Chemical warfare between microbes promotes biodiversity (original) (raw)
Abstract
Evolutionary processes generating biodiversity and ecological mechanisms maintaining biodiversity seem to be diverse themselves. Conventional explanations of biodiversity such as niche differentiation, density-dependent predation pressure, or habitat heterogeneity seem satisfactory to explain diversity in communities of macrobial organisms such as higher plants and animals. For a long time the often high diversity among microscopic organisms in seemingly uniform environments, the famous “paradox of the plankton,” has been difficult to understand. The biodiversity in bacterial communities has been shown to be sometimes orders of magnitudes higher than the diversity of known macrobial systems. Based on a spatially explicit game theoretical model with multiply cyclic dominance structures, we suggest that antibiotic interactions within microbial communities may be very effective in maintaining diversity.
The past 10 years have seen great progress in measuring bacterial diversity by the application of several molecular approaches (1). Many studies (2) conducted in very diverse microbial habitats have arrived at the same general conclusion: only a very small fraction (less than 1%) of the bacterial species present can be recovered by standard cultivation techniques, but the actual biodiversity in microbial communities is enormous in many cases. For a striking example, a 30-g soil sample from a Norwegian forest has been estimated to contain ≈500,000 species, although this estimate necessarily is based on many ad hoc assumptions (3, 4). It is difficult to see how such astronomical species numbers could fit into the conventional resource competition framework even if forest soil can be considered a highly structured habitat.
Widespread Antibiosis in Microbial Communities
Microorganisms very commonly produce and excrete antibiotic compounds that inhibit or kill sensitive strains from their own or closely related species. The excretion of antimicrobial substances is known to be widespread among bacteria (5), yeasts (6), and other fungi (7). Particularly well studied systems comprise some bacteriocins (antimicrobial toxins produced by bacteria) such as the colicins from Escherichia coli and nisins from lactic acid bacteria (8). Experimental data based on the analysis of strains in E. coli collections (9, 10) suggest that at least 35% of the strains are colicinogenic. Most strains were sensitive to at least one of the 20 different types of colicin tested, but multiple resistance was common, with 22% of the strains being resistant to all of them. Also, the killer phenotypes of Saccharomyces cerevisiae and a few other yeasts have been studied in depth (11). Estimates of killer activity among wild yeasts from various habitats suggest that between 5 and 30% of the strains can kill a standard sensitive Candida glabrata strain (6, 12).
Effects of Excreted Toxins on Diversity: Inconclusive Experimental Evidence
In view of the widespread occurrence of antibiosis in microbial communities, the diversity conundrum seems particularly confusing: multiple toxic environments are the least expected to maintain many different species. Pioneering work (13, 14) on the ecological and evolutionary aspects of antimicrobial toxin excretion has demonstrated that competition between a colicinogenic and a sensitive strain of E. coli results in the final exclusion of one or the other depending on their initial proportions. Theoretical work (15, 16) on competition between colicin-producing and -sensitive strains seems to be in line with these results, but other sets of empirical data show a very different picture: small-scale coexistence of toxin-producing and -sensitive strains has been reported in a number of natural and laboratory systems (6, 12, 17). One theoretical explanation for killer-sensitive polymorphism within a single species invokes microscale habitat segregation such that sensitive strains do better in poor-quality habitats, and toxin producers do better in rich habitats (18). However, because of the requirement of a very special habitat structure and spatial distribution of different toxicity types, this explanation seems unlikely to be of general validity.
Cyclic Dominance Hierarchy of Killer (K), Resistant (R), and Sensitive (S) Strains May Favor Polymorphism
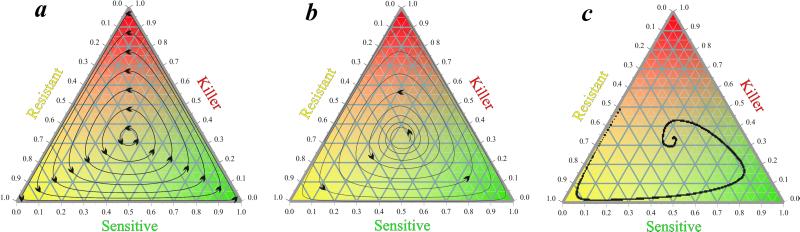
Another theoretical approach assumes an interplay of interference and resource competition (19–22), defining a cyclic dominance structure of K, R, and S strains within a species. The new element of this theory is the inclusion of resistance, a common empirical observation for bacteriocin systems left out from previous models altogether. R strains are immune to the corresponding toxin without actually producing it because of the damage or the loss of the toxin gene and the presence of intact immunity genes. Because toxin production involves a certain metabolic cost, R strains are assumed to be superior to Ks in resource competition. Similarly, S strains are supposed to outcompete Rs, because they do not pay the metabolic cost of producing the immunity factor. The remaining S-K interaction is settled to the advantage of the K strain by means of the interference competitive effect of toxic killing, a mechanism fundamentally different from resource competition, involving the active suppression of the concurrent population by means other than resource exhaustion. This cyclic interaction pattern (K beats S beats R beats K) can be cast naturally into the game theoretical framework of the rock-scissors-paper (RSP) game (23, 24). However, the mean-field (nonspatial) model for RSP dynamics admits neutrally stable periodic trajectories on the state space of the system at best (Fig. 1a), and assuming a small fitness cost of interaction makes the situation even worse: the system becomes unstable, with all trajectories approaching the margin of the state space gradually through all limits (Fig. 1b) such that eventually only one of the three strategies survives (24). The spatial (cellular automaton) version of the same model is very robustly stable (Fig. 1c; ref. 25).
Figure 1.
The single-toxin RSP game. (a) Periodic orbits of the simplest RSP game dynamics. (b) Divergent oscillations in the simple RSP game with a small interaction cost added. (c) Stability of the spatial (cellular automaton) version of the simple RSP game model.
An extension of the spatial cyclic exclusion approach to a multitype eco-evolutionary model (21) of the dynamics of nine different colicin plasmids in E. coli populations showed that a high diversity of colicin plasmid types is maintained easily in a single niche. This high diversity is expressed in one of two possible modes, dependent on the metabolic cost associated with resistance. For high resistance cost, persistent strains show “multitoxicity,” different strains maintaining different combinations of colicin genes. For low metabolic costs the community approaches the state of “hyperimmunity,” in which most bacteria are immune to most toxins but produce only a few, which is in good agreement with experimental data (9).
A Model of Antibiotics Excretion Shaping the Structure and Diversity of Microbial Communities
We have studied a spatially explicit model of a multispecies microbial community using a multitype RSP game implemented in a randomly updated cellular automaton. The arena is an environmentally uniform 180 × 180 square grid of cells with a toroidal topology. Each cell of the grid represents the site of a single microbial clone such as a bacterial colony that is characterized by its ability to produce particular antimicrobial toxin(s) and the corresponding resistance factors. All sites are always occupied, i.e., a colony can be replaced only by another one or remain locally persistent. The model specifies up to 14 different toxins, and for each one a clone is K (i.e., it can excrete the toxin and is also resistant to it), R (i.e., resistant to the toxin but unable to produce it), or S (i.e., unable to produce both the toxin and the resistance factor). Thus the maximum number of strains with different toxicity-resistance patterns is 314. Toxin production and resistance are assumed to be costly: metabolic costs are ordered as S < R < K. The update of a single cell is comprised of the following steps. First, with a probability m, the colony in the cell mutates a randomly chosen toxin type into the subsequent dominant state, i.e., K → R, R → S, and S → K. The first two transitions require the mutational inactivation of a gene and therefore are “normal” mutations. The transition from S to K requires the acquisition of a novel toxin production gene as well as the corresponding resistance gene and is probably best viewed as a horizontal transfer (e.g., by transformation) of a genetic element (e.g., a plasmid or virus RNA) carrying both genes or as some other rare sequence of events resulting in the evolution of a novel toxin system (26). When mutation does not occur the colony undergoes recombination, with probability r, with a neighboring colony that is chosen randomly from the four orthogonal neighbors. We have modeled recombination in the simplest possible way by not specifying any type of segregation; the strains simply incorporate the toxin and resistance genes of the other strain into their genome in addition to its own toxin and resistance genes, resulting in two identical colonies. If neither mutation nor recombination occurs, the two interacting colonies determine whether one colony is dominant over the other. Colony A interacting with a neighboring colony B will invade and replace B if (i) A can kill B, but B cannot kill A, (ii) neither can kill the other, but A has a smaller metabolic burden, or (iii) both can kill the other, but A has a smaller metabolic burden. The update rule for a single site can be represented in pseudocode as follows
Pick-Random-Site; if (chance < mutation_rate) Mutate-Random-Toxin-Type Else-If (chance < recombination_rate) Recombine-With-Random-Neighbor; Else Interact-With-Random-Neighbor:
It is clear that the time scale of an update step of the model is comparable to the time scale of local population dynamics; a complete colony replacement process can be accomplished during such a time unit. One generation is a number of such update steps equal to the number of sites in the grid such that on average each site is updated once every generation.
Results
“Frozen State” Quasi-Equilibrium.
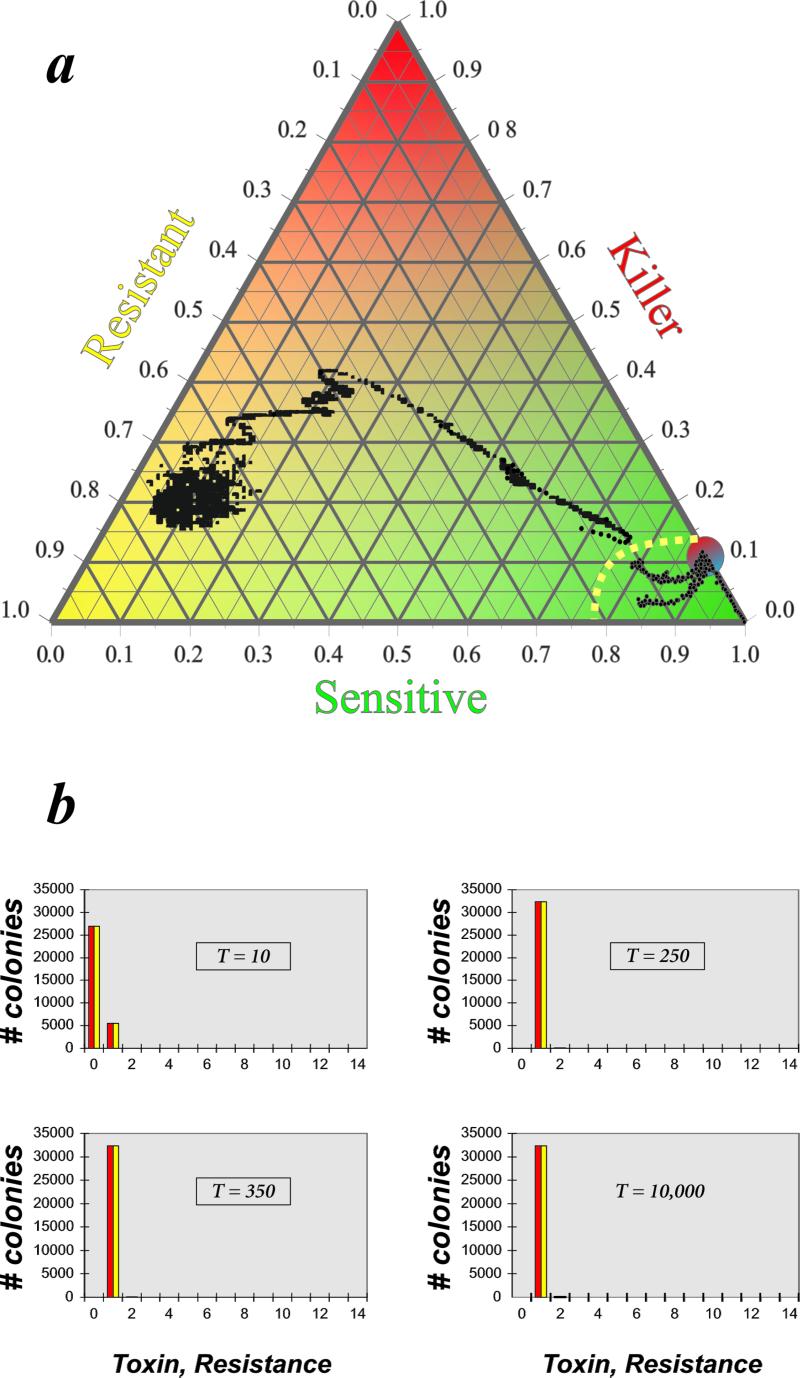
First we consider how a microbial community in which initially no toxin is excreted evolves toward a state of widespread toxin production. Starting from a random community close to the uniform all-S state [S(0) close to 1.0], allowing small S → K, K → R, and R → S mutation rates, and setting the metabolic cost of K (toxin production + resistance) equal to twice the cost of R (just resistance), the system evolves to a fine-grained distribution of small patches consisting of monotoxic strains. All 14 possible toxins persist in the system, but the great majority of the colonies carry just one toxin gene plus the corresponding resistance gene. Fig. 2 illustrates the main findings in this case. After a few hundreds of generations the spatial pattern appears frozen, with very little subsequent change going on. We interpret the quasi-frozen mosaic pattern as a “deadlock,” in which the great majority of neighboring colonies hold each other in check because their (single) toxin is either the same, in which case they are resistant to each other, or their toxins are different, in which case neither can invade the other because both have the same metabolic cost. Colonies that have acquired an additional toxin gene will spread within the monotoxic patch (group of neighboring cells containing identical colonies), in which they appeared because they are effective against the “parent” strain but then with a high probability will encounter a neighboring patch consisting of monotoxic colonies with a third type of toxin to which they are not resistant. These monotoxic neighbors will defeat the bitoxic mutant strain because of the higher metabolic burden the latter carries; the patch of the mutant is invaded by monotoxic colonies from all sides, and finally it disappears and the pattern becomes frozen again. Remarkably, the quasi-frozen spatial distribution of small patches of monotoxic strains does not occur in the simulations when the number, n, of possible different toxins is smaller than 4. A plausible explanation for this limit invokes the famous four-color-map theorem (27) of mathematical topology, which states that for any map four colors are sufficient to color neighboring countries always different. Our grid containing patches (coherent groups of identical monotoxic colonies) can be viewed as a map, on which each patch is a different country. With less than four different toxins, a (monotoxic) patch will with a high probability border to a patch of the same type. In other words, at start the grid then will actually contain a labyrinth-like percolation network of connected patches for each monotoxic type. Let us consider the n = 3 case. A bitoxic mutant A has a large ramified habitat to colonize, giving it more time to survive. During this longer survival time, also a neighboring patch carrying a different toxin has a good chance to obtain a second toxin to form mutant B. The two bitoxic strains play draw against each other, and the time for both to pick up the third toxin is again long enough. With four or more different toxins, the likelihood of forming extended patches is small from the outset (this is the point at which the four-color-map theorem becomes relevant), therefore mutations remain local, confined to relatively small patches, resulting in quick elimination by monotoxic neighbors.
Figure 2.
Multitoxin RSP game: frozen state. (a) Time series of the relative frequencies of K, R, and S phenotypes on an average toxin locus with different initial states. The yellow dotted line is the approximate “separatrix” between the “basins of attraction” for the frozen state and the generic hyperimmunity state. (b) Distribution of the numbers of K and R phenotypes per strain at different generations for the all-S initial state. Simulation type: 14 toxin loci; 180 × 180 grid; metabolic cost for killing is twice as large as for just resistance; all mutation rates are m = 10−5; no recombination (r = 0). T, time in generations.
Hyperimmunity Quasi-Equilibrium.
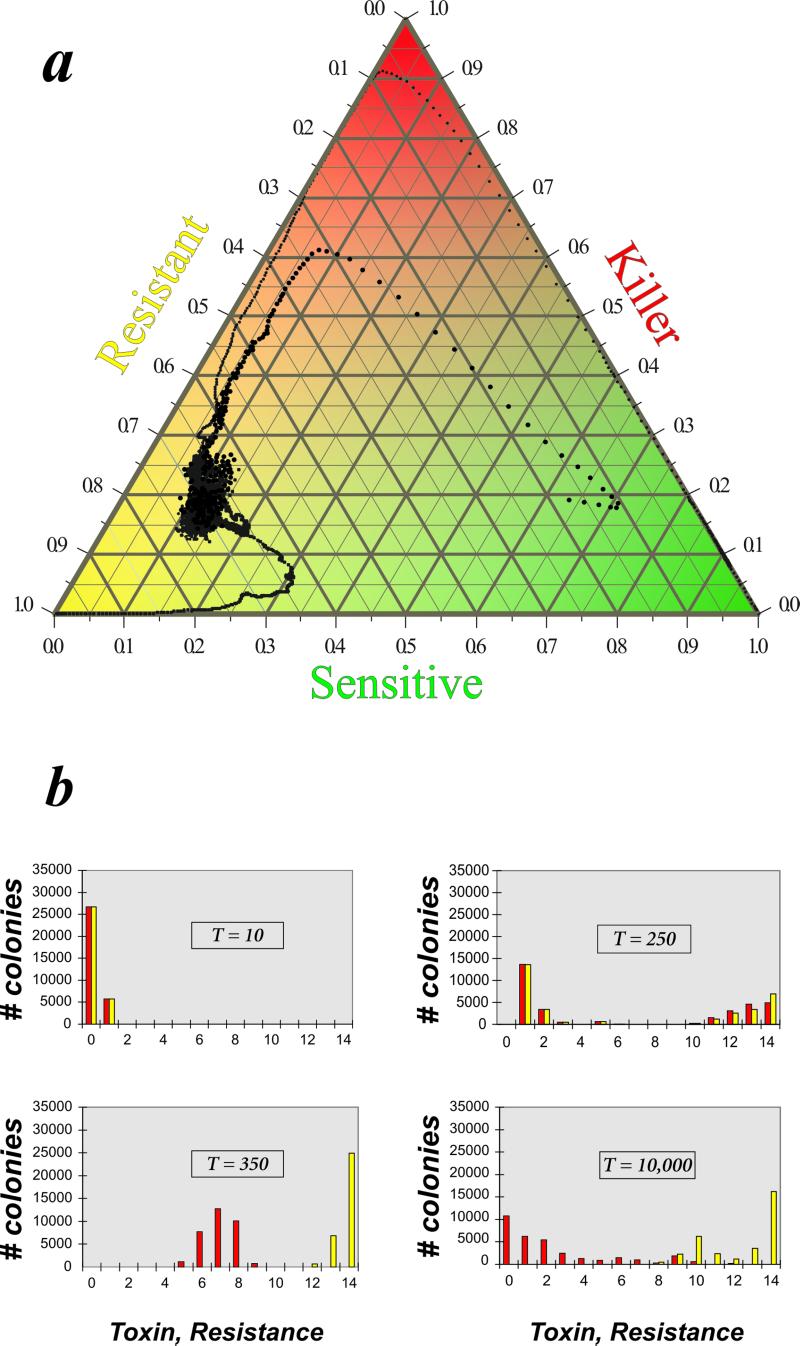
Starting the simulations from a community composition in which the average strain already possesses a few toxin and resistance genes produces convergence to a very different hyperimmunity (21) quasi-equilibrium characterized by low toxicity, high resistance, and a very dynamic pattern. Apparently with the average initial strain possessing more than one toxin plus resistance factor, the likelihood of a draw against all neighboring patches is so small that a frozen pattern is prevented from emerging. The patches persist long enough to collect more and more toxin + resistance genes by mutation, and the system evolves toward multiple toxicity. However, once this state has developed, losing some toxin genes while maintaining resistance to all the toxins becomes advantageous because of the reduction of metabolic costs. Eventually the system settles at the generic quasi-equilibrium at which the average colony harbors a few (typically 0–5 in the 14-toxin system) toxin genes and many (10–14) resistance genes (Fig. 3). The diversity of toxicity/resistance types is very high at the quasi-equilibrium; we have found up to a thousand different strains in a 180 × 180 grid, and this number increases with grid size. Although the average levels of toxicity and resistance remain within rather narrow limits, the spatial configuration of the community continues to change while the average patch size is much greater than in the frozen pattern.
Figure 3.
Multitoxin RSP game: hyperimmunity state. Time series (a) and phenotype distribution (b) with r = 10−3 recombination rate. All other parameters are as described for Fig. 2. T, time in generations.
Frozen State Prevented by Recombination and Fast Interference.
The frozen state is prevented also by frequent recombination. If the frequency of recombination events is higher than 10−4 per colony per generation, the system converges to the hyperimmune quasi-equilibrium. Recombination creates multiply toxic strains on the border of two different patches that can invade both parental patches, thus acquiring a relatively large territory. With a sufficiently high rate of recombination, this acquisition may be repeated frequently enough at the newly established borders with other patches to offset the invasion by strains with at least one different toxin and a smaller metabolic cost. In this way the average number of toxins per strain continues to rise until mutant strains appear with increasing numbers of resistances. Eventually the generic hyperimmune quasi-equilibrium is established with low-toxin and high-resistance multiplicity. This typical sequence of events is depicted in Fig. 3. Even the all-S start and negligible recombination are not enough to maintain the quasi-frozen monotoxicity state if the rate of exclusion is much slower for resource competition than it is for toxic killing (i.e., interference competition). If interference competition is at least four times faster than resource competition in excluding a competitor strain, the terminal state is always hyperimmunity.
Discussion
Several conclusions follow from these simulation studies. First, local interference competition resulting from the excretion of antibiotic compounds and the resource competitive effects caused by the associated metabolic costs may produce stable coexistence of huge numbers of different strains or species even in a temporally constant and spatially homogeneous environment if the fitness relations between K, R, and S types conform to those of the RSP game. The self-organized spatial pattern (28) of the system always plays a crucial role in defining the resulting community composition. Interestingly, the key idea underlying our work was proposed 25 years ago to explain observed patterns of sessile invertebrates on coral reefs. Jackson and Buss (29) suggested that a cyclic competitive hierarchy among species (A eliminates B, B eliminates C, but C eliminates A directly) may maintain diversity in space-limited systems in the absence of high levels of predation or physical disturbance. However, another study (30) failed to find evidence in support of this hypothesis. Apparently, in the following years ecologists lost interest in the idea, presumably because convincing examples were lacking. A second conclusion from our work is that the rates of processes such as recombination or horizontal genetic transfer, which enable the reallocation of genetic toxicity and resistance determinants among different strains, are relevant for the ultimate composition and spatial pattern of the system. With sufficiently frequent recombination (r > 10−4) a community evolves in which the average strain produces only a few toxins but is resistant to many, whereas for smaller r the system remains in the monotoxic frozen state in which the average strain produces just one toxin and is sensitive to all others. Recombination rate behaves as the control parameter of a phase transition here, with the critical value _r_c being close to 10−4.
Species Diversity in a Community or Polymorphic Species?
In our model the organisms (“strains”) are specified by their toxin-production profiles and their resistances and sensitivities to toxins. Depending on the rates of the evolution of novel toxin, resistance systems, and of their transmission between strains relative to the rates of genetic diversification in other traits and of speciation, different interpretations of the model strains are possible. At one extreme, every strain represents a different species in a community. This interpretation is applicable if novel toxicity systems appear at a very low rate and horizontal transmission between species is very rare. At the other extreme, all strains represent variants within a species. This interpretation applies in cases of relatively fast evolution of toxicity systems and high rates of between-strain transfer (either horizontally or by some recombination process). Intermediate interpretations apply to a community consisting of different species, in which species may share one or more toxin systems with some other species but also may exhibit some intraspecific polymorphism in toxin systems. The most natural interpretation of the quasi-frozen ensemble of strains in our model is a multispecies community with low rates of recombination between species. In the hyperimmunity state the many (≈1,000) different strains in the grid can be interpreted as belonging to the same species, but because plasmid transfer often also occurs between strains from different bacterial species, the game model also can be viewed as that of a multispecies bacterial community. It is interesting in this connection to contrast the available evidence on bacterial systems and yeast communities. Many bacterial systems are characterized by relatively frequent horizontal genetic transfer of toxin and resistance genes, particularly when these are located on conjugative plasmids, and the evidence on bacterial toxin systems indeed suggests that the majority of strains typically produces only a few toxins but are resistant to many (9). On the other hand, among yeast strains transfer of genes coding for toxin production and resistance (often located on symbiotic dsRNA virus-like particles) probably is very rare, and here the evidence, although not so extensive as for bacterial systems, points to monotoxicity and absence of multiple resistances (6, 11, 12). It would be very interesting to find out empirically whether yeast communities in fact may exhibit a spatially quasi-frozen mosaic pattern. The relative time scales of interference and resource competition can play a decisive role in shaping the outcome of interaction on the community level, the prediction of which also calls for experimental verification. It is fortunate that the class of organisms to which we think our model applies best, i.e., microorganisms, provides systems that are quite suitable for experimental testing under controlled conditions in the laboratory. This potential is testified by recent work on the relationships between diversity and productivity (31) and diversity and disturbance (32) using the bacterium Pseudomonas fluorescens. It will be a challenge to design experiments that can discriminate among various proposed explanations of diversity,whether they are based on environmental heterogeneity in some form or another (18) or should apply even in homogeneous environments (ref. 33 and the present model), although of course they do not exclude each other.
Acknowledgments
This work started in Collegium Budapest, Institute for Advanced Study, in 1995. We are grateful for the stimulating intellectual atmosphere of the Theoretical Biology Focus Group and the excellent library service of the Collegium. We thank Tom Starmer for discussion and information on killer yeasts. Funding has been provided by the Hungarian Scientific Research Foundation (T 25793), the Santa Fe Institute, the Research Council for Earth and Life Sciences (ALW), which is subsidized by the Netherlands Organization for Scientific Research (NWO), and the Netherlands Organization for Scientific Research Grant B 84-515.
Abbreviations
K
killer
R
resistant
S
sensitive
RSP
rock-scissors-paper
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 556.
References
- 1.Pace N R. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torsvik V, Goksøyr J, Daae F L. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dykhuizen D E. Antonie Leeuwenhoek. 1998;73:25–33. doi: 10.1023/a:1000665216662. [DOI] [PubMed] [Google Scholar]
- 5.Reeves P. The Bacteriocins. New York: Springer; 1972. [Google Scholar]
- 6.Starmer W T, Ganter P F, Aberdeen V, Lachance M-A, Phaff H J. Can J Microbiol. 1987;33:783–796. doi: 10.1139/m87-134. [DOI] [PubMed] [Google Scholar]
- 7.Berdy J. Adv Appl Microbiol. 1974;18:309–402. [PubMed] [Google Scholar]
- 8.James R, Lazdunski C, Pattus F, editors. Bacteriocins, Microcins, and Lantibiotics. New York: Springer; 1991. [Google Scholar]
- 9.Riley M A, Gordon D M. J Gen Microbiol. 1992;138:1345–1352. doi: 10.1099/00221287-138-7-1345. [DOI] [PubMed] [Google Scholar]
- 10.Achtman M A, Mercer A, Kusecek B, Pohl A, Heuzenroeder M, Aaronson W, Sutton A, Silver R P. Infect Immun. 1983;39:315–335. doi: 10.1128/iai.39.1.315-335.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wickner R B. Ann Rev Microbiol. 1992;46:437–375. doi: 10.1146/annurev.mi.46.100192.002023. [DOI] [PubMed] [Google Scholar]
- 12.Abranches J, Morais P B, Rosa C A, Mendonça-Hagler L C, Hagler A N. Can J Microbiol. 1997;43:328–336. doi: 10.1139/m97-046. [DOI] [PubMed] [Google Scholar]
- 13.Adams J, Kinney T, Thompson S, Rubin L, Helling R B. Genetics. 1979;91:627–637. doi: 10.1093/genetics/91.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chao L, Levin B R. Proc Natl Acad Sci USA. 1981;78:6324–6328. doi: 10.1073/pnas.78.10.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin B R. Philos Trans R Soc London B. 1988;319:459–472. doi: 10.1098/rstb.1988.0059. [DOI] [PubMed] [Google Scholar]
- 16.Iwasa Y, Nakamura M, Levin S A. Evol Ecol. 1998;12:785–802. [Google Scholar]
- 17.Ruiz-Barba J L, Cathcart D P, Warner P J, Jimenez-Diaz R. Appl Environ Microbiol. 1994;60:2059–2064. doi: 10.1128/aem.60.6.2059-2064.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank S A. Evol Ecol. 1994;8:369–386. [Google Scholar]
- 19.Durrett R, Levin S A. J Theor Biol. 1997;185:165–171. doi: 10.1006/jtbi.1996.0292. [DOI] [PubMed] [Google Scholar]
- 20.Durrett R, Levin S A. Theor Popul Biol. 1998;53:30–43. doi: 10.1006/tpbi.1997.1338. [DOI] [PubMed] [Google Scholar]
- 21.Pagie L, Hogeweg P. J Theor Biol. 1999;196:251–261. doi: 10.1006/jtbi.1998.0838. [DOI] [PubMed] [Google Scholar]
- 22.Szabó Gy, Czárán T. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Top. 2001;63:061904. doi: 10.1103/PhysRevE.63.061904. [DOI] [PubMed] [Google Scholar]
- 23.Maynard Smith J. Evolution and the Theory of Games. Cambridge, U.K.: Cambridge Univ. Press; 1982. [Google Scholar]
- 24.Hofbauer J, Sigmund K. Evolutionary Games and Population Dynamics. Cambridge, U.K.: Cambridge Univ. Press; 1998. [Google Scholar]
- 25.Frean M, Abraham E R. Proc R Soc London Ser B. 2001;268:1323–1327. doi: 10.1098/rspb.2001.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan Y, Riley M A. Trends Ecol Evol. 1997;12:348–351. doi: 10.1016/s0169-5347(97)01127-0. [DOI] [PubMed] [Google Scholar]
- 27.Appel K, Haken W. Illinois J Math. 1977;21:429–567. [Google Scholar]
- 28.Czárán T. Spatiotemporal Models of Population and Community Dynamics. London: Chapman & Hall; 1998. [Google Scholar]
- 29.Jackson J B C, Buss L. Proc Natl Acad Sci USA. 1975;72:5160–5163. doi: 10.1073/pnas.72.12.5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Connell J H. In: Coelenterate Ecology and Behavior. Mackie GO, editor. New York: Plenum; 1976. pp. 51–58. [Google Scholar]
- 31.Kassen R, Buckling A, Bell G, Rainey P B. Nature (London) 2000;406:508–512. doi: 10.1038/35020060. [DOI] [PubMed] [Google Scholar]
- 32.Buckling A, Kassen R, Bell G, Rainey P B. Nature (London) 2000;408:961–964. doi: 10.1038/35050080. [DOI] [PubMed] [Google Scholar]
- 33.Huisman J, Weissing F J. Nature (London) 1999;402:407–410. [Google Scholar]