications: Cross-linking was performed on cells resuspended in DMEM, 10% FCS for 15 min in the presence of 1% formaldehyde at room temperature. TFIIB (C-18, Santa-Cruz), RNAPolII (7C2, Euromedex), and H3 di-meK4, di-meK9 and tri-meK27 (Upstate Biotechnology) antibodies were used respectively at dilutions of 1/50, 1/500, 1/100, 1/100, and 1/500.
The immunoprecipitated (IP) DNA and a 1/100 dilution of the input DNA were analyzed by real-time PCR using SYBR Green Universal Mix and an ABI Prism 7700 (Perkin Elmer Applied Biosystems). Each PCR was run in triplicate to control for PCR variation. The primer sequences are provided as Supplemental Material. To standardize between experiments, we calculated the percentage of immunoprecipitation by dividing the average value of the IP by the average value of the corresponding input, both values first being normalized for dilution factors.
Quantitative RT-PCR
Total RNA isolated from cell cultures using RNable (Eurobio) was DNase-treated (Roche) and routinely verified by electrophoresis. Random-primed reverse transcription was performed at 42°C using Superscript II reverse transcriptase (Invitrogen). Control reactions lacking the RT enzyme were systematically verified for the absence of product. Quantitative real-time PCR measurements using SYBR Green Universal Mix were performed in triplicate, and Arpo P0 transcript levels were used to normalize between samples.
Acknowledgments
We thank the members of our laboratory for stimulating discussions, Philippe Clerc and Marc Lalande for critical reading of the manuscript, and Sandra Luikenhuis and Rudolf Jaenisch for the gift of the Tsix truncated ES cell lines. This work was supported by the Association pour le Recherche contre le Cancer (ARC) and the French Ministry of Research under the Action Concertée Incitative, contract number 032526. C.R. and P.A. are supported by the CNRS.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.341105.
References
- Bernstein B.E., Humphrey, E.L., Erlich, R.L., Schneider, R., Bouman, P., Liu, J.S., Kouzarides, T., and Schreiber, S.L. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. 99**:** 8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani B., Tonlorenzi, R., Simmler, M.-C., Dandolo, L., Arnaud, D., Capra, V., Grompe, M., Pizzuti, A., Muzni, D., Lawrence, C., et al. 1991. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351**:** 325-329. [DOI] [PubMed] [Google Scholar]
- Clemson C.M., McNeil, J.A., Willard, H.F., and Lawrence, J.B. 1996. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell. Biol. 132**:** 259-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debrand E., Chureau, C., Arnaud, D., Avner, P., and Heard, E. 1999. Functional analysis of the DXPas34 locus, a 3′ regulator of Xist expression. Mol. Cell. Biol. 19**:** 8513-8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier C., Goto, Y., Ballestar, E., Delaval, K., Hever, A.M., Esteller, M., and Feil, R. 2002. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 21**:** 6560-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E. 2004. Recent advances in X-chromosome inactivation. Curr. Opin. Cell Biol. 16**:** 247-255. [DOI] [PubMed] [Google Scholar]
- Heard E., Rougeulle, C., Arnaud, D., Avner, P., Allis, C.D., and Spector, D.L. 2001. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107**:** 727-738. [DOI] [PubMed] [Google Scholar]
- Huynh K.D. and Lee, J.T. 2003. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426**:** 857-862. [DOI] [PubMed] [Google Scholar]
- Johnston C.M., Nesterova, T.B., Formstone, E.J., Newall, A.E., Duthie, S.M., Sheardown, S.A., and Brockdorff, N. 1998. Developmentally regulated Xist promoter switch mediates initiation of X inactivation. Cell 94**:** 809-817. [DOI] [PubMed] [Google Scholar]
- Kay G.F., Barton, S.C., Surani, M.A., and Rastan, S. 1994. Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell 77**:** 639-650. [DOI] [PubMed] [Google Scholar]
- Kimura H., Tada, M., Hatano, S., Yamazaki, M., Nakatsuji, N., and Tada, T. 2002. Chromatin reprogramming of male somatic cell-derived Xist and Tsix in ES hybrid cells. Cytogenet. Genome Res. 99**:** 106-114. [DOI] [PubMed] [Google Scholar]
- Kimura H., Tada, M., Nakatsuji, N., and Tada, T. 2004. Histone code modifications on pluripotential nuclei of reprogrammed somatic cells. Mol. Cell Biol. 24**:** 5710-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.V., Dover, J., Wood, A., Schneider, J., Heidt, J., Boateng, M.A., Dean, K., Ryan, O.W., Golshani, A., Johnston, M., et al. 2003. The Paf1 complex is required for Histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol. Cell 11**:** 721-729. [DOI] [PubMed] [Google Scholar]
- Lee J.T. 2000. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell 103**:** 17-27. [DOI] [PubMed] [Google Scholar]
- Lee J.T. and Lu, N. 1999. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99**:** 47-57. [DOI] [PubMed] [Google Scholar]
- Lee T.I. and Young, R.A. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34**:** 77-137. [DOI] [PubMed] [Google Scholar]
- Lee J.T., Strauss, W.M., Dausman, J.A., and Jaenisch, R. 1996. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell 86**:** 83-94. [DOI] [PubMed] [Google Scholar]
- Lee J.T., Davidow, L.S., and Warshawsky, D. 1999. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 21**:** 400-404. [DOI] [PubMed] [Google Scholar]
- Lewis A., Mitsuya, K., Umlauf, D., Smith, P., Dean, W., Walter, J., Higgins, M., Feil, R., and Reik, W. 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 36**:** 1291-1295. [DOI] [PubMed] [Google Scholar]
- Luikenhuis S., Wutz, A., and Jaenisch, R. 2001. Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol. Cell. Biol. 21**:** 8512-8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald L.E., Paterson, C.A., and Kay, G.F. 1998. Bisulfite genomic sequencing-derived methylation profile of the Xist gene throughout early mouse development. Genomics 54**:** 379-386. [DOI] [PubMed] [Google Scholar]
- Mak W., Baxter, J., Silva, J., Newall, A.E., Otte, A.P., and Brockdorff, N. 2002. Mitotically stable association of polycomb group proteins Eed and Enx1 with the inactive X chromosome in trophoblast stem cells. Curr. Biol. 12**:** 1016-1020. [DOI] [PubMed] [Google Scholar]
- Mak W., Nesterova, T.B., de Napoles, M., Appanah, R., Yamanaka, S., Otte, A.P., and Brockdorff, N. 2004. Reactivation of the paternal X chromosome in early mouse embryos. Science 303**:** 666-669. [DOI] [PubMed] [Google Scholar]
- Marahrens Y., Panning, B., Dausman, J., Strauss, W., and Jaenisch, R. 1997. _Xist_-deficient mice are defective in dosage compensation but not spermatogenesis. Genes & Dev. 11**:** 156-166. [DOI] [PubMed] [Google Scholar]
- Mise N., Goto, Y., Nakajima, N., and Takagi, N. 1999. Molecular cloning of antisense transcripts of the mouse Xist gene. Biochem. Biophys. Res. Comm. 258**:** 537-541. [DOI] [PubMed] [Google Scholar]
- Morey C., Navarro, P., Debrand, E., Avner, P., Rougeulle, C., and Clerc, P. 2004. The region 3′ to Xist mediates X chromosome counting and H3 Lys-4 dimethylation within the Xist gene. EMBO J. 23**:** 594-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H., Robert, F., Young, R.A., and Struhl, K. 2003. Targeted recruitment of Set1 histone methylase by elongating pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11**:** 709-719. [DOI] [PubMed] [Google Scholar]
- Norris D.P., Patel, D., Kay, G.F., Penny, G.D., Brockdorff, N., Sheardown, S.A., and Rastan, S. 1994. Evidence that random and imprinted Xist expression is controlled by pre-emptive methylation. Cell 77**:** 41-51. [DOI] [PubMed] [Google Scholar]
- Okamoto I., Otte, A.P., Allis, C.D., Reinberg, D., and Heard, E. 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303**:** 644-649. [DOI] [PubMed] [Google Scholar]
- Panning B., Dausman, J., and Jaenisch, R. 1997. X chromosome inactivation is mediated by Xist RNA stabilization. Cell 90**:** 907-916. [DOI] [PubMed] [Google Scholar]
- Penny G.D., Kay, G.F., Sheardown, S.A., Rastan, S., Brockdorff, N., Avner, P., and Turner, B.M. 1996. Requirement for Xist in X chromosome inactivation. Nature 379**:** 131-137. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. 2000. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 25**:** 290-293. [DOI] [PubMed] [Google Scholar]
- Rougeulle C. and Avner, P. 2003. Controlling X-inactivation in mammals: What does the centre hold? Sem. Cell Dev. Biol. 14**:** 331-340. [DOI] [PubMed] [Google Scholar]
- ____. 2004. The role of antisense transcription in the regulation of X-inactivation. Curr. Top. Dev. Biol. 63**:** 61-89. [DOI] [PubMed] [Google Scholar]
- Rougeulle C., Navarro, P., and Avner, P. 2003. Promoter-restricted H3 Lys 4 di-methylation is an epigenetic mark for monoallelic expression. Hum. Mol. Genet. 12**:** 3343-3348. [DOI] [PubMed] [Google Scholar]
- Rougeulle C., Chaumeil, J., Sarma, K., Allis, C.D., Reinberg, D., Avner, P., and Heard, E. 2004. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol. Cell. Biol. 24**:** 5475-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T., Tada, T., and Takagi, N. 1996. Mosaic methylation of Xist gene before chromosome inactivation in undifferentiated female mouse embryonic stem and embryonic germ cells. Dev. Dyn. 205**:** 421-434. [DOI] [PubMed] [Google Scholar]
- Sado T., Wang, Z., Sasaki, H., and Li, E. 2001. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128**:** 1275-1286. [DOI] [PubMed] [Google Scholar]
- Sheardown S.A., Duthie, S.M., Johnston, C.M., Newall, A.E., Formstone, E.J., Arkell, R.M., Nesterova, T.B., Alghisi, G.C., Rastan, S., and Brockdorff, N. 1997. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell 91**:** 99-107. [DOI] [PubMed] [Google Scholar]
- Shibata S. and Lee, J.T. 2003. Characterization and quantitation of differential Tsix transcripts: Implications for Tsix function. Hum. Mol. Genet. 12**:** 125-136. [DOI] [PubMed] [Google Scholar]
- ____. 2004. Tsix transcription-versus RNA-based mechanisms in Xist repression and epigenetic choice. Curr. Biol. 14**:** 1747-1754. [DOI] [PubMed] [Google Scholar]
- Sims III R.J., Nishioka, K., and Reinberg, D. 2003. Histone lysine methylation: A signature for chromatin function. Trends Genet. 19**:** 629-639. [DOI] [PubMed] [Google Scholar]
- Takagi N. and Sasaki, M. 1975. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256**:** 640-642. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Kunath, T., Hadjantonakis, A.K., Nagy, A., and Rossant, J. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282**:** 2072-2075. [DOI] [PubMed] [Google Scholar]
- Umlauf D., Goto, Y., Cao, R., Cerqueira, F., Wagschal, A., Zhang, Y., and Feil, R. 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 36**:** 1296-1300. [DOI] [PubMed] [Google Scholar]
- Verdel A., Jia, S., Gerber, S., Sugiyama, T., Gygi, S., Grewal, S.I., and Moazed, D. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303**:** 672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T.H., Li, T., and Hoffman, A.R. 2004. Promoter-restricted histone code, not the differentially methylated DNA regions or antisense transcripts, marks the imprinting status of IGF2R in human and mouse. Hum. Mol. Genet. 13**:** 2233-2245. [DOI] [PubMed] [Google Scholar]
- Wutz A., Rasmussen, T.P., and Jaenisch, R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30**:** 167-174. [DOI] [PubMed] [Google Scholar]
- Xin Z., Allis, C.D., and Wagstaff, J. 2001. Parent-specific complementary patterns of histone H3 lysine 9 and H3 lysine 4 methylation at the Prader-Willi syndrome imprinting center. Am. J. Hum. Genet. 69**:** 1389-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]