Centrin 2 Stimulates Nucleotide Excision Repair by Interacting with Xeroderma Pigmentosum Group C Protein (original) (raw)
Abstract
Xeroderma pigmentosum group C (XPC) protein plays a key role in DNA damage recognition in global genome nucleotide excision repair (NER). The protein forms in vivo a heterotrimeric complex involving one of the two human homologs of Saccharomyces cerevisiae Rad23p and centrin 2, a centrosomal protein. Because centrin 2 is dispensable for the cell-free NER reaction, its role in NER has been unclear. Binding experiments with a series of truncated XPC proteins allowed the centrin 2 binding domain to be mapped to a presumed α-helical region near the C terminus, and three amino acid substitutions in this domain abrogated interaction with centrin 2. Human cell lines stably expressing the mutant XPC protein exhibited a significant reduction in global genome NER activity. Furthermore, centrin 2 enhanced the cell-free NER dual incision and damaged DNA binding activities of XPC, which likely require physical interaction between XPC and centrin 2. These results reveal a novel vital function for centrin 2 in NER, the potentiation of damage recognition by XPC.
Centrin was first identified as a small acidic calcium-binding protein in the flagellar apparatus of unicellular green algae (20, 21, 50). In Chlamydomonas reinhardtii, centrin is localized in nucleus-basal body connectors, distal striated fibers, and a flagellar transition zone (51, 65). Mutational analyses of centrin suggested that it may participate in localization or segregation of the basal body and play fundamental roles in microtubule severing (53, 54, 65). A centrin counterpart in Saccharomyces cerevisiae, Cdc31p, was shown to be localized to a half bridge of the spindle pole body, a functional homolog of the centrosome in higher eukaryotes, and to be necessary for spindle pole body duplication (6, 59). At least three centrin homologs have been identified in humans (11, 28, 35). The expression profiles of the three centrin genes are quite different. Expression of centrin 1 is restricted to the retina and testis, where ciliated cells are found (72). Moreover, centrin 1 mRNA can be specifically detected by reverse transcription-PCR during the differentiation of human nasal epithelial cells (26), whereas it is undetectable during the differentiation of tracheal epithelial cells (27). In contrast, centrin 2 and centrin 3 appear to be expressed ubiquitously. Centrin 2 mRNA increases during the differentiation of both nasal and tracheal epithelial cells, while centrin 3 mRNA increases only in nasal epithelial cells and is maintained at constant levels in tracheal epithelial cells (26, 27). In cultured human cells, a subpopulation of centrin 2 protein localizes to the centrosome, and phosphorylation of centrin 2 during the G2/M phase is required for centriole separation during centrosome duplication (29, 45). Furthermore, inhibition of centrin 2 expression by RNA interference in human cells causes failure of centriole duplication (52). On the other hand, it has been reported that most centrin 2 is unassociated with the centrosome and that roughly half of it is found in the nucleus, although its functions outside the centrosome have not been explored extensively (45).
Nucleotide excision repair (NER) is an important DNA repair pathway which eliminates a wide variety of base lesions, including UV-induced (6-4) photoproducts (6-4PP) and cyclobutane pyrimidine dimers (CPD). Impairment in NER activity has been associated with at least three human autosomal recessive genetic disorders, xeroderma pigmentosum (XP), Cockayne syndrome (CS), and trichothiodystrophy (TTD). So far, seven NER-deficient XP complementation groups (XPA through XPG), two CS groups (CS-A and CS-B), and one TTD group (TTD-A) have been identified, and in all cases, the responsible genes have been cloned (15, 17, 19). Mammalian NER consists of two subpathways, global genome NER (GG-NER), which operates throughout the genome, and transcription-coupled NER, which is specialized for the elimination of lesions from the transcribed strand of active genes. A major difference between the two modes of NER is apparent in the damage recognition step. In transcription-coupled NER, damage recognition is probably mediated by RNA polymerase II stalling at damage sites (66). In GG-NER, the XPC protein, a product of one of the XP complementation group genes, plays a key role in the recognition of DNA lesions (48, 61, 64, 70). Despite this initial difference, subsequent repair reactions are thought to be shared by both modes. Duplex DNA around the damaged site is unwound by helicase activities of TFIIH in the presence of XPG, XPA, and replication protein A (RPA) (12, 13, 39, 71). This open complex formation is a prerequisite for dual incision by two structure-specific endonucleases, XPF-ERCC1 and XPG, at sites 5′ and 3′ of the lesion, respectively (32, 42, 58). The resulting gap is filled by DNA polymerase (δ or ɛ), and the resulting nick is rejoined by DNA ligase I (for a review, see reference 8).
XPC in vivo is a heterotrimeric complex including one of two mammalian homologs of S. cerevisiae Rad23p (HR23A or HR23B) and centrin 2 (2, 30, 57). Although most XPC is bound to HR23B rather than to HR23A, the two Rad23p homologs are functionally redundant with respect to their GG-NER functions (40, 43, 62). In the absence of both HR23 proteins, XPC is markedly destabilized in vivo as well as in vitro, and thus, the HR23 proteins are effectively essential for efficient GG-NER (2, 5, 40, 43). A recombinant XPC-HR23B heterodimer exhibits specific binding affinities for various types of lesions, including 6-4PP, and has been successfully used to reconstitute a cell-free NER reaction (1, 3, 31). Centrin 2 is thus dispensable for NER at least in vitro, so the biological significance of its presence in the XPC complex remains to be elucidated, although the physical stability of XPC-HR23B may increase slightly upon binding to centrin 2 (2). To clarify the precise function of centrin 2 in GG-NER, we have identified a centrin 2-binding domain of XPC and generated a mutant XPC that does not physically interact with centrin 2. Functional analyses of the mutant protein unveiled a novel function for centrin 2 in stimulating NER.
MATERIALS AND METHODS
Cell lines and cultures.
Simian virus 40-transformed cells, a normal human fibroblast cell line (WI38 VA13), an XPC-deficient human cell line (XP4PASV), and stable XP4PASV transformants were cultured at 37°C in a humidified atmosphere of 5% CO2. All cell lines were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. HighFive cells were cultured at 27°C in Ex-cell 405 medium (JRH Biosciences).
Preparation of cell lysates.
For immunoblot analyses and binding experiments with XPC complex subunits, cells (typically in 60-mm dishes) were washed twice with ice-cold phosphate-buffered saline and lysed on ice for 60 min with 500 μl NP lysis buffer (25 mM Tris-HCl [pH 8.0], 1 mM EDTA, 10% glycerol, 1% Nonidet P-40 [NP-40], 0.25 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT], and a protease inhibitor cocktail [Complete; Roche Diagnostics]) containing 0.3 M NaCl. Cell lysates were scraped into a microcentrifuge tube, and the dishes were washed with 500 μl of the same buffer, which was included with the recovered lysates. Soluble extracts were obtained by centrifugation for 10 min at 20,000 × g. The resulting pellets were washed twice with the same buffer and resuspended in 500 μl NP lysis buffer containing 0.3 M NaCl with the aid of sonication.
For examination of expression levels of XPC in stable XP4PASV transformants (Fig. 1A), cells were lysed with 200 μl of a solution containing 50 mM Tris-HCl (pH 8.0), 0.6% sodium dodecyl sulfate (SDS), and 1 mM DTT. The lysates were heated at 95°C for 10 min and mixed with 1 ml buffer containing 0.1 M sodium phosphate (pH 7.2), 20 mM EDTA, 1% NP-40, and 0.25 M NaCl. Final whole-cell extracts were obtained by centrifugation for 90 min at 50,000 × g (Beckman TLA 45 rotor). Protein concentrations of the supernatant fractions were determined according to the method of Schaffner and Weissmann (55), with bovine serum albumin (BSA) as a standard.
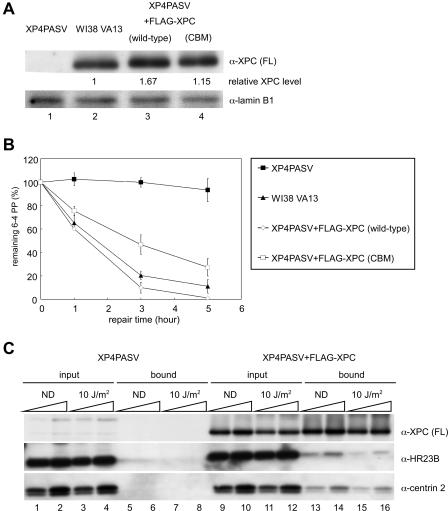
FIG. 1.
NER activity and complex formation of FLAG-tagged XPC stably expressed in XP4PASV cells. (A) XP4PASV cells were transfected with a construct stably expressing FLAG-tagged XPC, wild type or CBM. Cell lysates were prepared from one clone of each of the isolated transformants as well as from parental XP4PASV and control WI38 VA13 cells. Six micrograms of protein was subjected to immunoblotting with an anti-XPC (α-XPC) (FL) antibody. Lamin B1 was detected as a loading control. The amount of XPC in each lane was quantified, normalized to the lamin B1 signal, and indicated as a relative value compared to that of endogenous XPC in WI38 VA13 cells. (B) The indicated cells were irradiated with UV at 10 J/m2 and incubated for various times at 37°C in the presence of 6 mM thymidine. The levels of 6-4PP remaining in genomic DNA were quantified with a lesion-specific antibody. Mean values and standard errors were calculated from the results from at least two independent experiments. (C) A stable transformant expressing FLAG-XPC (wild type) and parental XP4PASV cells were irradiated with UV at 10 J/m2 or mock irradiated (ND). After further incubation at 37°C for 1 h, soluble extracts were subjected to immunoprecipitation with anti-FLAG antibody beads. XPC complex subunits in the extract (input) or in the immunoprecipitated fraction (bound) were detected by immunoblotting with indicated antibodies. Each lane contained 0.5 or 1% of the input extract and 5 or 10% of the bound fraction. α, anti.
Transient overexpression of XPC protein and binding assays.
Wild-type and mutant human XPC proteins, which were tagged with FLAG or glutathione _S_-transferase (GST) at the amino terminus, were transiently overexpressed from pCAGGS derivatives in XP4PASV cells (41). Typically, 4 μg of each expression construct was introduced into a 100-mm dish of XP4PASV cells with Lipofectamine Plus reagent (Invitrogen). Soluble cell extracts were prepared 20 h posttransfection as described above.
Soluble cell extracts (900 μl) containing overexpressed XPC proteins were incubated overnight at 4°C with 12.5 μl anti-FLAG M2 agarose (for FLAG-tagged XPC; Sigma) or 25 μl glutathione-Sepharose 4 fast flow (FF) (for GST-tagged XPC; Amersham Biosciences) beads. The beads were collected and washed twice with NP lysis buffer containing 0.3 M NaCl, three times with NP lysis buffer containing 1 M NaCl, and once with NP lysis buffer containing 0.3 M NaCl. Proteins retained on the beads were then eluted at 4°C for 2 h with 100 μl NP lysis buffer containing 0.3 M NaCl and 500 μg/ml FLAG peptide (Sigma) or 10 mM glutathione. The eluates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE), followed by immunoblot analysis.
Measurement of in vivo repair rates of UV-induced 6-4PPs.
Cells were cultured to 80% confluence in 100-mm dishes and maintained at 37°C for 2 h in medium containing 6 mM thymidine to prevent dilution of the lesions by DNA replication. Cells were then irradiated with UV (10 J/m2, under a germicidal lamp with a 254-nm peak) and cultured in the presence of 6 mM thymidine for various times to allow DNA repair. Genomic DNA was purified with the QIAamp DNA blood mini kit (QIAGEN), and the levels of remaining 6-4PP were determined by an enzyme-linked immunosorbent assay using a lesion-specific monoclonal antibody as previously described (37).
Establishment of stable cell lines expressing FLAG-tagged XPC protein.
cDNA encoding FLAG-XPC (wild type or mutant) was cloned into the vector pIREShyg (Clontech), from which FLAG-XPC and the hygromycin B resistance gene were transcribed as a single mRNA. Twenty micrograms of each construct was linearized and electroporated into a 100-mm dish of XP4PASV cells using a Gene Pulser II (Bio-Rad). Stable transformants were selected in the presence of 200 μg/ml hygromycin B (Invitrogen). The hygromycin B concentration in the culture medium was then gradually reduced. With the bicistronic expression system, the expression levels of FLAG-XPC naturally dropped to certain levels corresponding to the reduced selective pressure. This procedure made it rather easy to isolate clones that express appropriate levels of FLAG-XPC.
Preparation of recombinant proteins.
Centrin 2 was purified essentially according to the methods of Araki et al. (2), except that a HiTrap phenyl-FF (high sub) column (1 ml; Amersham Biosciences) was substituted for the phenyl-Superose step.
For preparation of the XPC-HR23B complex, recombinant FLAG-tagged XPC (wild type or mutant) and hexahistidine-tagged HR23B (HR23B-His) were purified separately. Recombinant baculoviruses expressing FLAG-XPC were generated with the Bac-to-Bac expression system (Invitrogen). Twenty 150-mm dishes of HighFive cells were infected with recombinant baculovirus at a multiplicity of infection of 10 and incubated at 27°C for 3 days. The cells were harvested, washed twice with ice-cold phosphate-buffered saline, and suspended in an eightfold volume of NP lysis buffer containing 0.3 M NaCl. After incubation on ice for 30 min, the soluble fraction was obtained by centrifugation at 20,000 × g for 30 min and dialyzed against buffer A (20 mM sodium phosphate [pH 7.8], 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.25 mM PMSF, and 0.1 M NaCl). Insoluble materials were removed by further centrifugation at 150,000 × g for 30 min, and the clarified extract was loaded onto a HiPrep 16/10 heparin FF column (Amersham Biosciences) equilibrated with buffer B (20 mM sodium phosphate [pH 7.8], 10% glycerol, 1 mM EDTA, 0.1 mM DTT, 0.25 mM PMSF, and 0.01% Triton X-100) containing 0.1 M NaCl. The column was successively washed with buffer B containing 0.1, 0.3, and 1 M NaCl, and the 1 M NaCl fraction was loaded onto an anti-FLAG M2 agarose column (7 ml) equilibrated with buffer B containing 0.3 M NaCl. After being washed with buffer B containing 1 M NaCl and then with buffer C (buffer B minus EDTA) containing 0.3 M NaCl, bound proteins were eluted with buffer C containing 0.3 M NaCl and 100 μg/ml FLAG peptide. To concentrate proteins and change the buffer, the proteins were loaded onto a HiTrap heparin high-performance (HP) column (1 ml; Amersham Biosciences) equilibrated with buffer C containing 0.3 M NaCl and eluted with buffer D (20 mM sodium phosphate [pH 7.8], 10% glycerol, 0.01% Triton X-100, 10 mM β-mercaptoethanol, 0.25 mM PMSF) containing 1 M NaCl. To reconstitute the heterodimeric complex, purified FLAG-XPC was mixed with the same amount (by weight) of HR23B-His and incubated on ice for 2 h. The mixture was then loaded onto a HiTrap chelating HP column (1 ml; Amersham Biosciences) prebound with nickel ions and equilibrated with buffer D containing 0.3 M NaCl and 20 mM imidazole. The column was washed with the same buffer, and bound proteins were eluted with buffer D containing 0.3 M NaCl and 100 mM imidazole. The eluate was loaded onto a Mini S PC 3.2/3 column (Amersham Biosciences) equilibrated with buffer D containing 0.3 M NaCl. After being washed with the same buffer, the reconstituted complex was eluted with a linear gradient of 0.3 to 1 M NaCl in buffer D. The peak fractions were pooled and dialyzed against buffer B containing 0.3 M NaCl and 20% sucrose.
Other human NER proteins, including XPA with an amino-terminal FLAG tag (FLAG-XPA) and heterotrimeric RPA, were also expressed in HighFive cells, and soluble extracts were prepared as for FLAG-XPC. Initial purification of FLAG-XPA was carried out using heparin-Sepharose and anti-FLAG M2 agarose columns as described above for FLAG-XPC. The eluate was further purified with a hydroxyapatite column (Econo-Pac CHT-II; Bio-Rad), essentially as described previously (24).
For purification of RPA, the soluble extract was loaded onto a HiPrep 16/10 DEAE FF column (Amersham Biosciences) equilibrated with buffer E (25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 10% glycerol, 0.01% Triton X-100, 1 mM DTT, 0.25 mM PMSF) containing 0.1 M NaCl. Bound proteins were eluted stepwise with buffer E containing 0.6 and 1 M NaCl. RPA recovered in the 0.6 M NaCl fraction was further purified using HiTrap Blue HP and RESOURCE Q columns (Amersham Biosciences) essentially as described previously (18).
Human XPF-ERCC1-His (1) and XPG (42) were expressed in insect cells and purified as described previously. Both proteins were further purified by passing through a Superdex 200 PC 1.6/30 column (Amersham Biosciences) equilibrated with buffer E containing 0.3 M NaCl. TFIIH was purchased from Protein One.
Binding assay.
Purified FLAG-XPC-HR23B-His complex (1 μg) was incubated on ice for 2 h with the same amount (by weight) of centrin 2 in 30 μl buffer B containing 0.3 M NaCl. Twenty-five microliters of anti-FLAG M2 agarose beads was added, and incubation was continued on ice for 2 h with occasional mixing. The beads were washed twice with buffer B containing 0.3 M NaCl, three times with buffer B containing 1 M NaCl, and once with buffer B containing 0.3 M NaCl. Proteins retained on the beads were eluted at 4°C for 30 min with 50 μl buffer B containing 0.3 M NaCl and 500 μg/ml FLAG peptide. The samples were subjected to SDS-PAGE and detected by immunoblotting with antibodies specific for each subunit of the XPC complex.
Nucleotide excision repair assay.
An internally 32P-labeled DNA substrate containing a site-specific 6-4PP was prepared as described previously (63). The DNA substrate (∼100,000 cpm) was incubated at 30°C for 1 h in 25 μl of a reaction mixture containing 40 mM HEPES-KOH (pH 7.8), 70 mM NaCl, 0.5 mM DTT, 5 mM MgCl2, 0.5 mM EDTA, 2 mM ATP, 20 μM concentrations (each) of dATP, dGTP, and dTTP, 8 μM dCTP, 22.5 mM creatine phosphate (di-Tris; Sigma), creatine phosphokinase (1.25 μg, type I; Sigma), BSA (9 μg), XP3BE (group C) whole-cell extract (100 μg protein), and the indicated amounts of the XPC-HR23B complex. For reconstituted NER reaction mixtures (15 μl), FLAG-XPA (25 ng), XPF-ERCC1-His (12.5 ng), RPA (100 ng), XPG (12 ng), TFIIH (134 ng), and the indicated amounts of the XPC-HR23B complex and centrin 2 were included instead of the XP3BE extract and the NaCl concentration was adjusted to 100 mM. Before addition of DNA substrates, the reaction mixtures were preincubated at 30°C for 10 min. DNA samples were purified and subjected to 10% denaturing PAGE, followed by autoradiography.
Electrophoretic mobility shift assay.
32P-labeled blunt-ended DNA fragments containing a site-specific lesion were prepared as described previously (63). Binding reactions were preformed at 30°C for 30 min in a 10-μl solution containing 20 mM sodium phosphate (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, BSA (1 μg), 0.01% Triton X-100, 5% glycerol, covalently closed circular plasmid DNA (0.5 ng), 32P-labeled substrate DNA (0.35 nM), and the indicated amounts of purified XPC-HR23B and centrin 2. The resulting reaction mixtures were subjected to 4% native PAGE as described previously (63), except that the glutaraldehyde fixation step was omitted. Gels were dried and exposed to X-ray film (X-OMAT; Kodak) at −80°C with intensifying screens. The percentage of substrate bound to the XPC protein complex was quantified using the BAS2500 bioimaging analyzer (Fujifilm).
Other materials and methods.
Anti-XPC (FL), anti-hHR23B (60), anti-XPC (C-terminal) (43), anti-centrin 2 (2), and anti-6-4PP (6-4M2) (37) antibodies were obtained as described previously. Anti-lamin B1 antibody was purchased from Santa Cruz Biotechnology. For immunoblotting, proteins were separated by SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Immobilon-P; Millipore). Detection was performed by chemiluminescence using appropriate alkaline phosphatase-conjugated secondary antibodies (Sigma) and CDP-Star (Roche Diagnostics) as a substrate. Quantification was carried out with the luminescent imaging analyzer (LAS-1000 plus; Fujifilm) and accompanying software (ImageGauge v3.45), and blots were also exposed to X-ray film. The XP3BE whole-cell extract was prepared according to a previously described method (63). Concentrations of purified proteins were determined by the Bradford method (7) using a reagent purchased from Bio-Rad and BSA as a standard.
RESULTS
Centrin 2 is expressed in excess relative to XPC.
To obtain insights into the functions of centrin 2 in GG-NER, we first examined the in vivo behavior of centrin 2 in the XPC complex. To specifically examine centrin 2 associated with XPC, we isolated a stably transformed cell line that expressed FLAG-tagged XPC protein in the XPC-deficient XP4PASV background. Total cell lysates prepared from the transformed cells, designated as XP4PASV+FLAG-XPC (wild-type), as well as from simian virus 40-transformed normal human fibroblasts, WI38 VA13, were subjected to immunoblot analyses, which revealed that the isolated transformant expressed a level of FLAG-XPC that was ∼1.7 times greater than the level of endogenous XPC expressed by WI38 VA13 cells (Fig. 1A, compare lane 3 with lane 2). We assessed the GG-NER activity of these transformed cells by measuring the rate of 6-4PP removal after UV irradiation. The overall level of 6-4PP in genomic DNA was quantified with a lesion-specific monoclonal antibody. As shown in Fig. 1B, the transformant displayed similar but slightly faster kinetics of 6-4PP repair than did WI38 VA13 cells, indicating that the exogenously expressed FLAG-XPC was functional. To examine the subunit composition of the XPC complex, transformed and control cells were UV irradiated (at 10 J/m2) or mock irradiated and FLAG-XPC was immunoprecipitated with anti-FLAG antibody beads. We confirmed that HR23B and centrin 2 were not bound to anti-FLAG beads when lysates from parental XP4PASV cells were used (Fig. 1C, lanes 5 to 8). In agreement with previous findings (60, 69), only a small percentage of HR23B was coprecipitated with FLAG-XPC. Whereas FLAG-XPC appeared to be concentrated by immunoprecipitation, the intensity of HR23B in the bound fraction was much weaker than in the input fraction (compare lanes 13 and 14 with lanes 9 and 10). Quantitative analyses revealed that 5% of total HR23B was complexed with FLAG-XPC in these cells. Likewise, centrin 2 also appeared to be present in a large excess over XPC, and approximately 10% of total centrin 2 was associated with FLAG-XPC. In addition, centrin 2 was normally expressed in XPC-deficient parental cells (lanes 1 to 4), which is consistent with its functions in processes other than GG-NER. The observed stoichiometry of these subunits did not change significantly after UV irradiation (compare lanes 15 and 16 with lanes 13 and 14). Although some XPC underwent posttranslational modification upon UV irradiation (lanes 11, 12, 15 and 16) (63a), no detectable change in electrophoretic mobility was observed for HR23B and centrin 2.
Determination of the centrin 2-interacting domain of XPC.
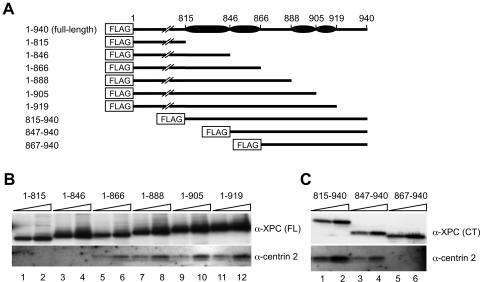
We next mapped the centrin 2-interacting domain of XPC. Centrin 2 belongs to a calmodulin superfamily, most of whose members interact with α-helical regions of partner proteins (44, 73). In analyses of several XPC truncation mutants, we found that a C-terminal region of XPC comprising 222 amino acids (719 to 940) is sufficient for binding to centrin 2 (data not shown). Secondary structure analysis with the PHD program (49) suggested the presence of several α-helices in this region, as shown in Fig. 2A. Based on these results, we constructed a set of truncated XPC proteins (Fig. 2A). These mutant proteins were N-terminally tagged with the FLAG motif and transiently overexpressed in XP4PASV cells. FLAG-tagged proteins were immunoprecipitated from soluble cell extracts, and endogenous centrin 2 coprecipitated with the truncated XPC protein was then detected by immunoblotting (Fig. 2B and C). Although the mutant XPC (amino acids 1 to 866), which lacks the C-terminal 74 amino acids, still bound centrin 2 (Fig. 2B, lanes 5 and 6), centrin 2 was not coprecipitated with more truncated proteins (Fig. 2B, lanes 1 to 4). Among N-terminal deletion mutants, only XPC (amino acids 867 to 940) failed to interact with centrin 2 (Fig. 2C), indicating that the putative α-helical region defined by residues 847 to 866 is necessary for centrin 2 binding. The amino terminus of this short domain was fused to the GST tag and overexpressed in XP4PASV cells. When the soluble cell extract was subjected to the GST pull-down assay, centrin 2 was found to coprecipitate with the GST fusion protein (Fig. 3B, lanes 4 to 6) but not with GST alone (lanes 1 to 3). Therefore, these 20 amino acid residues are not only necessary but also sufficient for interaction of XPC with centrin 2.
FIG. 2.
Determination of the centrin 2-binding domain of XPC. (A) Schematic representation of truncated XPC proteins fused to an N-terminal FLAG tag. The solid ovals indicate predicted α-helical regions. (B and C) Truncated XPC proteins shown in panel A were transiently overexpressed in XP4PASV cells. Solubilized FLAG-XPC proteins were immunoprecipitated with anti-FLAG antibody beads and detected by immunoblotting with anti-XPC (FL or C-terminal [CT]) antibodies (upper panels). The bound proteins were eluted with FLAG peptide as described in Materials and Methods, and 10 and 20% of each eluate were loaded in parallel. Coprecipitated centrin 2 was also visualized by immunoblotting (lower panel). α, anti.
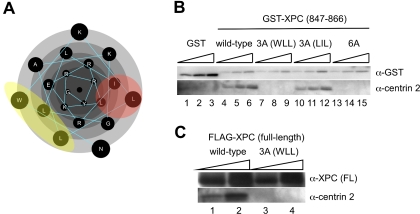
FIG. 3.
Generation of a mutant XPC protein that cannot interact with centrin 2. (A) Amino acids comprising the centrin 2-binding domain (847-NWKLLAKGLLIRERLKRRYG-866) are presented as a helical wheel plot with Protean software (Lasergene; DNASTAR, Inc.). Mutated amino acids are shaded in yellow and red. (B) The centrin 2-binding domain of XPC (847 to 866) and its mutant versions were fused to an N-terminal GST tag and overexpressed in XP4PASV cells. Soluble cell extracts were subjected to GST pull-down assays, and GST fusion proteins and coprecipitated centrin 2 were detected by immunoblotting with anti-GST and anti-centrin 2 antibodies, respectively. Ten, 20, and 30% of each eluate were loaded in parallel. (C) FLAG-tagged full-length XPC, either wild-type or bearing the 3A (WLL) mutation, was overexpressed in XP4PASV cells. Solubilized FLAG-XPC proteins were immunoprecipitated using an anti-FLAG antibody, and precipitated fractions were subjected to SDS-PAGE followed by immunoblotting with an anti-XPC (FL) or anti-centrin 2 antibody. Five and 15% of each eluate were loaded in parallel. α, anti.
Generation of mutant XPC proteins that cannot bind centrin 2.
To generate mutant XPC proteins that cannot interact with centrin 2, we sought to identify amino acids in the 847 to 866 domain that are important for interaction. Because the heterotrimeric XPC complex is stable even in the presence of high salt concentrations during purification (up to 1.5 M KCl) (2), complex formation is conjectured to be mediated mainly by hydrophobic interactions. When we plotted amino acids in the centrin 2-binding domain of XPC as a helical wheel, we noted at least two hydrophobic surfaces (Fig. 3A). One consists of the W848, L851, and L855 residues, and the other consists of the L850, I857, and L861 residues. To examine the roles of these hydrophobic residues, each set of three amino acids was replaced by alanine, generating mutants designated 3A (WLL) and 3A (LIL). Another mutant in which all 6 amino acids were changed to alanine (6A) was also constructed. The centrin 2-binding domains harboring these mutations were tagged with the GST motif and overexpressed in XP4PASV cells. Their ability to interact with centrin 2 was tested by the GST pull-down assay (Fig. 3B, lanes 7 to 15). Although the 3A (LIL) mutant retained centrin 2-binding activity, the 3A (WLL) and 6A mutants apparently did not. These results suggest that the hydrophobic surface defined by the W848, L851, and L855 residues is important for interaction with centrin 2. Furthermore, we also expressed FLAG-tagged full-length XPC protein containing the 3A (WLL) mutations, which was subjected to immunoprecipitation (Fig. 3C). The results of this experiment indicate that the 3A (WLL) mutations also prevent the full-length protein from interacting with centrin 2. Hereafter, the full-length XPC protein harboring the 3 amino acid substitutions is designated XPC (CBM) (_c_entrin 2-_b_inding _m_utant).
Reduced in vivo NER activity of XPC (CBM).
To identify possible roles for centrin 2 binding in GG-NER in vivo, we established stable transformants of XP4PASV cells expressing FLAG-tagged XPC (CBM) (Fig. 1A). One isolated clone was found to express a level of FLAG-XPC (CBM) that was ∼1.2 times greater than the level of endogenous XPC expressed by WI38 VA13 cells (compare lane 4 with lane 2). To assess GG-NER activity, these cells were irradiated with UV at 10 J/m2 and cultured at 37°C for various time periods to measure the kinetics of 6-4PP repair. As shown in Fig. 1B, repair of 6-4PP was significantly retarded in the transformant expressing FLAG-XPC (CBM). In cells expressing wild-type FLAG-XPC, the levels of residual 6-4PP were 10% ± 4.6% and 1% ± 0.5% at 3 and 5 h postirradiation, respectively, whereas 47% ± 8.3% and 27% ± 7.5% of the lesions remained at the same time points in XPC (CBM) cells. This difference in the rate of 6-4PP repair is not due to the different expression levels of FLAG-XPC because another clone that expressed a level of FLAG-XPC (CBM) that was ∼2.2 times greater than the level of endogenous XPC expressed by WI38 VA13 cells still showed slower 6-4PP repair (data not shown). These results raised the possibility that the presence of centrin 2 in the XPC complex affects the efficiency of GG-NER, although it is not essential for repair.
XPC (CBM) exhibits diminished NER activity in vitro.
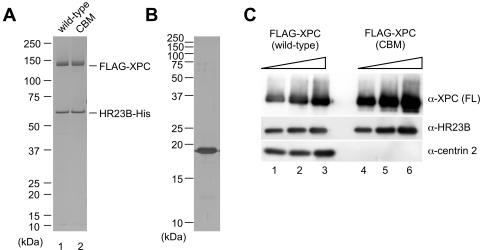
To determine whether centrin 2 plays direct roles in NER reactions, we carried out in vitro experiments. FLAG-tagged XPC (wild type or CBM) was expressed in insect cells and purified to reconstitute in vitro a heterodimer with hexahistidine-tagged HR23B (HR23B-His). The 3 amino acid substitutions in XPC (CBM) did not interfere with binding to HR23B (Fig. 4A). Each of the XPC heterodimers was then incubated with purified centrin 2, which was separately expressed in Escherichia coli. When FLAG-XPC was immunoprecipitated with anti-FLAG antibody beads, centrin 2 was found to coprecipitate with wild-type XPC (Fig. 4C, lanes 1 to 3) but not with XPC (CBM) (lanes 4 to 6), as expected.
FIG. 4.
In vitro complex formation of purified XPC (CBM). (A and B) Purified recombinant XPC-HR23B-His complex (100 ng) containing wild-type FLAG-XPC (lane 1) or FLAG-XPC (CBM) (lane 2) and purified centrin 2 (1 μg) was subjected to SDS-PAGE and silver stained. (C) One microgram of purified centrin 2 was incubated with 1 μg of the FLAG-XPC-HR23B-His complex, either wild type (lane 1 to 3) or CBM (lane 4 to 6), and then pulled down with anti-FLAG antibody beads. After unbound proteins were washed out, 2, 4, or 8% of the bound proteins was subjected to SDS-PAGE and detected by immunoblotting using antibodies recognizing individual subunits. α, anti.
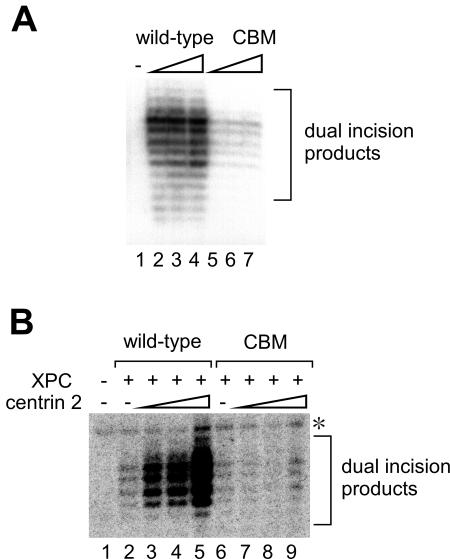
To examine the effects of centrin 2 on NER, we carried out in vitro dual-incision assays using a whole-cell extract from XPC-deficient XP3BE cells. Like the XP4PASV cell extract analyzed in Fig. 1C, this extract also contained free centrin 2, which allowed reconstitution of the heterotrimeric complex upon the inclusion of XPC-HR23B (data not shown). An internally 32P-labeled double-stranded circular DNA containing a single 6-4PP was incubated with XP3BE cell extract, with or without the XPC-HR23B complex (wild type or CBM) (Fig. 5A). As anticipated, dual incision products containing the photolesion were detected when the wild-type XPC complex was included (lanes 2 to 4). The XPC (CBM) complex, on the other hand, was proficient for damage excision but was significantly less efficient than the wild-type XPC complex (lanes 5 to 7).
FIG. 5.
Centrin 2 stimulates cell-free NER incision. (A) NER dual-incision assays were carried out with a crude extract from XP3BE cells and a 32P-labeled DNA substrate containing a single 6-4PP. Purified FLAG-XPC-HR23B-His complexes, wild type (lanes 2 to 4) or CBM (lanes 5 to 7), were included as indicated. The concentrations of the XPC complex were 0.22 nM (lanes 2 and 5), 0.44 nM (lanes 3 and 6), and 0.88 nM (lanes 4 and 7), respectively, while XPC complex was omitted from the reaction mixture (lane 1) as a negative control. Purified DNA samples containing dual-incision products were subjected to 10% denaturing PAGE. (B) NER dual-incision assays with purified NER factors involving FLAG-XPC-HR23B-His complexes, wild type (8.8 nM, lanes 2 to 5) or CBM (2.2 nM, lanes 6 to 9). Various concentrations of purified centrin 2 (2.2, 4.4, and 8.8 nM) were also included where indicated. As a negative control, both XPC and centrin 2 were omitted (lane 1). The asterisk indicates a nonspecific band that appeared in an XPC-independent manner. +, present; −, absent.
Although the observed differences in NER activity may be due to the ability of XPC to bind to centrin 2, it is also possible that the amino acid substitutions in XPC (CBM) may compromise interactions with NER factors or other factors present in the crude extract. To confirm the direct involvement of centrin 2 in NER, we performed dual-incision assays with purified NER factors. The internally labeled 6-4PP substrate was incubated with five essential NER factors (XPA, TFIIH, XPF-ERCC1, XPG, and RPA) in the presence or absence of the XPC-HR23B complex (wild type or CBM) and various concentrations of centrin 2 (Fig. 5B). In this system, both wild-type and mutant XPC complexes supported dual incision near the 6-4PP site, and incision was undetectable in their absence (Fig. 5B, compare lanes 2 and 6 with lane 1). When centrin 2 was added to these reactions, dual incision mediated by the wild-type XPC complex was greatly enhanced (lanes 3 to 5). In marked contrast, the efficiency of XPC (CBM) complex-dependent incision was not influenced by the inclusion of centrin 2 (lanes 7 to 9). Therefore, centrin 2 plays a direct stimulatory role in NER, for which complex formation with XPC appears to be required.
Centrin 2 stimulates DNA binding of XPC.
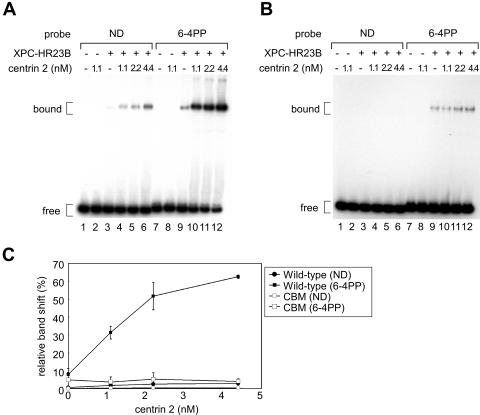
To obtain insights into the mechanism by which centrin 2 stimulates NER, we examined its influence on the binding of DNA by the XPC complex. For this purpose, we carried out electrophoretic mobility shift assays using a 32P-labeled 180-bp DNA fragment with or without a site-specific 6-4PP. In agreement with our previous findings, the XPC-HR23B heterodimer exhibited a specific binding affinity for 6-4PP, which was almost unaffected by the amino acid substitutions in XPC (CBM) (Fig. 6A and B, compare lane 9 with lane 3). Intriguingly, the damage-specific binding of the wild-type XPC complex was dramatically enhanced by the addition of centrin 2 (Fig. 6A, lanes 9 to 12, and C, quantitative data). While complex formation with nondamaged DNA was also stimulated 4-fold in the presence of centrin 2, binding to the 6-4PP probe was enhanced up to 20-fold under the same conditions. On the other hand, little stimulation was observed for the XPC (CBM) complex (Fig. 6B and C). Based on these data, we conclude that complex formation with centrin 2 reinforces the lesion-binding activity of XPC, which can account for the stimulatory effect of centrin 2 in NER.
FIG. 6.
Centrin 2 enhances the DNA binding activity of XPC. (A and B) Electrophoretic mobility shift assays were carried out with purified FLAG-XPC-HR23B-His complexes, wild type (4.4 nM) (A) or CBM (1.1 nM) (B), and a 32P-labeled, 180-bp probe (0.35 nM) with or without a single 6-4PP. Various concentrations of centrin 2 were also included as indicated. The resulting DNA-protein complexes were resolved by 4% native PAGE. (C) The percentages of the shifted DNA substrates were quantified for panels A and B and plotted. Mean values and standard errors were calculated from the results from at least two independent experiments. +, present; −, absent.
DISCUSSION
Complex formation of centrin 2 with XPC.
In this study, we showed that, like HR23B, centrin 2 is normally expressed even in the absence of XPC and that as little as 10% of total centrin 2 is included in XPC complexes in XP4PASV+FLAG-XPC cells (Fig. 1). It has been reported that less than 10% of total centrin 2 in human KE37 lymphoblast cells is localized to the centrosome, while 50% is recovered in the nuclear fraction upon cell fractionation. These data indicate that most centrin 2 is not associated with either the centrosome or XPC, suggestive of multiple functions beyond centrosome duplication. Upon UV irradiation, the stoichiometry of XPC, HR23B, and centrin 2 is almost unaltered, suggesting that centrin 2 may constitutively form a stable complex with XPC and that it is not inducibly incorporated into this complex when DNA is damaged. Furthermore, centrin 2 in the XPC complex does not show a detectable shift in electrophoretic mobility when cells are irradiated with UV, indicating that extensive posttranslational modification, such as phosphorylation, does not occur. However, it is still possible that a minor centrin 2 fraction dissociates transiently from XPC during the NER reaction or that it may undergo posttranslational modification. Further studies are necessary to understand the in vivo behaviors of centrin 2 in the context of NER.
Centrin 2-binding domain of XPC.
A database that contains known calmodulin target sequences and their properties was recently developed which allows calmodulin-binding sites within given amino acid sequences to be predicted (73). As centrin 2 belongs to the calmodulin superfamily, Popescu et al. (46) used this database and reported that centrin 2 binds to the region of XPC corresponding to amino acids 847 to 863. We independently mapped the centrin 2-binding domain of XPC based on a biochemical binding assay using a series of deletion mutants. Our data indicate that the putative α-helical region from amino acid residues 847 to 866 is sufficient for the interaction with centrin 2, largely consistent with the results of Popescu et al. (46). It has been proposed that many calmodulin-binding sequences adopt an amphiphilic α-helical structure that consists of a positively charged surface and a hydrophobic surface (44). Calmodulin-binding sequences have been typically classified into four groups: 1-10, 1-14, 1-16, and IQ motifs (73). The centrin 2-binding sequence of XPC is indeed predicted to form a basic amphiphilic α-helical structure and is categorized as a 1-5-8-14 motif, a subclass of the 1-14 motif (Fig. 7A). Thus far, there are two other members of this group for which the molecular structures have been solved as complexes with calmodulin, smooth muscle myosin light chain kinase (smMLCK) and skeletal muscle myosin light chain kinase (skMLCK) (22, 34). In both cases, interaction with calmodulin is mediated via hydrophobic interactions (Fig. 7A), and the binding regions are wrapped by two EF-hand domains of calmodulin. Although the positions of hydrophobic amino acids that may participate in interaction with calmodulin are conserved among members of this group, including XPC, only one hydrophobic surface, including W848, L851, and L855 of XPC, appears to be important for centrin 2 binding. In agreement with this, only the C-terminal half of centrin 2 may be required for interaction with XPC (10, 31, 46). Although the primary amino acid sequence of the centrin 2-binding domain of XPC is classified as a 1-14 motif, the structural basis of complex formation may be different than that for typical calmodulin complexes like smMLCK and skMLCK. On the other hand, two recently solved calmodulin complex structures indicated that two EF-hand domains in calmodulin interact with individual α-helices in partner proteins (9, 56). Therefore, the N-terminal half of centrin 2 may interact with a different site in XPC or with other factors, although such interactions may be much weaker than those investigated in this study.
FIG. 7.
Sequence alignments of the centrin 2-binding domain. (A) Alignment of amino acid sequences of the centrin 2-binding domain of XPC and the calmodulin-binding sites of smMLCK and skMLCK, both of which are classified as 1-5-8-14 motifs. Typical positions of hydrophobic amino acids for this motif are indicated by asterisks. The amino acids that are responsible for the interaction with centrin 2 or with calmodulin are indicated in red. (B) Evolutionary conservation of amino acid sequences of the putative centrin 2-binding domain among XPC homologs of human (Hs), mouse (Mm), Drosophila melanogaster (Dm), Arabidopsis thaliana (At), and Saccharomyces cerevisiae (Sc). The ClustalW alignment of amino acid sequences was made with MacVector software (Accelrys, Inc.). Identical and similar amino acids with respect to the human sequence are indicated by solid and shaded boxes, respectively. The positions of the three amino acids that are involved in interaction with centrin 2 are indicated by red boxes.
The amino acid sequences of the centrin 2-binding site appear to be well conserved among XPC homologs in various species (Fig. 7B). Notably, the three amino acids comprising the hydrophobic interaction surface are conserved almost completely. Therefore, the NER function of centrin might have evolved in lower eukaryotes, although complex formation between Rad4p and Cdc31p in yeast has not yet been demonstrated.
Centrin 2 stimulates NER both in vivo and in vitro.
To investigate the function of centrin 2 in NER, we constructed the mutant XPC (CBM), which does not interact with centrin 2 (Fig. 3C). As reported by several groups (2, 4, 38), centrin 2 is not essential for cell-free NER, so that XPC (CBM) appears to retain a basal activity in the reconstituted NER system involving only purified proteins (Fig. 5B). However, addition of centrin 2 results in a marked increase in dual-incision products containing 6-4PP in the presence of wild-type XPC but not in the presence of XPC (CBM). These data indicate that centrin 2 plays a direct stimulatory role in NER that requires physical interaction between centrin 2 and XPC. Previous experiments using UV-irradiated simian virus 40 minichromosomes as substrates showed that in vitro NER activity is not detectably affected by the presence or absence of centrin 2 (2). This discrepancy might be due to differences in assay conditions because the balance among protein factors may vary in reconstitutions of the multicomponent reaction.
It is well documented that the XPC-HR23B heterodimeric complex is involved in damage recognition for NER and that it specifically binds to various lesions, such as 6-4PP (5, 25, 61, 63). Our present study shows that the DNA binding activity of XPC-HR23B is significantly reinforced by the addition of centrin 2, for which physical interaction between XPC and centrin 2 is essential (Fig. 6). Electrophoretic mobility shift assays showed that centrin 2 stimulates the binding of XPC-HR23B to DNA containing 6-4PP by up to 20-fold and enhances binding to nondamaged DNA by 4-fold. The estimated equilibrium dissociation constant of the interaction between XPC-HR23B and the 6-4PP DNA is reduced from 5.1 × 10−8 to 2.5 × 10−9 M by the addition of saturating concentrations of centrin 2. In the case of nondamaged DNA, the equilibrium dissociation constant is reduced from 6.0 × 10−7 to 1.5 × 10−7 M under the same conditions. Complex formation with centrin 2 thus appears to augment not only the affinity of binding to DNA alone but also damage specificity to some extent. Although the XPC-HR23B heterodimer alone poorly recognizes CPD (63, 64), we observed specific (but nonetheless low) binding to CPD following the addition of centrin 2 (data not shown). This enhancement of lesion binding may be primarily responsible for the observed stimulation of cell-free NER by centrin 2. As centrin 2 itself does not bind to DNA (Fig. 6A, lane 2), it is likely that it induces a conformational change in XPC that contributes to the elevated affinity of the latter for damaged DNA. On the other hand, TFIIH interacts directly with XPC (3, 74), and the TFIIH-binding domain of XPC maps to the C terminus (816 to 940) (68). Because this domain includes the centrin 2-binding site, it is conceivable that complex formation with centrin 2 may affect interaction with TFIIH. However, in vitro binding experiments using purified factors did not reveal significant effects of centrin 2 on the XPC-TFIIH interaction (data not shown), although it remains possible that centrin 2 facilitates the recruitment and/or entry of TFIIH to lesion sites by stabilizing the XPC-DNA complex and/or by enhancing XPC-induced DNA bending (23).
Compared to the marked stimulatory effect of centrin 2 on in vitro NER, the contribution of the XPC-centrin 2 interaction to the in vivo repair rate of 6-4PP is less pronounced (Fig. 1B). We recently found that XPC physically interacts with UV-DDB, another damage recognition complex involved in GG-NER (63a). Although UV-DDB is not essential for cell-free NER, it displays a much higher binding affinity than XPC for both 6-4PP and CPD (5, 16, 47, 67) and appears to promote damage recognition through the recruitment of XPC, at least for CPD (14). Therefore, in vivo GG-NER activity may not be simply proportional to the damage binding ability of XPC, particularly in the presence of UV-DDB. The possible roles of centrin 2 in the XPC-UV-DDB interaction are now under investigation.
As described above, centrin 2 is a member of the calmodulin superfamily and has calcium-binding activity. Although in the absence of calcium, centrin 2 still binds to a peptide corresponding to amino acids 847 to 863 of XPC, affinity is increased by a factor of 28 in the presence of saturating concentrations of calcium (46). The hydrophobic patch of centrin 2 may be exposed in the presence of calcium (10, 31). Because our data strongly indicate that centrin 2 interacts with XPC via hydrophobic interactions (Fig. 3 and 7), centrin 2 may exist as a calcium-bound form in the XPC complex. Notably, the XPC heterotrimer appears to be very stable during purification, although EDTA was included in all buffers at 1 mM or higher concentrations (2, 30). This suggests that calcium ions, if present, may be tightly bound within the complex. A potential role for calcium in the NER reaction has not been extensively examined. To address this role, it may be necessary to purify centrin 2 in a calcium-free form and test it in reconstitution of the XPC complex and in cell-free NER.
In conclusion, we have unveiled a novel function for centrin 2, the stimulation of GG-NER by promoting the damage recognition activity of XPC via complex formation. Although centrin 2 has been supposed to be dispensable for NER, at least in vitro, it plays a vital role in damage recognition beyond mere complex stabilization. This is the first example that shows a functional alteration in a target protein induced by centrin 2 binding. It has been recently reported that the absence of centrin 2 in Arabidopsis thaliana results in enhanced homologous recombination (36), suggesting that centrin 2 may have multiple functions in the nucleus. It remains to be determined why centrin 2 is commonly involved in apparently unrelated functions such as centrosome duplication, NER, and homologous recombination. A fascinating possibility is that centrin 2 may link DNA repair with the cell division apparatus, thereby regulating cellular damage responses such as checkpoints and apoptosis. It has been reported that UV irradiation results in an elevation of cytoplasmic calcium, which may be involved in the induction of apoptosis (for a review, see reference 33). Further detailed studies on the NER function and intracellular behaviors of centrin 2 will shed light on its additional functions in DNA repair as well as in damage-induced signal transduction.
Acknowledgments
We thank all the members of the Cellular Physiology Laboratory for helpful discussions and encouragement. We also thank Y. Ichikawa and R. Nakazawa (Bioarchitect Research Group, RIKEN) for DNA sequencing.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by the Human Frontier Science Program, and by Solution Oriented Research for Science and Technology (SORST) from the Japan Science and Technology Agency. This work was also supported by the Bioarchitect Research Project of RIKEN. R.N. was supported by a fellowship from the Center of Excellence (COE) of the Japan Society for the Promotion of Science.
REFERENCES
- 1.Araki, M., C. Masutani, T. Maekawa, Y. Watanabe, A. Yamada, R. Kusumoto, D. Sakai, K. Sugasawa, Y. Ohkuma, and F. Hanaoka. 2000. Reconstitution of damage DNA excision reaction from SV40 minichromosomes with purified nucleotide excision repair proteins. Mutat. Res. 459**:**147-160. [DOI] [PubMed] [Google Scholar]
- 2.Araki, M., C. Masutani, M. Takemura, A. Uchida, K. Sugasawa, J. Kondoh, Y. Ohkuma, and F. Hanaoka. 2001. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 276**:**18665-18672. [DOI] [PubMed] [Google Scholar]
- 3.Araújo, S. J., E. A. Nigg, and R. D. Wood. 2001. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol. Cell. Biol. 21**:**2281-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araújo, S. J., F. Tirode, F. Coin, H. Pospiech, J. E. Syvaoja, M. Stucki, U. Hübscher, J.-M. Egly, and R. D. Wood. 2000. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 14**:**349-359. [PMC free article] [PubMed] [Google Scholar]
- 5.Batty, D., V. Rapic'-Otrin, A. S. Levine, and R. D. Wood. 2000. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol. 300**:**275-290. [DOI] [PubMed] [Google Scholar]
- 6.Baum, P., C. Furlong, and B. Byers. 1986. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc. Natl. Acad. Sci. USA 83**:**5512-5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72**:**248-254. [DOI] [PubMed] [Google Scholar]
- 8.de Laat, W. L., N. G. J. Jaspers, and J. H. J. Hoeijmakers. 1999. Molecular mechanism of nucleotide excision repair. Genes Dev. 13**:**768-785. [DOI] [PubMed] [Google Scholar]
- 9.Drum, C. L., S. Z. Yan, J. Bard, Y.-Q. Shen, D. Lu, S. Soelaiman, Z. Grabarek, A. Bohm, and W. J. Tang. 2002. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature 415**:**396-402. [DOI] [PubMed] [Google Scholar]
- 10.Durussel, I., Y. Blouquit, S. Middendorp, C. T. Craescu, and J. A. Cox. 2000. Cation- and peptide-binding properties of human centrin 2. FEBS Lett. 472**:**208-212. [DOI] [PubMed] [Google Scholar]
- 11.Errabolu, R., M. A. Sanders, and J. L. Salisbury. 1994. Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J. Cell Sci. 107**:**9-16. [DOI] [PubMed] [Google Scholar]
- 12.Evans, E., J. Fellows, A. Coffer, and R. D. Wood. 1997. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 16**:**625-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, E., J. G. Moggs, J. R. Hwang, J.-M. Egly, and R. D. Wood. 1997. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 16**:**6559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitch, M. E., S. Nakajima, A. Yasui, and J. M. Ford. 2003. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J. Biol. Chem. 278**:**46906-46910. [DOI] [PubMed] [Google Scholar]
- 15.Friedberg, E. C. 2001. How nucleotide excision repair protects against cancer. Nat. Rev. Cancer 1**:**22-33. [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara, Y., C. Masutani, T. Mizukoshi, J. Kondo, F. Hanaoka, and S. Iwai. 1999. Characterization of DNA recognition by the human UV-damaged DNA-binding protein. J. Biol. Chem. 274**:**20027-20033. [DOI] [PubMed] [Google Scholar]
- 17.Giglia-Mari, G., F. Coin, J. A. Ranish, D. Hoogstraten, A. Theil, N. Wijgers, N. G. Jaspers, A. Raams, M. Argentini, P. J. van der Spek, E. Botta, M. Stefanini, J.-M. Egly, R. Aebersold, J. H. J. Hoeijmakers, and W. Vermeulen. 2004. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 36**:**714-719. [DOI] [PubMed] [Google Scholar]
- 18.Henricksen, L. A., C. B. Umbricht, and M. S. Wold. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269**:**11121-11132. [PubMed] [Google Scholar]
- 19.Hoeijmakers, J. H. J. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411**:**366-374. [DOI] [PubMed] [Google Scholar]
- 20.Huang, B., A. Mengersen, and V. D. Lee. 1988. Molecular cloning of cDNA for caltractin, a basal body-associated Ca2+-binding protein: homology in its protein sequence with calmodulin and the yeast CDC31 gene product. J. Cell Biol. 107**:**133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, B., D. M. Watterson, V. D. Lee, and M. J. Schibler. 1988. Purification and characterization of a basal body-associated Ca2+-binding protein. J. Cell Biol. 107**:**121-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikura, M., G. M. Clore, A. M. Gronenborn, G. Zhu, C. B. Klee, and A. Bax. 1992. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256**:**632-638. [DOI] [PubMed] [Google Scholar]
- 23.Jani ć ijevi ć , A., K. Sugasawa, Y. Shimizu, F. Hanaoka, N. Wijgers, M. Djurica, J. H. J. Hoeijmakers, and C. Wyman. 2003. DNA bending by the human damage recognition complex XPC-HR23B. DNA Repair 2**:**325-336. [DOI] [PubMed] [Google Scholar]
- 24.Jones, C. J., and R. D. Wood. 1993. Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry 32**:**12096-12104. [DOI] [PubMed] [Google Scholar]
- 25.Kusumoto, R., C. Masutani, K. Sugasawa, S. Iwai, M. Araki, A. Uchida, T. Mizukoshi, and F. Hanaoka. 2001. Diversity of the damage recognition step in the global genomic nucleotide excision repair in vitro. Mutat. Res. 485**:**219-227. [DOI] [PubMed] [Google Scholar]
- 26.Laoukili, J., E. Perret, S. Middendorp, O. Houcine, C. Guennou, F. Marano, M. Bornens, and F. Tournier. 2000. Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J. Cell Sci. 113**:**1355-1364. [DOI] [PubMed] [Google Scholar]
- 27.LeDizet, M., J. C. Beck, and W. E. Finkbeiner. 1998. Differential regulation of centrin genes during ciliogenesis in human tracheal epithelial cells. Am. J. Physiol. 275**:**L1145-L1156. [DOI] [PubMed] [Google Scholar]
- 28.Lee, V. D., and B. Huang. 1993. Molecular cloning and centrosomal localization of human caltractin. Proc. Natl. Acad. Sci. USA 90**:**11039-11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutz, W., W. L. Lingle, D. McCormick, T. M. Greenwood, and J. L. Salisbury. 2001. Phosphorylation of centrin during the cell cycle and its role in centriole separation preceding centrosome duplication. J. Biol. Chem. 276**:**20774-20780. [DOI] [PubMed] [Google Scholar]
- 30.Masutani, C., K. Sugasawa, J. Yanagisawa, T. Sonoyama, M. Ui, T. Enomoto, K. Takio, K. Tanaka, P. J. van der Spek, D. Bootsma, et al. 1994. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 13**:**1831-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matei, E., S. Miron, Y. Blouquit, P. Duchambon, I. Durussel, J. A. Cox, and C. T. Craescu. 2003. C-terminal half of human centrin 2 behaves like a regulatory EF-hand domain. Biochemistry 42**:**1439-1450. [DOI] [PubMed] [Google Scholar]
- 32.Matsunaga, T., D. Mu, C.-H. Park, J. T. Reardon, and A. Sancar. 1995. Human DNA repair excision nuclease. Analysis of the roles of the subunits involved in dual incisions by using anti-XPG and anti-ERCC1 antibodies. J. Biol. Chem. 270**:**20862-20869. [DOI] [PubMed] [Google Scholar]
- 33.McConkey, D. J., and S. Orrenius. 1997. The role of calcium in the regulation of apoptosis. Biochem. Biophys. Res. Commun. 239**:**357-366. [DOI] [PubMed] [Google Scholar]
- 34.Meador, W. E., A. R. Means, and F. A. Quiocho. 1992. Target enzyme recognition by calmodulin: 2.4 Å structure of a calmodulin-peptide complex. Science 257**:**1251-1255. [DOI] [PubMed] [Google Scholar]
- 35.Middendorp, S., A. Paoletti, E. Schiebel, and M. Bornens. 1997. Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc. Natl. Acad. Sci. USA 94**:**9141-9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molinier, J., C. Ramos, O. Fritsch, and B. Hohn. 2004. CENTRIN2 modulates homologous recombination and nucleotide excision repair in Arabidopsis. Plant Cell. 16**:**1633-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mori, T., M. Nakane, T. Hattori, T. Matsunaga, M. Ihara, and O. Nikaido. 1991. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem. Photobiol. 54**:**225-232. [DOI] [PubMed] [Google Scholar]
- 38.Mu, D., C.-H. Park, T. Matsunaga, D. S. Hsu, J. T. Reardon, and A. Sancar. 1995. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 270**:**2415-2418. [DOI] [PubMed] [Google Scholar]
- 39.Mu, D., M. Wakasugi, D. S. Hsu, and A. Sancar. 1997. Characterization of reaction intermediates of human excision repair nuclease. J. Biol. Chem. 272**:**28971-28979. [DOI] [PubMed] [Google Scholar]
- 40.Ng, J. M. Y., W. Vermeulen, G. T. J. van der Horst, S. Bergink, K. Sugasawa, H. Vrieling, and J. H. J. Hoeijmakers. 2003. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 17**:**1630-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108**:**193-199. [DOI] [PubMed] [Google Scholar]
- 42.O'Donovan, A., A. A. Davies, J. G. Moggs, S. C. West, and R. D. Wood. 1994. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature 371**:**432-435. [DOI] [PubMed] [Google Scholar]
- 43.Okuda, Y., R. Nishi, J. M. Y. Ng, W. Vermeulen, G. T. J. Van Der Horst, T. Mori, J. H. J. Hoeijmakers, F. Hanaoka, and K. Sugasawa. 2004. Relative levels of the two mammalian Rad23 homologs determine composition and stability of the xeroderma pigmentosum group C protein complex. DNA Repair 3**:**1285-1295. [DOI] [PubMed] [Google Scholar]
- 44.O'Neil, K. T., and W. F. DeGrado. 1990. How calmodulin binds its targets: sequence independent recognition of amphiphilic α-helices. Trends Biochem. Sci. 15**:**59-64. [DOI] [PubMed] [Google Scholar]
- 45.Paoletti, A., M. Moudjou, M. Paintrand, J. L. Salisbury, and M. Bornens. 1996. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 109**:**3089-3102. [DOI] [PubMed] [Google Scholar]
- 46.Popescu, A., S. Miron, Y. Blouquit, P. Duchambon, P. Christova, and C. T. Craescu. 2003. Xeroderma pigmentosum group C protein possesses a high affinity binding site to human centrin 2 and calmodulin. J. Biol. Chem. 278**:**40252-40261. [DOI] [PubMed] [Google Scholar]
- 47.Reardon, J. T., A. F. Nichols, S. Keeney, C. A. Smith, J.-S. Taylor, S. Linn, and A. Sancar. 1993. Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts: T[c,_s_]T, T[t,_s_]T, T[6-4]T, and T[Dewar]T. J. Biol. Chem. 268**:**21301-21308. [PubMed] [Google Scholar]
- 48.Riedl, T., F. Hanaoka, and J.-M. Egly. 2003. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 22**:**5293-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rost, B. 1996. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 266**:**525-539. [DOI] [PubMed] [Google Scholar]
- 50.Salisbury, J. L., A. Baron, B. Surek, and M. Melkonian. 1984. Striated flagellar roots: isolation and partial characterization of a calcium-modulated contractile organelle. J. Cell Biol. 99**:**962-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salisbury, J. L., A. T. Baron, and M. A. Sanders. 1988. The centrin-based cytoskeleton of Chlamydomonas reinhardtii: distribution in interphase and mitotic cells. J. Cell Biol. 107**:**635-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salisbury, J. L., K. M. Suino, R. Busby, and M. Springett. 2002. Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12**:**1287-1292. [DOI] [PubMed] [Google Scholar]
- 53.Sanders, M. A., and J. L. Salisbury. 1989. Centrin-mediated microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 108**:**1751-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanders, M. A., and J. L. Salisbury. 1994. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 124**:**795-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaffner, W., and C. Weissmann. 1973. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal. Biochem. 56**:**502-514. [DOI] [PubMed] [Google Scholar]
- 56.Schumacher, M. A., A. F. Rivard, H. P. Bachinger, and J. P. Adelman. 2001. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature 410**:**1120-1124. [DOI] [PubMed] [Google Scholar]
- 57.Shivji, M. K. K., A. P. M. Eker, and R. D. Wood. 1994. DNA repair defect in xeroderma pigmentosum group C and complementing factor from HeLa cells. J. Biol. Chem. 269**:**22749-22757. [PubMed] [Google Scholar]
- 58.Sijbers, A. M., W. L. de Laat, R. R. Ariza, M. Biggerstaff, Y.-F. Wei, J. G. Moggs, K. C. Carter, B. K. Shell, E. Evans, M. C. de Jong, S. Rademakers, J. de Rooij, N. G. J. Jaspers, J. H. J. Hoeijmakers, and R. D. Wood. 1996. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 86**:**811-822. [DOI] [PubMed] [Google Scholar]
- 59.Spang, A., I. Courtney, U. Fackler, M. Matzner, and E. Schiebel. 1993. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J. Cell Biol. 123**:**405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugasawa, K., C. Masutani, A. Uchida, T. Maekawa, P. J. van der Spek, D. Bootsma, J. H. J. Hoeijmakers, and F. Hanaoka. 1996. HHR23B, a human Rad23 homolog, stimulates XPC protein in nucleotide excision repair in vitro. Mol. Cell. Biol. 16**:**4852-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugasawa, K., J. M. Y. Ng, C. Masutani, S. Iwai, P. J. van der Spek, A. P. M. Eker, F. Hanaoka, D. Bootsma, and J. H. J. Hoeijmakers. 1998. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2**:**223-232. [DOI] [PubMed] [Google Scholar]
- 62.Sugasawa, K., J. M. Y. Ng, C. Masutani, T. Maekawa, A. Uchida, P. J. van der Spek, A. P. M. Eker, S. Rademakers, C. Visser, A. Aboussekhra, R. D. Wood, F. Hanaoka, D. Bootsma, and J. H. J. Hoeijmakers. 1997. Two human homologs of Rad23 are functionally interchangeable in complex formation and stimulation of XPC repair activity. Mol. Cell. Biol. 17**:**6924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sugasawa, K., T. Okamoto, Y. Shimizu, C. Masutani, S. Iwai, and F. Hanaoka. 2001. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 15**:**507-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63a.Sugasawa, K., Y. Okuda, M. Saijo, R. Nishi, N. Matsuda, G. Chu, T. Mori, S. Iwai, K. Tanaka, and F. Hanaoka. 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121**:**387-400. [DOI] [PubMed] [Google Scholar]
- 64.Sugasawa, K., Y. Shimizu, S. Iwai, and F. Hanaoka. 2002. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair 1**:**95-107. [DOI] [PubMed] [Google Scholar]
- 65.Taillon, B. E., S. A. Adler, J. P. Suhan, and J. W. Jarvik. 1992. Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J. Cell Biol. 119**:**1613-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tornaletti, S., and P. C. Hanawalt. 1999. Effect of DNA lesions on transcription elongation. Biochimie 81**:**139-146. [DOI] [PubMed] [Google Scholar]
- 67.Treiber, D. K., Z. Chen, and J. M. Essigmann. 1992. An ultraviolet light-damaged DNA recognition protein absent in xeroderma pigmentosum group E cells binds selectively to pyrimidine (6-4) pyrimidone photoproducts. Nucleic Acids Res. 20**:**5805-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uchida, A., K. Sugasawa, C. Masutani, N. Dohmae, M. Araki, M. Yokoi, Y. Ohkuma, and F. Hanaoka. 2002. The carboxy-terminal domain of the XPC protein plays a crucial role in nucleotide excision repair through interactions with transcription factor IIH. DNA Repair 1**:**449-461. [DOI] [PubMed] [Google Scholar]
- 69.van der Spek, P. J., A. Eker, S. Rademakers, C. Visser, K. Sugasawa, C. Masutani, F. Hanaoka, D. Bootsma, and J. H. J. Hoeijmakers. 1996. XPC and human homologs of RAD23: intracellular localization and relationship to other nucleotide excision repair complexes. Nucleic Acids Res. 24**:**2551-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Volker, M., M. J. Mone, P. Karmakar, A. van Hoffen, W. Schul, W. Vermeulen, J. H. J. Hoeijmakers, R. van Driel, A. A. van Zeeland, and L. H. Mullenders. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8**:**213-224. [DOI] [PubMed] [Google Scholar]
- 71.Wakasugi, M., J. T. Reardon, and A. Sancar. 1997. The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J. Biol. Chem. 272**:**16030-16034. [DOI] [PubMed] [Google Scholar]
- 72.Wolfrum, U., and J. L. Salisbury. 1998. Expression of centrin isoforms in the mammalian retina. Exp. Cell Res. 242**:**10-17. [DOI] [PubMed] [Google Scholar]
- 73.Yap, K. L., J. Kim, K. Truong, M. Sherman, T. Yuan, and M. Ikura. 2000. Calmodulin target database. J. Struct. Funct. Genomics 1**:**8-14. [DOI] [PubMed] [Google Scholar]
- 74.Yokoi, M., C. Masutani, T. Maekawa, K. Sugasawa, Y. Ohkuma, and F. Hanaoka. 2000. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 275**:**9870-9875. [DOI] [PubMed] [Google Scholar]