Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members (original) (raw)
Abstract
We report an in-depth computational study of the protein sequences and structures of the superfamily of archaeo-eukaryotic primases (AEPs). This analysis greatly expands the range of diversity of the AEPs and reveals the unique active site shared by all members of this superfamily. In particular, it is shown that eukaryotic nucleo-cytoplasmic large DNA viruses, including poxviruses, asfarviruses, iridoviruses, phycodnaviruses and the mimivirus, encode AEPs of a distinct family, which also includes the herpesvirus primases whose relationship to AEPs has not been recognized previously. Many eukaryotic genomes, including chordates and plants, encode previously uncharacterized homologs of these predicted viral primases, which might be involved in novel DNA repair pathways. At a deeper level of evolutionary connections, structural comparisons indicate that AEPs, the nucleases involved in the initiation of rolling circle replication in plasmids and viruses, and origin-binding domains of papilloma and polyoma viruses evolved from a common ancestral protein that might have been involved in a protein-priming mechanism of initiation of DNA replication. Contextual analysis of multidomain protein architectures and gene neighborhoods in prokaryotes and viruses reveals remarkable parallels between AEPs and the unrelated DnaG-type primases, in particular, tight associations with the same repertoire of helicases. These observations point to a functional equivalence of the two classes of primases, which seem to have repeatedly displaced each other in various extrachromosomal replicons.
INTRODUCTION
In almost all currently known DNA replication systems, the initiation of replication requires a free hydroxyl group to which nucleotides are transferred by DNA polymerases for the synthesis of a new strand (1,2). Several distinct solutions for this requirement have been described:
- All cellular life forms and many DNA viruses, phages and plasmids use a primase to synthesize a short RNA primer with a free 3′ OH group that is subsequently elongated by a DNA polymerase (2).
- The retroelements (including retroviruses) employ a tRNA that primes DNA replication by providing a free 3′ OH that is used for elongation by the reverse transcriptase (1).
- In adenoviruses and the φ29 family of bacteriophages, a hydroxyl group is provided by the side-chain of an amino acid of the genome-attached protein (the terminal protein) to which nucleotides are added by the DNA polymerase to form a new strand (3).
- In several families of DNA viruses, such as parvoviruses, geminiviruses, circoviruses, and many phages and plasmids that adopt a rolling circle replication (RCR) model, the RCR endonuclease (RCRE) creates a nick in one of the DNA strands. The 5′ end of the nicked strand is transferred to a tyrosine residue on the nuclease, and the free 3′ OH group is elongated by a DNA polymerase for the new strand synthesis (4,5).
Although all known cellular replication systems utilize an RNA primer for DNA replication initiation, two structurally distinct and non-homologous versions of primases catalyze primer synthesis. In all bacteria, the primase involved in DNA replication, belongs to the DnaG superfamily and contains a catalytic domain of the TOPRIM fold (6,7). The TOPRIM fold contains an α/β core with four conserved strands in a Rossmann-like topology and is also found in the catalytic domains of topoisomerase Ia, topoisomerase II, the OLD-family nucleases and DNA repair proteins related to RecR (6). In the archaeo-eukaryotic lineage, the principal primase involved in replication contains a highly derived version of the RNA recognition motif (RRM) fold (8,9). A catalytic domain based on an RRM-like scaffold is also present in viral RNA-dependent RNA polymerases, reverse transcriptases, cyclic nucleotide generating cyclases, and DNA polymerases of the A/B/Y families that are involved in DNA replication and repair (9–17). This fundamental dichotomy between the primases in the archaeo-eukaryotic and bacterial lineages is paralleled in several other components of DNA replication. In particular, the principal replicative polymerase and the gap-filling polymerase are very distinct and, apparently, non-homologous between the archaeo-eukaryotes and bacteria. Similarly, the principal replicative helicases are non-orthologous between these two primary divisions of cellular life. In contrast, several other components of the DNA replication machinery, such as the DNA ligases, PCNA, clamp-loader ATPases, Topoisomerase Ia and RNAse HII, are orthologous between archaeo-eukaryotes and bacteria (18,19).
Comparative analysis of proteins containing the TOPRIM domain provides substantial clues as to the early stages of evolution of this class of proteins. Specifically, the TOPRIM domain of DnaG is also found in the archaeal DnaG ortholog, which is conserved in all archaeal species, and in topoisomerase Ia and topoisomerase II, which are present in both archaea and bacteria (6,20). This suggests that distinct versions of the TOPRIM domain which were, respectively, ancestors of the DnaG-type primases and the topoisomerases were already present in the last universal common ancestor (LUCA) (6,18). However, given that the archaeal DnaG proteins have been shown to associate with the RNA-degrading exosome complex (20), it is unclear if the ancestral form of DnaG in LUCA was involved in RNA metabolism or DNA replication.
The origin of the archaeo-eukaryotic primase (AEP) superfamily is more obscure. In addition to the archaea, eukaryotes and baculoviruses, a divergent member of the AEP superfamily was detected in diverse bacteria, where it appears to function, along with homologs of the Ku protein and ATP-dependent DNA ligases, in a DNA repair system involved in non-homologous end-joining (NHEJ) (21–23). The phyletic profile of the cellular members of the AEP superfamily suggests that it was not represented in LUCA and was recruited for primer synthesis only at the base of the archaeo-eukaryotic lineage, with subsequent acquisition by bacteria via horizontal gene transfer (HGT).
Recently, a novel family of AEPs called the prim-pol, which is sporadically present in crenarchaeal and Gram-positive bacterial plasmids, has been described (24,25). The prim-pols catalyze both a DNA polymerase and a primase reaction (hence the name). They are often fused to Superfamily III helicases or are encoded by genes in the neighborhood of those encoding such helicases (24,25). These primases and the associated helicases have been proposed to form the replication initiation complex of the respective plasmids (24). Crystal structures of the prim-pols showed that they shared a structure and several identically positioned catalytic residues with the archaeal replicative primase, indicating that the prim-pols were divergent members of the AEP superfamily (25). The presence of both DNA polymerase and RNA polymerase activities in the prim-pols suggests a primitive state, possibly, resembling early, transitional DNA replication systems where the same enzyme catalyzed both initiation and elongation (24).
Given our long-standing interest in understanding the nature of early DNA replication systems, the discovery of the prim-pols and the report of their crystal structure prompted us to re-investigate these systems from the standpoint of the origins of the AEP. With this objective, we performed a comprehensive study of the phyletic distribution and evolutionary affinities of the prim-pols in relation to the other members of the AEP superfamily and the larger class of nucleic acid polymerases and cyclases, which also contain the catalytic palm domain with an RRM-like fold. In addition, we were interested in the provenance of two other non-cellular primases of uncertain evolutionary affinities, such as, the herpesvirus UL52 primases and the RepA-like primases of ColE2-like plasmids (26–29).
In this study, we expand the AEP superfamily to include the UL52-like primases of herpesviruses, the N-terminal domains of the D5-like protein of the nucleo-cytoplasmic large DNA viruses (NCLDV) and phages, the RepA-like primases of the ColE2 group of plasmid, a previously unknown family of predicted eukaryotic cellular primases, and several other primase-helicase proteins from bacteriophages, predicted transposons and plasmids. Our analysis points to the existence of at least 13 distinct families in the AEP superfamily. Structural comparisons show that the RCREs and the DNA replication origin-binding domains of papovaviruses are the closest relatives of the AEPs among the RRM-like nucleic acid palm domains. This suggests that both nucleotidyl transferase and nuclease activity evolved in the same class of proteins, which is reminiscent of the evolution of primase and nuclease activities in the TOPRIM fold (6).
MATERIALS AND METHODS
The non-redundant (NR) database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda) was searched using the BLASTP program (30). Iterative database searches were conducted using the PSI-BLAST program with either a single sequence or an alignment used as the query, with the PSSM inclusion expectation (E) value threshold of 0.01 (unless specified otherwise); the searches were iterated until convergence. Hidden Markov models (HMMs) were built from alignments using the hmmbuild program and searches carried out using the hmmsearch program from the HMMer package (31). For all searches with compositionally biased proteins, the statistical correction for this bias was employed (32). Identification and statistical evaluation of conserved motifs in multiple protein sequences were performed using the Gibbs sampling method as implemented in the MACAW program (33,34). Multiple alignments were constructed using the T_Coffee, MUSCLE and PCMA programs, followed by manual correction based on the PSI-BLAST results (35–37). Similarity-based clustering of proteins was carried out using the BLASTCLUST program (ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html). All large-scale sequence and structure analysis procedures were carried out using the TASS package, which operates similar to the SEALS package (38).
Protein secondary structure was predicted using a multiple alignment to generate a HMM and PSSM and using this information for the Jpred program to produce a final structural prediction with 76% or greater accuracy (39,40). Protein structure manipulations were performed using the Swiss-PDB viewer program and the ribbon diagrams were constructed using the MOLSCRIPT program (41,42). For structural comparisons, the DALI/FSSP and VAST programs were used (43–45). The studies on clustering based DALI _Z_-scores have suggested that _Z_-scores >10 are characteristic of obvious relationships, such as those between two closely related proteins of the same family. Between _Z_-scores 10 and 6, typically, the relationships correspond to more distant relationships that might be recovered through sequence profile analysis and searches using HMMs. _Z_-scores <3 fall in the realm of remote structural relationships and require additional analysis, such as comparisons of topologies to make further inference regarding these relationships (43,44).
Phylogenetic analysis was carried out using the maximum-likelihood, neighbor-joining and minimum evolution (least squares) methods (46–48). Gene neighborhoods were identified by searching the completed genome sequences and whole genome shot-gun sequences with a custom-written script. The current list of completed and shot-gun genomes sequences can be accessed from the genomes division of the Entrez retrieval system (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Genome).
RESULTS AND DISCUSSION
Structural analysis and higher order relationships of the AEP catalytic domain
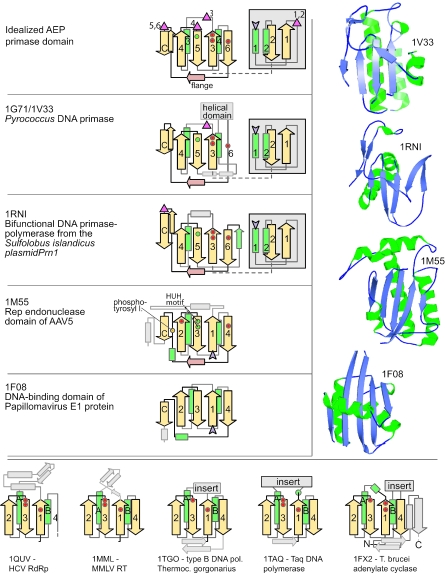
Analysis of the crystal structure of the prim-pol module from Sulfolobus plasmid pRN1 showed that it shares a structural core with the archaeo-eukaryotic replicative primases (25). This shared core corresponds to two successive structural modules. The N-terminal module contains a (αβ)2 unit for which there is no equivalent structure in the PDB database, while the C-terminal unit contains the previously reported derived version of the RRM-like fold that is also seen in the catalytic palm module of other polymerases (Figure 1). These two units define the shared catalytic core of the AEP superfamily and are packed against each other, with the active sites residues lying in the space between the two units. Thus, this shared structure includes at least six conserved strands (two from the N-terminal unit and four from the RRM-like fold) and four helices (two from each unit). Hereinafter, they are referred to as strands 1–6 and helices 1–4 (Figure 1) when the catalytic module of the AEPs is discussed. The RRM-like unit of the AEPs shows some minor deviations from the prototype RRM structure in that helix-4 (equivalent to the second helix of the prototype RRM-like fold) is completely distorted to form a strand in the prim-pols, and strand 6 (the fourth strand of the prototype RRM-like fold) is distorted in some members of the superfamily to acquire a helical character (Figure 1). The prim-pols and cellular primases also share a characteristic extended, flange-like structure after core strand 6 that is perpendicular to the plane of the RRM sheet and leads into a hairpin, which packs anti-parallel to strand 3 of the RRM-like unit (Figure 1).
Figure 1.
Topology diagrams and structures of AEP-type primases and related proteins. Strands are shown as arrows with the arrowhead on the C-terminal side and numbered 1 through 4 or 6, respectively, according to the conventions used in the text. Helices are shown as green rectangles, non-conserved elements in faint gray. Purple arrowheads mark the protein's C-terminus. The location of catalytically important residues is indicated by colored circles (green—histidine, red—acidic, the yellow circle represents a tyrosine residue that becomes covalently attached to the 5′ phosphate of a cleaved DNA strand in RCRE). The topology diagram at the top of the figure is an idealization and is not derived from an actual structure. It shows the positions of the Zn-clusters found in various members of the AEP superfamily as discussed in the text. The N-terminal (αβ)2 units of primase and primpol (in gray box) pack against the beta sheet of the palm fold but they are drawn here as a ‘slide-out’ for clarity of presentation. The structures of selected AEP-type primases are shown in the right hand panel. They are in the same orientation as the topology diagrams with the flange strand running above the plane of the beta sheet. The bottom panel shows topology diagrams of palm domain proteins from outside the AEP primase group for comparison purposes. The structural and topology diagrams were derived from the following PDB IDs: 1V33, 1G71 (8), 1RNI (25), 1M55 (59), 1F08 (54), 1QUV (92), 1MML (93), 1TGO (94), 1TAQ (95) and 1FX2 (96).
Three motifs, a hhhDhD motif in strand 3, an sxH motif in strand 5 (where ‘s’ is a small residue) and an h- (‘-’ is an acidic residue) in strand 6, are shared by both the prim-pols and the archaeo-eukaryotic cellular primases, and our analysis (see below) showed that they are strongly conserved across the entire, extended AEP superfamily (Figure 2). Site-directed mutagenesis has shown that these residues are essential for catalysis, with the DxD motif being involved in binding a magnesium ion (25,49). This suggests that, like in many other polymerases, Mg2+ is central for the catalytic activity of the AEPs. It has been proposed that the AEP superfamily might be related to the PolX superfamily (50), which includes nucleotidyl transferases, such as terminal transferase, Poly(A) polymerase and tRNA CCA-adding enzymes (51). However, a comparison of the available structures of the AEP and PolX superfamilies shows that the PolX superfamily contains a fold unrelated to the RRM-like fold, even though the two superfamilies possess similar sets of acidic metal-chelating residues. Furthermore, the overall arrangement of the catalytic residues in the AEPs shows certain differences from that seen in other polymerases, which possess similar metal-chelating motifs. Specifically, in the AEPs, the first aspartate of the DxD motif, the histidine residue in strand 5 and the conserved aspartate in strand 6 are positioned at the same level, within hydrogen-bonding distance, in the central sheet of the RRM (Figure 1). This arrangement of a central residue, which is flanked by an acidic and a basic residue, and its strong conservation in all catalytically active members of the AEP superfamily, suggests that during catalysis there is an interaction between these residues. No equivalent of the second highly conserved acidic residue from strand 6 is seen in any of the other polymerases that utilize the canonical di-metal mechanism. This might imply a deviation from the typical di-metal-ion-based nucleotidyltransferase reaction proposed for several DNA and RNA polymerases (52). Experiments have also suggested that residues in the flange are involved in nucleotide-binding in the archaeal primases (50). However, there are no conserved residues in the flange, so these associations with the nucleotides might be lineage-specific.
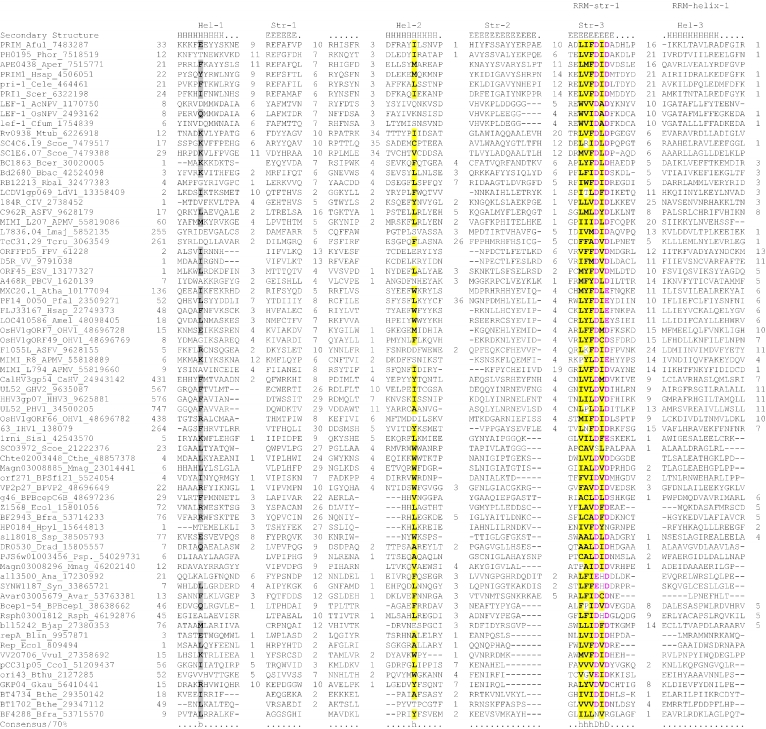
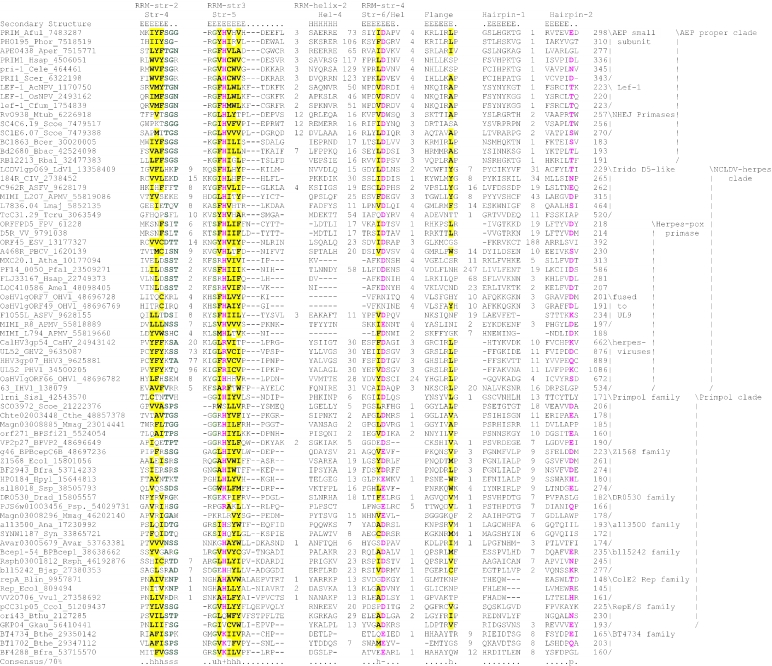
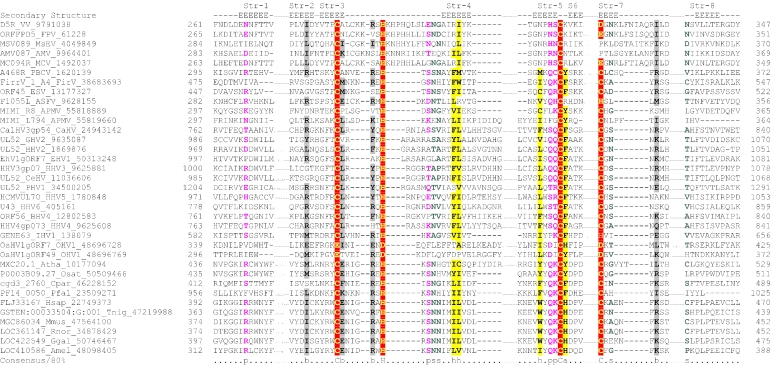
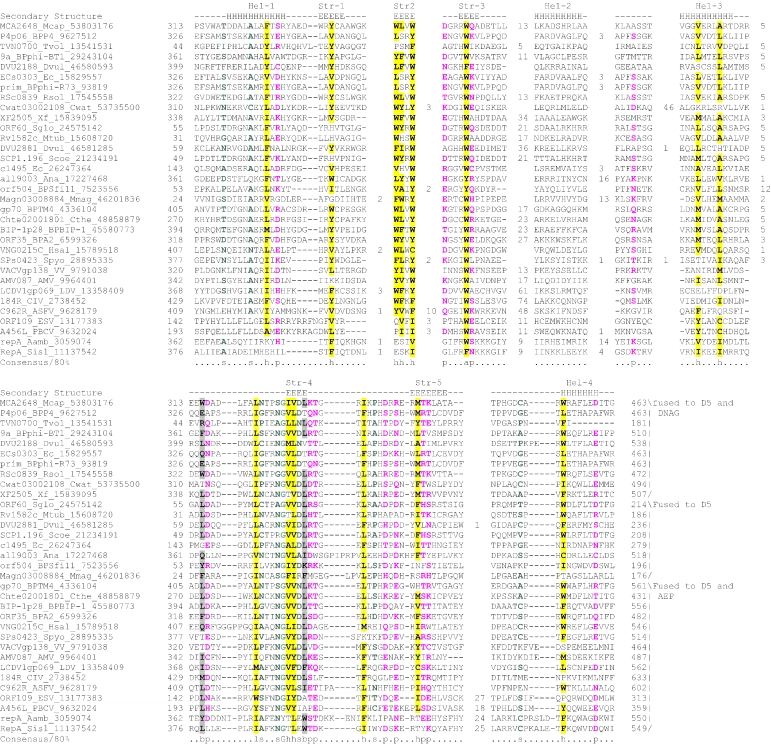
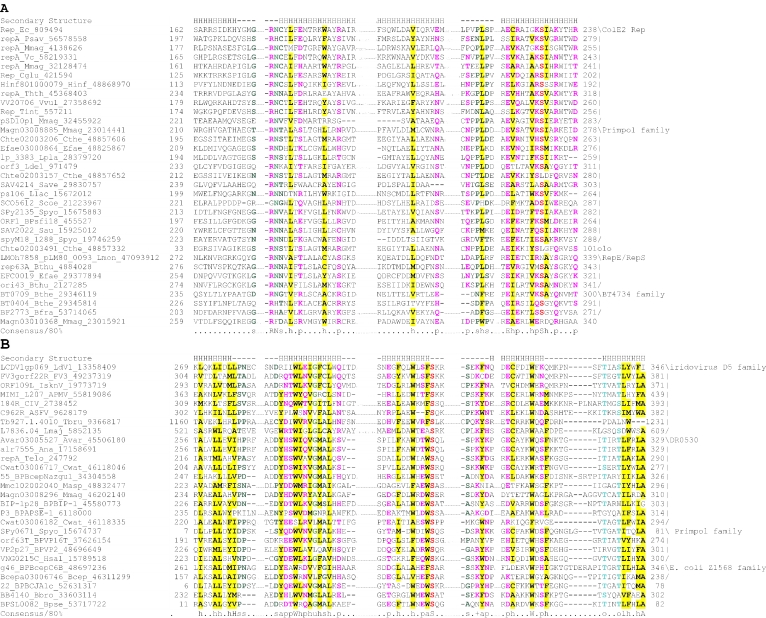
Figure 2.
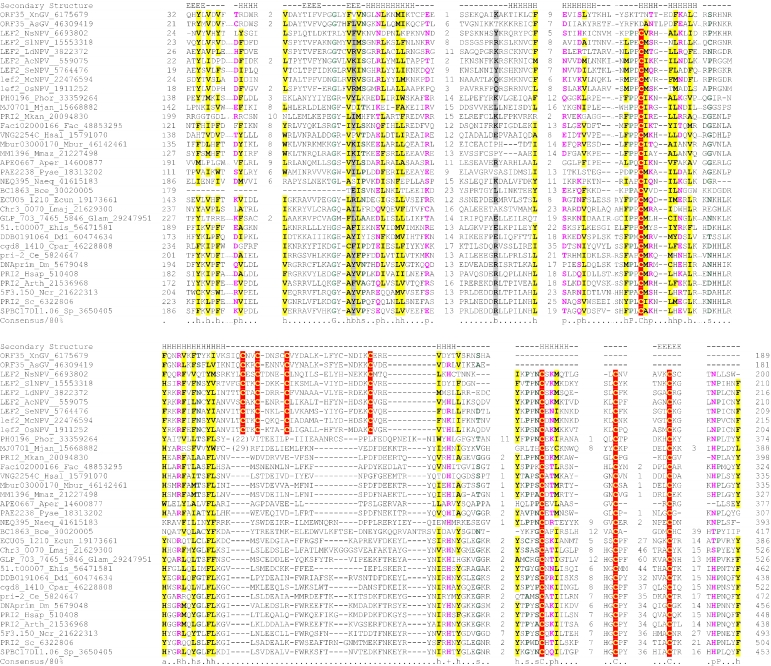
Multiple sequence alignment of the AEP superfamily. Proteins are designated by their gene names, species abbreviations and GenBank IDs separated by underscores. Columns of amino acids are colored based on their side chain properties and conservation in the alignment; 70% conservation was used to calculate the consensus. Poorly conserved, large inserts are replaced by the corresponding number of residues. The secondary structure shown above the alignment was derived from the crystal structures of the archaeal primase (PDB ID: 1g71) and the primpol protein (PDB ID: 1rni). Strands and helices are denoted above the alignment by E and H, respectively. The coloring scheme and consensus abbreviations are as follows: h, hydrophobic residues (ACFILMVWY), shaded yellow; b, big residues (LIYERFQKMW), shaded gray; s, small residues (AGSVCDN) and u, tiny residues (GAS), colored green; p, polar residues (STEDKRNQHC); +, basic residues (HRK) and -, acidic residues (DE), colored magenta. Species abbreviations are as follows: AMV: Amsacta moorei entomopoxvirus; APMV: Acanthamoeba polyphaga mimivirus; ASFV: African swine fever virus; Aamb: Acidianus ambivalens; AcNPV: Autographa californica nucleopolyhedrovirus; Aful: Archaeoglobus fulgidus; Amel: Apis mellifera; Ana: Nostoc sp.; Aper: Aeropyrum pernix; AsGV: Agrotis segetum granulovirus; Atha: Arabidopsis thaliana; Avar: Anabaena variabilis; BHV4: Bovine herpesvirus 4; BP315.5: Streptococcus pyogenes phage 315.5; BPA2: Lactobacillus casei bacteriophage A2; BPAPSE-1: Acyrthosiphon pisum bacteriophage APSE-1; BPAT3: Bacteriophage phi AT3; BPBCJA1c: Bacillus clarkii bacteriophage BCJA1c; BPBIP-1: Bordetella phage BIP-1; BPBcep1: Burkholderia cenocepacia phage Bcep1; BPBcepC6B: Burkholderia cepacia complex phage BcepC6B; BPBcepNazgul: Burkholderia cepacia phage BcepNazgul; BPN15: Bacteriophage N15; BPP4: Bacteriophage P4; BPSA: Bacteriophage PSA; BPSFi18: Streptococcus thermophilus bacteriophage SFi18; BPSfi11: Streptococcus thermophilus bacteriophage Sfi11; BPSfi21: Streptococcus thermophilus bacteriophage Sfi21; BPTM4: Mycobacteriophage TM4; BPVP16T: Vibrio parahaemolyticus phage VP16T; BPVP2: Vibriophage VP2; BPadh: Lactobacillus bacteriophage phi adh; BPmi7-9: Lactococcus phage mi7-9; BPphi-BT1: Bacteriophage phi-BT1; BPphi-R73: Bacteriophage phi-R73; BPphi105: Bacteriophage phi-105; BPphi31: Lactococcus bacteriophage phi31; BPphiHSIC: Listonella pelagia phage phiHSIC; BPphig1e: Bacteriophage phig1e; Bbac: Bdellovibrio bacteriovorus; Bbro: Bordetella bronchiseptica; Bcep: Burkholderia cepacia; Bcer: Bacillus cereus; Bfra: Bacteroides fragilis; Bjap: Bradyrhizobium japonicum; Blic: Bacillus licheniformis; Blin: Brevibacterium linens; Bpse: Burkholderia pseudomallei; Bthe: Bacteroides thetaiotaomicron; Bthu: Bacillus thuringiensis; CIV: Chilo iridescent virus; CaHV: Callitrichine herpesvirus 3; Ccol: Campylobacter coli; CeHV: Cercopithecine herpesvirus 9; Cele: Caenorhabditis elegans; Cfum: Choristoneura fumiferana; Cglu: Corynebacterium glutamicum; Cpar: Cryptosporidium parvum; Cthe: Clostridium thermocellum; Cwat: Crocosphaera watsonii; Ddes: Desulfovibrio desulfuricans; Ddis: Dictyostelium discoideum; Dmel: Drosophila melanogaster; Drad: Deinococcus radiodurans; Dvul: Desulfovibrio vulgaris; EHV1: Equid herpesvirus 1; ESV: Ectocarpus siliculosus virus; Ecoli: Escherichia coli; Ecun: Encephalitozoon cuniculi; Efae: Enterococcus faecalis; Efae: Enterococcus faecium; Ehis: Entamoeba histolytica; FPV: Fowlpox virus; FV3: Frog virus 3; Faci Ferroplasma acidarmanus; FirV: Feldmannia irregularis virus a; GHV2: Gallid herpesvirus 2; Ggal: Gallus gallus; Gkau: Geobacillus kaustophilus; Glam: Giardia lamblia; HHV2: Human herpesvirus 2; HHV3: Human herpesvirus 3; HHV4: Human herpesvirus 4; HHV5: Human herpesvirus 5; HHV6: Human herpesvirus 6B; Hinf: Haemophilus influenzae; Hpyl: Helicobacter pylori; Hsal: Halobacterium salinarum; Hsap: Homo sapiens; IHV1: Ictalurid herpesvirus 1; IsknV: Infectious spleen and kidney necrosis virus; LdNPV: Lymantria dispar nucleopolyhedrovirus; LdV1: Lymphocystis disease virus 1; Ldel: Lactobacillus delbrueckii; Linn: Listeria innocua; Llac: Lactococcus lactis; Lmaj: Leishmania major; Lmon: Listeria monocytogenes; Lpla: Lactobacillus plantarum; MCV: Molluscum contagiosum virus subtype 1; Masp: Magnetococcus sp.; Mbur: Methanococcoides burtonii; McNPV: Mamestra configurata nucleopolyhedrovirus B; Mcap: Methylococcus capsulatus; Mjan: Methanocaldococcus jannaschii; Mkan: Methanopyrus kandleri; Mmag: Magnetospirillum magnetotacticum; Mmaz: Methanosarcina mazei; Mmus: Mus musculus; MsEV: Melanoplus sanguinipes entomopoxvirus; Msp.: Micrococcus sp.; Mtub: Mycobacterium tuberculosis; Ncra: Neurospora crassa; Nequ: Nanoarchaeum equitans; NsNPV: Neodiprion sertifer nucleopolyhedrovirus; OHV1: Ostreid herpesvirus 1; OsNPV: Orgyia pseudotsugata multicapsid nucleopolyhedrovirus; Osat: Oryza sativa; PBCV: Paramecium bursaria Chlorella virus 1; PHV1: Psittacid herpesvirus 1; Pfal: Plasmodium falciparum; Phor: Pyrococcus horikoshii; Psav: Pseudomonas savastanoi; Psp.: Polaromonas sp.; Pyae: Pyrobaculum aerophilum; Rbal: Rhodopirellula baltica; Rnor: Rattus norvegicus; Rsol: Ralstonia solanacearum; Rsp.: Rhodococcus sp.; Rsph Rhodobacter sphaeroides; Saur: Staphylococcus aureus; Save: Streptomyces avermitilis; Scer: Saccharomyces cerevisiae; Scoe: Streptomyces coelicolor; SeNPV: Spodoptera exigua nucleopolyhedrovirus; Sglo: Streptomyces globisporus; Sisl: Sulfolobus islandicus; SlNPV: Spodoptera litura nucleopolyhedrovirus; Spom: Schizosaccharomyces pombe; Spyo: Streptococcus pyogenes; Ssp: Synechocystis sp.; Ssui: Streptococcus suis; Syn: Synechococcus sp.; Tbru: Trypanosoma brucei; Tcru: Trypanosoma cruzi; Telo: Thermosynechococcus elongatus; Tint: Thiobacillus intermedius; Tnig: Tetraodon nigroviridis; Tsp.: Thiobacillus sp.; Tthe: Thermus thermophilus; Tvol: Thermoplasma volcanium; VV: Vaccinia virus; Vcho: Vibrio cholerae; Vvul: Vibrio vulnificus; Xcam: Xanthomonas campestris; Xfas: Xylella fastidiosa; XnGV: Xestia c-nigrum granulovirus.
To investigate the higher order relationships of the prim-pols and the AEPs, we searched the PDB database for similar structures using the DALI program with the AEP (PDB ID: 1g71) as query. This search recovered several hits with significant _Z_-scores suggesting a genuine structural relationship. The best hits included the prim-pols (_Z_-score: 5.2), the N-terminal endonuclease domain (of the RCR superfamily) of the Rep protein of adeno-associated virus (AAV) (PDB ID: 1m55, _Z_-score: 4.1) and, with a less significant _Z_-score, the origin-binding domain (OBD) of the SV40 T-antigen (PDB ID: 2tbd, _Z_-score: 3.5). Further DALI searches with the OBD of the SV40 T-antigen retrieved the OBD of the E1 protein of the papillomaviruses (PDB ID: 1f08, _Z_-score: 7.6) and the RCR nuclease domain of the geminivirus replication initiation protein (PDB ID: 1l2m, _Z_-score: 5.5). Reciprocal DALI searches with each of these structures consistently recovered a similar group of proteins as best-hits with comparable _Z_-scores. A comparison of the common structural unit shared by the members of the AEP superfamily, the RCREs and the OBDs of papavoviruses showed, not unexpectedly, that it encompassed an RRM-like fold domain. Beyond this basic core, these proteins also shared some unique features, such as, the extended flange that runs perpendicular to the direction of the sheet of the RRM-like fold and a downstream strand that packs anti-parallel to the second strand of the RRM-like core (Figure 1). To assess the significance of this distinct shared structural feature, we systematically surveyed all proteins with an RRM-like fold in the PDB database (see ferredoxin fold in the SCOP database, http://scop.mrc-lmb.cam.ac.uk/scop/). The C-terminal flange is typically not encountered in other RRM-like fold proteins, with the potential exception of the C-terminal extensions observed in the RRM-like folds of polypyrimidine-tract-binding protein (PTB) (53) and the C-terminal domain of the eukaryotic EF-1γ (EF-1γC). PTB is a member of the classic RNA-binding RRM (RBD) superfamily, and the region corresponding to the C-terminal extension is missing in several closely related RBD proteins, such as sex-lethal, HnRNPA1 and splicing factor U1A. Examination of this region in the PTBs suggests that the C-terminal extension resembling the flange and the downstream strand is derived from a poorly conserved low-complexity region in the C-terminus. Thus, this feature of the PTB structure probably emerged late in the evolution of the RBD superfamily, and its specific similarity to the AEPs, RCREs and OBDs is, most likely, coincidental. The restricted phyletic profile of the eukaryote-specific translation factor eEF-1γC and the lack of a close relationship in sequence or structure to any other members of the RRM-like fold make it hard to assess the possibility of its specific relationship with the AEPs, RCREs and OBDs.
In addition to the shared structural features, the AEPs, RCREs and OBDs also have obvious common functional characteristics: members of each of these families are principally involved in DNA replication initiation, albeit in three distinct fashions (54,55). Specifically, the OBDs of the papilloma and polyoma viruses bind the origin of DNA replication and recruit additional viral and host components to initiate replication (54,55). Similarly, the RCREs introduce a nick at the origin of replication, following which the 5′ end of the nicked strand is transferred to a tyrosine on the protein and the free 3′ OH group is elongated by other components of the DNA replication apparatus (4,5). Finally, the AEPs are bona fide polymerases involved in the RNA primer synthesis. Hence, we propose that the common structural feature of the RCREs, the OBDs of polyoma and papillomaviruses, and the AEP superfamily is a shared derived character (synapomorphy) that separates them from other polymerases and cyclases with a similar RRM-like scaffold. Together with the above results of structural comparisons, this leads us to conclude that these three protein families evolved from a common ancestral protein that was already involved in initiation of DNA replication. The OBDs and the RCREs have a helix after the flange that leads into the C-terminal strand, whereas the primases and prim-pols have an additional strand that forms a hairpin with this C-terminal strand (Figure 1). This suggests that the OBDs are more closely related to or derived from the RCREs (Figure 3). While several structural studies have noted similarities and proposed a monophyletic origin for the polyoma-, papilloma- viral and OBDs and the geminiviral RCREs, the relationship of these to the AEP superfamily (53,56), to our knowledge, has not been elucidated to date.
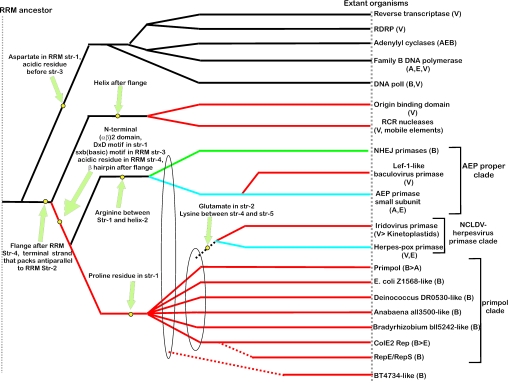
Figure 3.
Inferred evolutionary history of the AEP superfamily. The overall topology of the phylogram was derived using synapomorphies and clustering based on DALI _Z_-scores. Synapomorphies that unify a set of lineages are indicated next to the filled yellow circles. The ellipses indicate large assemblages within which individual lineages show a generic relationship. Broken lines indicate an uncertainty with respect to the exact point of origin of a lineage. Archaeal and eukaryotic (including viral) branches are colored blue, bacterial branches are colored green, branches that include predominantly proteins from plasmids, phages and mobile elements are colored red. Ancestral branches and branches outside the AEP superfamily are in black. The phyletic distribution is shown in brackets: B, Bacteria; A, Archaea; E, Eukaryotes; V, Viruses; > represents a proposed lateral transfer.
A structural alignment of the AEPs and the RCREs indicates that, in these two families, a conserved catalytic histidine located in the third strand of the RRM-like unit is congruently positioned (Figure 1). However, the histidine of the RCREs is directly involved in coordinating a magnesium or zinc ion, whereas the histidine of the AEPs interacts with a conserved aspartate that is involved in coordinating the catalytic divalent magnesium ion (25,56,57). The RCREs additionally have another N-terminal histidine in the third strand, forming the characteristic HXH motif, and a conserved tyrosine downstream of the flange (5,53) (Figure 1). In contrast, the primases have the distinct DxD motif in the first strand of the RRM-like unit and a conserved aspartate in the fourth strand (Figure 1). Thus, it appears that the RCREs and the AEPs evolved from a common ancestor that acted on DNA, with at least one shared active site residue retained in both families. Subsequently, each family acquired a specific constellation of functionally important residues, which was differentially fine-tuned for their distinctive metal ion affinities and activities. While the details of the residues involved in DNA-binding differ in the AEPs, RCREs and OBDs, the general shape of the surface of the molecule that contacts DNA appears to be conserved (25,54,58,59). Thus, the divergence of these three families from an ancestral DNA-binding RRM-like unit apparently was accompanied by specific innovations in the DNA-binding motifs, whereas the ancestral binding surface remained intact.
Besides the primases, the catalytic domains of the palm-type RRM fold are characteristic of the A, B and Y (DinB) families of DNA polymerases, certain novel predicted DNA polymerases of archaea and bacteria (the MJ1672-like proteins), phage DNA-dependent-RNA polymerases, RNA-dependent RNA polymerases of RNA viruses, reverse transcriptases and nucleotide cyclases (9–15). All these proteins seem to share a catalytic mechanism of di-metal-ion-mediated nucleotide transfer, whereby two acidic residues located at the end of the first strand and between the second and third strands of the RRM-like unit, respectively, chelate two divalent cations (Figure 1). The position of the divalent metal and at least one of the acidic residues that in the first strand of the RRM-like unit (the second aspartate in the DxD motif of AEPs), which is involved in coordinating the Mg2+, is almost identical between the AEPs and the catalytic palm module of the other polymerases and cyclases (9) (Figure 1). This suggests that the catalytic domain of the AEPs and the palm domain of the polymerases and cyclases have a common origin (Figure 3). Extrapolating from the activities of the members of this assemblage, it seems plausible that their common ancestor had at least a rudimentary nucleic acid polymerase activity which required an aspartate and a metal ion. In view of this conclusion, the monophyletic relationship between the AEPs, the RCREs and the non-catalytic OBDs (see above) seems somewhat paradoxical, given the lack of polymerase activity in the latter two groups of proteins. The simplest scenario reconciling these findings is that the RCREs and the OBDs were derived from a common ancestor with the AEPs, which was a polymerase, with the RCREs evolving the nuclease activity to displace the polymerase one and the OBDs losing the catalytic activity altogether. Alternatively, it cannot be ruled out that the common ancestor of the AEP-RCR-OBD assemblage and the palm-domain polymerases was merely a nucleic-acid-binding protein which used the conserved acidic residue to coordinate a divalent cation to aid in nucleic-acid-binding. Polymerase activity, then, would have been independently acquired in multiple descendent lineages, while the ancestral RCRE acquired nuclease activity.
Sequence analysis and detection of novel members of the AEP superfamily
Prior to the present study, the AEP superfamily included the replicative primases from archaea, eukaryotes and baculoviruses, the ligase- and Ku-associated DNA repair primases of several bacteria and the prim-pols (21,22,25). We were interested in the phyletic diversity of the prim-pol-like proteins and in detecting putative new, perhaps, divergent members of the AEP superfamily. For this purpose, we performed PSI-BLAST searches using as starting points several known primases and prim-pol proteins; the searches were iterated to convergence. Potential new members of the AEP superfamily that were detected in these searches with statistically significant _E_-values were used as starting points for transitive PSI-BLAST searches. Other proteins that had a DxD and sxb (b:basic, H/K/R) motif, similar to those in the primases, but were recovered with borderline _E_-values (E ∼ 0.05), were also further examined in transitive searches to assess the conservation of these motifs in their orthologs. In these cases, the relationship to the primases was also further evaluated using the Gibbs sampling algorithm for motif detection. For example, iterative searches with the Nanoarchaeum equitans primase NEQ395 (gi: 41615183) retrieved, in addition to previously known AEP superfamily members, the recently reported primase fused to the MCM domain (BC1863) (60) from the Bacillus cereus phage phBC6A51 (first pass, _E_-value: 3 × 10−3), a primase fused to a reverse transcriptase in Bdellovibrio, Bd2680 (Iteration 2, _E_-value: 3 × 10−4) and a primase from Bacteriophage RM 378 (Rm378p005, Iteration 6, _E_-value: 4 × 10−3). A PSI-BLAST search with the Sulfolobus pRN1 prim-pol domain (gi: 42543570) as the query retrieved, at significant _E_-values, AEP domains in proteins from Ferroplasma, the RepE/S proteins from Gram-positive bacteria, and uncharacterized proteins from a wide range of bacteria and phages, including Low-GC Gram-positive bacteria, actinomycetes, Thermus, Deinococcus, cyanobacteria and a few proteobacteria (Iterations 2–7, _E_-values: 10−6–10−44). In many of these proteins, the AEP module was fused to a D5-like Superfamily-III (SFIII) helicase. Transitive searches conducted with the Anabaena protein Avar03005527 containing the AEP module (gi: 45506180), which was detected in the above searches, retrieved the MIMI_L207 protein of the Mimivirus in the first pass of the BLAST search (_E_-value: ∼10−4) and the N-terminal domains of D5 helicase proteins from Iridoviruses and African Swine fever virus, and the Tb927.1.4010 protein from Trypanosoma brucei in the second and third iterations (_E_-value: ∼10−4–10−11). In further iterations of this search, hits to proteins from the vertebrate iridoviruses (Frog Virus 3 D5, Iteration 3, _E_-value: 10−20) were also obtained. The alignments produced in these searches encompassed regions highly conserved within the AEP domain, including the sxH motif.
Further transitive searches seeded with the N-terminal domain of the D5 protein of iridoviruses, which was found to be homologous to the AEP catalytic module, recovered the Paramecium bursaria Chlorella Virus (PBCV) A468R-like proteins from phycodnaviruses. In turn, PSI-BLAST searches with the PBCV A468R retrieved the N-terminal region of the D5-like proteins from poxviruses, the UL9 helicase-containing protein from ASFV and mimivirus, at least two other proteins of the mimivirus, several uncharacterized eukaryotic proteins (e.g. human FLJ33167), ORF7 and ORF49 from the Ostreid herpesvirus, and ORF63 from the Ictalurid herpesvirus. All these proteins share the DxD and the sxH motif characteristic of the AEP superfamily. Notably, searches initiated with the N-terminal regions of ORF7 and ORF49 of Ostreid herpesvirus also retrieved the primases of vertebrate herspesviruses with borderline _E_-value. A closer inspection showed that herpesvirus primases contained well-conserved equivalents of the DxD motif and the sxH motif, although the histidine was replaced with an arginine in most of these proteins. The ColE2 Rep primases were also recovered by similar transitive searches. A motif search using the Gibbs sampling algorithm detected two statistically significant 7 amino acid motifs (P ≤ 10−9) in the N-terminal regions of the D5 proteins of the NCLDV, the herpesviruses primases, AEPs, prim-pols and the ColE2 Rep proteins. These motifs corresponded to a region encompassing strand 3 with the DxD signature (the first strand of the RRM-like unit) and another region corresponding to strand 5 with the sxH signature. In an independent procedure, we constructed a seed alignment of the AEP modules from the proteins detected in our initial searches and derived an HMM from it. This HMM was used to progressively search the viral and cellular proteomes to detect potential new members of the superfamily. All proteins with statistically significant similarity to the HMM were then added to the alignment, a new HMM was constructed and the search resumed. Using this procedure, we recovered nearly the same group of proteins as recovered in the PSI-BLAST searches, thereby supporting the validity of the detected relationships.
To further investigate these relationships, we used similarity-based single-linkage clustering (the BLASTCLUST program) to classify the known and newly detected members of the AEP superfamily. Multiple alignments were constructed for each of the identified groups using the T-coffee program. A nearly complete congruence was seen in the arrangement of the predicted secondary structure elements and catalytic residues in all groups. Taken together, these observations indicate that the N-terminal domain of the D5-like proteins of poxviruses, ASFV, iridoviruses and the mimivirus, the A468R-like proteins of phycodnaviruses, the herpesvirus primases, the eukaryotic homologs of these viral proteins and the plasmid ColE2 Rep proteins were novel members of the AEP superfamily. The individually aligned clusters were further unified into a super-alignment using as anchors the conserved motifs and secondary structure elements derived from the structural alignment of the prim-pol and primases. The alignment was further refined using the GIBBS sampling search and the PSI-BLAST search results. All members of the extended AEP superfamily contained the structural core consisting of the N-terminal (αβ)2 unit and the RRM-like unit and bearing the active site residues (see above), with the characteristic C-terminal extension forming the flange and the distal strand (Figure 2). Residues that were consistently conserved across all the families included a hhhDhD (h: hydrophobic), motif in strand 3, an sss (s: small) motif at the end of strand 4, a uh+hhh motif (+: basic residue, mostly histidine but an arginine or lysine in some families; for details see below) in strand 5, a h- motif (-: acidic, mostly aspartate) in strand 6 (which can sometimes adopt a helical configuration) and a polar residue in the second strand of the terminal hairpin (Figure 2).
Evolutionary classification of the AEP superfamily
To identify the major clades within the AEP superfamily, the multiple alignment (Figure 2) was examined for distinct sequence signatures characteristic of subsets of the superfamily members. Single linkage clustering using BLAST-scores with the BLASTCLUST program was employed to identify sub-groups and probable orthologous lineages. Finally, at the level of high sequence similarity, such as within an orthologous group or a tight cluster of paralogs, conventional phylogenetic tree analysis using maximum-likelihood, neighbor-joining and minimum evolution methods was performed to unravel the evolutionary history of each such group. In the case of fast-evolving proteins of viruses and extra-chromosomal elements, contextual information from domain architectures was also used as a phylogenetic marker. Using these approaches, the AEP superfamily was classified into 13 major families, of which 12 could be further grouped into three higher-order clades (Figure 3).
The AEP proper clade
This major clade includes three families, namely, the classical AEP small subunits, the Lef-1 like primases of baculoviruses and the bacterial NHEJ primases associated with the Ku protein and the ATP-dependent ligases, and is unified by several synapomorphies (Table 1 and Figure 2). Recent genome sequencing projects added several new members to this family. These include the B.cereus phage phBC6A51 protein BC1863, the Bdellovibrio protein Bd2680 and the RB12213 protein from Rhodopirellula. These proteins are closer in sequence to the archaeal versions of the AEP superfamily, but are not specifically related to a representative from any one archaeal species. BC1863 is fused to an MCM domain (60) and the gene for RB12213 is adjacent to the dnaB gene of Rhodopirellula (Figure 4). An MCM gene is also found next to the primase gene (MJECS07) of the small extrachromosomal element of Methanocaldococcus jannaschii (Figure 4). This pattern is characteristic of the diverse primase-helicase associations seen in other plasmids and phages (see below) (Figure 4). The Bdellovibrio Bd2680 primase is fused to a reverse transcriptase and might be involved in synthesizing a primer for the latter (Figure 4). The classical AEP family is generally uncharacteristic of bacteria. The few members of this family that are encoded on bacterial chromosomes are likely to be relatively late transfers from phage or plasmid replicons.
Table 1.
The major clades of the AEP superfamily and their distinctive features
| Families | Phyletic distribution | Synapomorphies and specific comments |
|---|---|---|
| AEP proper clade | Arginine between strand 1 and helix-2, small residue between helix-2 and strand 2, charged residue (mostly glutamate) in strand 3 and a basic residue preceding strand 5 | |
| NHEJ primases family | Proteobacteria, Actinomycetes, Parachlamydia, Low-GC Gram-positive bacteria | Most members are fused to or in the gene neighborhood of a DNA ligase and a predicted nuclease |
| Lef-1 like baculovirus primase family | Baculoviruses | Arginine between helix-1 and strand 1, histidine between strand 1 and helix-2, acidic residue between helix-2 and strand 2 |
| AEP primase small subunit primase family | Archaea, eukaryotes, B.cereus phage phBC6A51_, Bdellovibrio_, Rhodopirellula | Most eukaryotes have a single primase small subunit with duplications in a few species, such as C.elegans, Entamoeba histolytica and Cryptococcus neoformans |
| NCLDV/Herpesvirus primase clade | Glutamate [present as an Exb (b: big, mostly hydrophobic) motif] in strand 2, a lysine in the turn between strands 4 and 5, a hydrophobic residue before helix-3 and a polar residue at the end of strand 4 | |
| Iridovirus primase family | Iridoviruses, ASFV, mimivirus and kinetoplastids | Fused to PriCT-2 domain |
| Herpes-pox primase family | Poxviruses, phycodnaviruses, ASFV, mimivirus, vertebrates, Ciona, Apis, plants, apicomplexans, Dictyostelium | Fused to a C-terminal strand rich zinc ribbon-like domain that has 3 cysteines and one histidine |
| Prim-pol clade | Proline residue in strand 1 | |
| Prim-pol family | Bacterial and phage proteins in Low-GC Gram-positive, Actinomycetes, Thermus, Trichodesmium, Bacteroides, Magnetospirillum, Vibriophages, Halobacterium, Methanosarcina and crenarchaeal plasmids | Threonine residue at the beginning of strand 4 and a ‘sS’ motif (s: small, mostly Proline) at the beginning of the first strand of the terminal hairpin. Most members are fused to a PriCT-1 domain |
| E.coli Z1568-like family | O157:H7 strain of E.coli, Desulfovibrio, Burkholderia cepacia complex phage BcepC6B, ε-proteobacteria, Corynebacterium glutamicum, Bacteroides fragilis, Streptococcus pneumoniae and cyanobacterial plasmids | Polar residue in strand-2 (aspartate or asparagine), Ps (s: small, mostly asparagine) motif preceding the flange, polar residue (mostly asparagine) in the first strand of the terminal hairpin and a hPhp (p: polar) motif preceding the terminal strand |
| Deinococcus DR0530-like family | Deinococcus, α- and β-proteobacteria, phages of Burkholderia and Bordetella, cyanobacterial plasmids and Picrophilus torridus | Members of this family lack the conserved histidine in strand 5 and instead have a polar (often, basic) residue, conserved glutamate instead of the aspartate residue in strand 6, HP motif in the first strand of the terminal hairpin. Most members fused to PriCT-2 |
| Anabaena all3500-like family | Cyanobacteria, Streptococcus pyogenes, Bacillus clarkii bacteriophage BCJA1c | Asparagine residue in strand 2, basic residue at the end of strand 6, basic residue in the first strand of the C-terminal hairpin |
| Bradyrhizobium bll5242-like family | Bradyrhizobium, Rhodobacter, Desulfotalea, Burkholderia cepacia phages Bcep43, Bcep1 and Bcep781 | Basic residue between strand 1 and helix-2, threonine N-terminal to strand 2, Proline N-terminal to helix-3. Strand with a Gah (a: aromatic, h: hydrophobic) motif between helix-4 and strand 6 |
| ColE2 Rep-like family | Proteobacteria, Actinomycetes and Thermus | Proline after helix-1, oss motif (o: serine or threonine, s: small mostly aspartate or asparagine) in strand 1, Ps (s: small, mostly asparagine) motif in strand 4, lysine in the flange, HxW motif in the first strand of the hairpin. Fused to Pri-CT1 and HTH domains at the C-terminus |
| RepE/RepS family | Conjugative plasmids pXO2 of Bacillus anthracis and E.faecalis RE25, pTS1 plasmid of Treponema denticola, plasmids from Low-GC Gram-positive bacteria and cyanobacteria | Fused to a wHTH at the C-terminus |
| Families not associated with any particular clade | ||
| BT4734-like family | Bacteroides | Basic residue in helix-1, lysine instead of the conserved histidine in strand 5, conserved aspartate in the terminal strand of the hairpin |
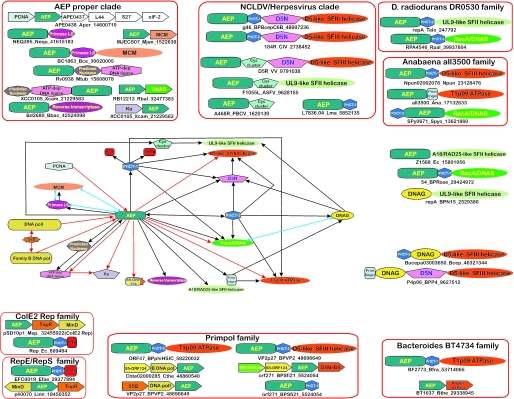
Figure 4.
Ordered graph of domain architectures and genome contexts. Each vertex represents a domain and the edges represent a contextual association. Domain combinations are shown as black arrows, with the arrow pointing from the N-terminus to the C-terminus of the multi-domain protein. Gene neighborhood associations are shown as red arrows with the arrows pointing in the 5′–3′ direction of the coding sequence. The blue lines with boxed ends represent experimentally observed functional associations. Domain architectures and gene neighborhood organizations are shown around the ordered graph. Where possible, these are organized into clade- or family-specific groups enclosed in an orange box. Proteins or genes that are depicted as domain architectures or operon clusters are denoted by the standard notation as in Figure 2. The species abbreviations are as in Figure 2. Genes with conserved neighborhoods are shown as boxed arrows with the arrow pointing in the 5′–3′ direction of the coding sequence. The C-terminal tail motif of the SFI-ORF24 proteins is represented by an orange extension in the domain representation.
All archaeal and eukaryotic members of the AEP small subunit family and the baculoviral Lef-1 family appear to form a functional complex with the respective orthologs of the eukaryotic primase large subunit (yeast Pri2p and baculovirus Lef-2 protein) (61–64). Notably, in N.equitans and BC1863, the AEP is fused to the ortholog of the large subunit (Figure 4). While at least one ortholog of the large subunit is present in archaeal, eukaryotic, phBC6A51 and baculoviral genomes, no homologs of the large subunit were detected in the other bacterial, plasmid and phage replicons, suggesting that these small-subunit homologs might function independently of the large subunit.
We also detected several previously unnoticed members of the bacterial NHEJ primase family scattered among proteobacteria, actinomycetes, Parachlamydia and several Low-GC Gram-positive bacteria (Table 1). As described previously, most of the bacterial NHEJ primases are fused to a DNA ligase and a predicted nuclease located N-terminally to the primase domain (21,22) (Figure 4). Solo versions are typically encoded in the same neighborhood with genes for DNA ligases and Ku homologs (22,23) (Figure 4). The patchy phyletic pattern of this system, with highly conserved representatives present in various, phylogenetically distant bacteria, strongly suggests that they disseminated via horizontal gene transfer. This is reminiscent of some other DNA repair operons that have widely disseminated among prokaryotes (15,22,65,66).
Prim-pol-like clade
This clade includes proteins from a wide range of bacteria, bacteriophages, plasmids and a few archaea (Table 1 and Figure 2). With the exception of two families, proteins in this clade also have one of two distantly related C-terminal α-helical modules which we designate the Primase-C Terminal-1 (PriCT-1) and Primase-C Terminal-2 (PriCT-2) domains (Figure 4). Based on the presence of conserved residues, domain architecture and genome contexts, seven distinct families are discernible in this clade, which include the prim-pol proper family, the Z1568-like family, the D.radiodurans DR0530-like family, the Anabaena all3500–like family, the Bradyrhizobium bll5242-like family, the ColE2 Rep-like family and the RepE/RepS family (Figure 3).
The Prim-pol proper family typified by the primase-DNA polymerase domain of the crenarchaeal RepA-like proteins is found only in prokaryotes and their viruses (Table 1 and Figure 2). The presence of these proteins in several phages suggests that the extensive, sporadic dissemination in bacteria was mediated by lysogenic phages and plasmids. In Streptomyces and Clostridium thermocellum, there is a lineage-specific expansion of solo prim-pols with ∼8–13 members. Interestingly, in six members of this family in Streptomyces and the Tfus020011 protein of Thermobifida, the second asparate of the DxD motif is mutated and, in the Streptomyces members, the conserved histidine in strand 5 is mutated as well, suggesting that these proteins are inactive primase derivatives (Figure 2). Such abundance of apparently inactive versions is reminiscent of inactivated transposases that are often encountered in several multicopy transposons from various genomes (67). One may speculate that these AEP expansions in Streptomyces and Clostridium represent a novel class of DNA transposons. The prim-pols might function as primases and, possibly, also as DNA polymerases during replicative transposition of these putative novel mobile elements. The presence of apparently inactivated version of the prim-pol domain in some of these putative transposons suggests that the primase activity could be supplied in trans by the active versions. Experimental study of this system would be of considerable value because it might uncover a novel mode of transposon propagation.
The Z1568-like family is sporadically found in bacteria and their plasmids (Table 1 and Figure 2). The proteins of this family typically contain a PriCT-1 domain immediately C-terminal to the AEP module (Figure 4). Members of this family in ε-proteobacteria and cyanobacterial plasmids have a conserved glutamate in place of the aspartate in strand 6. The ε-proteobacterial versions also have an asparagine residue replacing the second aspartate in the DxD motif (Figure 2).
D.radiodurans DR0530-like family is sporadically distributed in prokaryotes (Table 1); members of this family lack the conserved histidine in strand 5 and instead have a polar (often, basic) residue (Figure 2). They have a conserved glutamate instead of the aspartate residue in strand 6 and share an HP motif in the first strand of the terminal hairpin. Most members of this family (with the exception of the Picrophilus PTO0356 protein) are fused to a PriCT-2-like domain. The small Anabaena all3500–like family contains members showing fusions to both PriCT-1 and PriCT-2 domains (Table 1) (Figures 2 and 4). In some versions, such as the SPy0671-like protein of Streptococcus pyogenes and the Bacillus clarkii bacteriophage BCJA1c, the PriCT-2 domain is atypically present N-terminal to the AEP module (Figure 4). The Bradyrhizobium bll5242-like family is another small group with a restricted phyletic pattern in bacteria and phages (Table 1 and Figure 2).
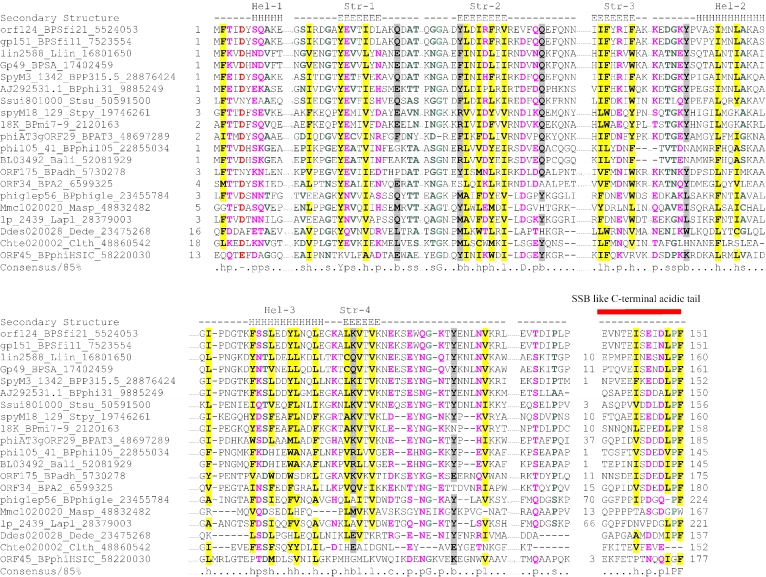
Members of the ColE2 Rep family include primases of ColE2 family plasmids that are mainly present in proteobacteria, a few actinobacteria and Thermus (Table 1 and Figure 2). They are characterized by a fusion to a helix–turn–helix (HTH) domain to the C-terminus of the PriCT-1 domain, which is essential for origin-binding (68) (Figure 3). It has been shown that Rep proteins are involved in the synthesis of a 3 nt RNA primer during the initiation of plasmid replication (28,29).
Proteins of the RepE/RepS primase family (Table 1 and Figure 2) are fused to a winged HTH domain, which flanks the C-terminus of the primases (Figure 4). The domain architectures and gene neighborhoods (see below and Figure 4) of these AEPs suggest that they might be distantly related to the Rep family of the ColE2 plasmids, although they share very few sequence features of the prim-pol clade.
The NCLDV-herpesvirus primase clade
Members of this clade include predicted primases detected in the NCLDV, herpesviruses, kinetoplastids and a novel eukaryotic subfamily exemplified by the human protein FLJ33167 (hereinafter we refer to these proteins as the Eukprim2 for Eukaryotic primase, version 2). The synapomorphies of this clade include two strongly conserved residues, a glutamate [present as an Exb (b: big, mostly hydrophobic) motif] in strand 2 and a lysine in the turn between strands 4 and 5, a hydrophobic residue before helix-3 and a polar residue at the end of strand 4 (Figure 2). Superposition of these conserved residues onto the available structures of the AEP superfamily proteins suggests that the conserved lysine and glutamate are positioned close to the active site and might have a role in substrate interaction, such as binding the nucleotide backbone. Based on the domain architectures, this clade can be further classified into two families, the iridovirus primase family and the herpes-pox primase family.
The iridovirus primase family is characterized by a C-terminal fusion of the primase domain to the α-helical PriCT-2 domain (Figure 4). Members of the family include the D5-like proteins from iridoviruses and ASFV (C962R), the mimivirus MIMI_L207 protein and the Leishmania L7836.04-like protein from the kinetoplastids. In addition to the PriCT-2 domain, the iridovirus and ASFV proteins are fused to D5N and D5 helicase domains. The mimivirus MIMI_L207 is a neighbor of the D5-like helicase in the genome, which implies a functional association. The kinetoplastid L7836.04-like proteins, while closely related to other members of this family, lack the characteristic, conserved glutamate in strand 2, although they retain the conserved lysine between strands 4 and 5 (Figure 2). This is the second putative primase of the kinetoplastids, in addition to the typical eukaryotic primase.
The Herpes-pox primase family is characterized by the presence of a conserved C-terminal β-strand-rich region in place of the PriCT domains (Figure 4). In most proteins of this family, this region has three conserved cysteines and a histidine, and its secondary structure pattern resembles a highly derived Zn-ribbon (Figure 5). Members of this family include the A468R-like proteins from phycodnaviruses, the N-terminal region of the poxvirus D5 proteins, F1055L from ASFV, the mimivirus proteins MIMI_R8, MIMI_L537 and MIMI_L794, the herpesvirus primases and the eukaryotic Eukprim2 proteins. Several members of this family also show fusions and associations with different ATPases (Figure 4). The poxvirus N-terminal primase domains are fused to a D5 helicase of SFIII (separated by the D5N domain), whereas the ASFV F1055L, the mimivirus MIMI_R8 and the Ostreid herpesvirus primases, ORF7 and ORF49, are fused to a UL9-like helicase of SFII (Figure 4). Similarly, the third AEP encoded by the Ostreid herpesvirus, ORF66, is adjacent to a gene encoding a UL9-like helicase (ORF67). These fusion proteins with the AEPs and UL9-like helicases in the NCLDV and herpesviruses are closely related and suggest gene exchange between these two clades of large DNA viruses. The vertebrate herpesvirus primases of this family (UL52/70) are characterized by a replacement of the conserved histidine in strand 5 by an arginine (Figure 2). This substitution is also seen in the divergent gene 63 product of the fish (Ictalurid) herpesviruses. In contrast, the primases of the herpesviruses of the bivalves, the Ostreid herpesvirus ORF7, ORF49 and ORF66, show a typical sxH signature in strand 5 (Figure 2). Moreover, the Ostreid herpesvirus proteins lack several inserts shared by the vertebrate herpesviruses (see below), suggesting that these inserts and the substitution of the conserved histidine to arginine occurred after the radiation of vertebrate herpesviruses from the common ancestor with the mollusk herpesviruses. Apparently, the vertebrate herpesvirus primases evolved rapidly after this divergence. Alternatively, the primase of the Ostreid Herpesvirus might have been displaced by a version from the NCLDVs or the cellular Eukprim2.
Figure 5.
Multiple alignment of the zinc ribbon-like domain located C-terminal to the AEP domain in poxvirus and herpesvirus primases and the Eukprim2 family. The coloring scheme, consensus abbreviations, secondary structure representations and species abbreviations are as in Figure 2. The residues predicted to be involved in metal binding are shaded red. Poorly conserved short inserts seen in some sequences are shown with a reduced font size.
The Eukprim2 proteins are present in vertebrates, Ciona intestinalis, Apis mellifera (honey bee), plants, apicomplexans and the slime mold Dictyostelium. The apicomplexan versions are closely related to plant ones (Figure 2), suggesting that they were acquired from the algal endosymbiont of the apicomplexan ancestor. This unusual distribution of Eukprim2 suggests that the gene for this protein was acquired early in the evolution of the eukaryotic crown group, lost independently on multiple occasions in the fungi and animals, such as Caenorhabditis elegans and Drosophila. The Eukprim2 is not closely related to the principal eukaryotic primases (the AEP small subunit family discussed above). Together with the observation that the maximum diversity of the AEPs is seen in viruses and other mobile elements, this suggests that Eukprim2 was acquired from a viral or plasmid source by an early eukaryote.
The ultimate origin of the NCLDV-herpesvirus primase clade, which is restricted in its distribution to eukaryotes and their viruses, probably, can be traced to bacteriophages or bacterial proteins that have a PriCT-2 domain fused to the AEP core. An ancestral version of this AEP present in an ancient, large eukaryotic DNA virus acquired the clade-specific glutamate and arginine residues. This was, in all likelihood, the precursor of the version that propagated in the NCLDV and herpesvirus genomes, although the details of the exchanges of the AEP and associated domains between these two large groups of DNA viruses remain obscure. The greater diversity of the domain in the NCLDVs suggests that herpesviruses might have acquired their extant primase from a member of the NCLDV class. This scenario further implies that the PriCT-2 domain was displaced by a β-strand-rich domain containing a cysteine cluster in the Herpes-pox primase family. The Eukprim2 and the kinetoplastid L7836.04-like proteins, which show a scattered distribution among eukaryotic lineages, were probably acquired from viral sources on two occasions independently. No role for this protein has been reported in any of the well-studied eukaryotic replication initiation systems. The patchy phyletic profile of these proteins (Table 1) is in contrast to the core components of the replication apparatus (18) and argues against an indispensable role in replication. It seems more likely that these proteins function as primases in a novel DNA repair pathway and/or in specialized DNA amplification systems in specific cell types. The presence of multiple primases in the mimivirus and ASFV is intriguing (Table 1). Given their large genomes, the NCLDV might have greater selective pressures for DNA repair than smaller DNA viruses. Furthermore, since most of the NCLDVs replicate in the cytoplasm or in independent compartments, they might not have ready access to cellular DNA repair enzymes. Hence, they might deploy distinct primases for the initiation of DNA synthesis during replication and repair. It is also possible that these viruses have multiple replication origins, which are initiated by different primases or that their leading and lagging strands are initiated by distinct primases.
Members of the Bacteroides specific BT4734-like family do not have any sequence specific features to warrant their inclusion in any one of the above families (Table 1, Figure 2). At least 5 members of a total of 11 detected in Bacteroides thetaiotaomicron are potentially inactive suggesting that, similar to the members of the prim-pol family discussed above, these proteins might belong to a novel type of transposon. Of the 11 proteins of this family in B.thetaiotaomicron, at least 5 are fused to a D5-like helicase and the remaining six are either fused to or in the neighborhood of a T1p09-like Superfamily III helicase. Some versions of the latter variety are also fused to a PriCT-1 domain.
Domain architectures of proteins containing the AEP domain
The domain architectures and conserved gene neighborhoods of the AEPs throw considerable light on the diverse functional associations of these proteins in replication systems. While the domain architectures of cellular proteins of this superfamily are rather stereotypical, the architectures observed in viruses and extrachromosomal elements show considerable diversity (69). We depicted these associations as an ordered graph with domains forming the nodes of the graph and the architectural or gene-neighborhood connections forming the edges (Figure 4). The most striking contextual theme that emerged from this analysis was the association of members of the AEP superfamily with diverse ATPases of the P-loop NTPase fold. In particular, AEPs are fused to three distinct groups of AAA+ ATPases: (i) the D5-like SFIII helicases of the NCLDVs and other viruses (70,71), (ii) the phage T1 T1p09-like highly derived SFIII helicases and (iii) the MCM family of AAA+ ATPase in the BC1863 protein of the B.cereus phage phBC6A51 (60,70). Analogous to eukaryotic and archaeal cells, the phage MCM ATPase is likely to function as the principal helicase in the initiation of replication (60,72). The AEPs are also fused to members of a subfamily of phage DnaB helicases, which belong to the RecA ATPase class (73), and two distinct groups of superfamily II helicases, the UL9 and A18R/Rad25-like helicases (Figure 4). Helicases of the RecA and AAA+ superfamilies typically form hexameric rings around the DNA and are known as ring helicases, whereas the UL9 and A18R/Rad25-like SFII helicases do not form rings (66,74,75). Hence, the AEPs appear to be able to function in conjunction with both types of helicases that have substantially different mechanisms of DNA unwinding.
With the exception of the AEP proper clade, in all other families of the AEP superfamily, the most common fusion of the primase domain is with D5-like SFIII helicases. This suggests that the D5-like helicases were the original functional partners of the AEPs, other than those of the AEP proper clade. However, in most of these families, there are at least a few representatives with fusions to other helicases (Figure 4). For example, the T1p09-like highly derived SFIII helicases are fused to the AEPs in some members of the prim-pol proper, DR0530-like, all3500-like and BT4734 families. The DnaB/RecA-like ATPases are fused to AEPs of the prim-pol proper family in the mycobacteriophage rosebush gp54-like proteins, and in the SPOA0086 protein from Silicibacter, to the DR0530-like family in RPA4546 protein from Rhodopseudomonas, and to the all3500-like family in SPy0671 in Streptococcus pyogenes and the Bacillus clarkii bacteriophage BCJA1c. The UL9-like SFII helicases are fused to AEPs of the DR0530-like family (in cyanobacterial plasmid pUH4; gi: 247792) and in members of the Herpes-pox primase family. Furthermore, in different members of the Bradyrhizobium bll5242-like family, the AEP is associated with distinct versions of the D5-like ATPases, which are only distantly related to each other. This diversity of domain fusions suggests that there have been several independent associations between helicases and AEPs, which appear to have evolved via displacement of one type of helicase by another in viral and plasmid genomes.
Interestingly, analysis of the domain architectures of multi-domain proteins containing the TOPRIM primase domain also revealed fusions to D5-like, DnaB-like and UL9-like helicases, which are closely related to the helicases associated with the AEPs (Figure 4). In particular, in the gp37 protein of phage N15, the TOPRIM primase is fused to a UL9 helicase, whereas in the Bcep02006798 protein of the bacterium Burkholderia fungorum, it is fused to a C-terminal D5-like helicase. These combinations of unrelated primases with the same repertoire of helicases suggest that the primases of the AEP and TOPRIM superfamilies are functionally equivalent or at least highly similar. These independently derived analogous architectures also suggest a strong mechanistic coupling of the earliest stages of DNA synthesis, such as, unwinding of the double-stranded template at the initiation site and primer synthesis. Thus, in the genomes of extra-chromosomal elements and phages, a remarkably diverse array of combinations of the enzymes catalyzing these two steps had been explored during evolution. In many cases, the D5-like ATPase domain is preceded by another conserved globular domain, the D5 N-terminal (D5N) domain. The D5N domain is also found as a solo protein encoded by genes in the vicinity of those coding for D5-like helicases (e.g. SpyM3_1340 from Streptococcus phage 315.5). This domain is predicted to adopt an α+β fold and is characterized by the presence of conserved aromatic residues (mostly tryptophan) in strands 2 and 3, and a Ghhpxp (p polar, the first polar residue is, mostly, an aspartate or asparagine) motif in strand 4 (Figure 6). The role of the D5N domain remains unclear, but its strict association with the D5-like proteins suggests that it might be specifically involved in some aspect of substrate recognition by these helicases, which function with either AEP or TOPRIM-fold primases.
Figure 6.
Multiple alignment of the D5N domain. The coloring scheme, consensus abbreviations, secondary structure representations and species abbreviations are as in Figure 2.
In addition to the helicases, AEPs as well as the TOPRIM domain primases form associations with several non-catalytic domains. Perhaps, the most notable of these are the PriCT-1 and PriCT-2 domains, which are typically fused to the C-terminus of the AEP domain of the prim-pol, the BT4734-like family or NCLDV-herpesvirus primases (Figure 4). The PriCT-1 domain was also detected as a stand-alone protein, e.g. Chte02003491 of Clostridium thermocellum, and the PriCT-2 domain is also found as an N-terminal fusion to the TOPRIM domain, e.g. in the Bcep02006798 protein of B.fungorum (Figure 4). The two domains seem to be distantly related as iterative database searches starting from the PriCT-1 domain sequences recover the PriCT-2 domains, albeit with relatively high _E_-values (data not shown). This is supported by the nearly identical secondary structure predictions for the PriCT-1 and PriCT-2, which indicate a core of four conserved helices (Figure 7). The two domains share several conserved hydrophobic residues in the helices across the alignment. The members of the Herpes-pox primase family from various NCLDVs and herpesviruses contain a Zn ribbon-like domain in place of the PriCT domains (Figure 5). Experiments in herpesvirus UL52 have shown that mutations in this Zn-chelating domain severely compromise DNA-binding and primase activity, suggesting that this domain is involved in the recognition of initiation sites (76).
Figure 7.
Multiple alignment of the (A)PriCT-1 and PriCT-2 (B) domains. The coloring scheme, consensus abbreviations, secondary structure representations and species abbreviations are as in Figure 2. Furthermore, alcohol side chain containing residues (ST) are colored blue and denoted by an ‘o’ and aliphatic residues (LIV) are shaded yellow. Equivalent helices in PriCT-1 and PriCT-2 have been aligned with each other. Poorly conserved short inserts seen in some sequences are shown with a reduced font size.
Some members of the all3500-like family contain a primase-type Zn-finger fused with the AEP domain, which resembles the architecture of the bacterial DnaG primases (Figure 4). In the latter, the Zn-finger targets the catalytic TOPRIM domain to the primer initiation sites (77,78), and, most likely, it performs the same function in association with the AEP domain. In particular, these domains might help in specific cis interaction of the primases with the replication initiation sites of their cognate replicons. The congruence of these architectures further supports the functional equivalence of the TOPRIM and AEP superfamily primases and suggests that these primase domains might have repeatedly displaced each other in various replication systems, particularly, those of extra-chromosomal elements, while associating with the same set of DNA-binding domains. Given the domain architectures in which the TOPRIM and the AEP primases occupy equivalent positions in the same polypeptide with respect to the other domains (Figure 4), it even seems likely that some of these displacements occurred in situ, within the same gene.
Members of the AEP proper clade show fusions to neither PriCT domains nor the β-strand-rich Zn-chelating C-terminal domain. However, in the BC1863 protein of the B.cereus bacteriophage phBC6A51 and in the AEP of the archaeon N.equitans (NEQ395), a region homologous to the large subunit of the cellular primases and baculovirus Lef-2 is fused to the C-terminus of the AEP domain. This consists of two distinct, conserved domains (Figure 8). The N-terminal domain is largely alpha-helical and culminates in the first conserved cysteine (Figure 8). The C-terminal domain, which is predicted to have two conserved helices and two strands, contains three conserved cysteine residues. These four cysteine residues are probably involved in coordinating a metal ion (Figure 8). The Lef-2 proteins of the baculoviruses contain another potential α-helical Zn-cluster with four conserved cysteines between the N- and C-terminal domains. All the other members of the primase large subunit family contain an unrelated α-helical region between the conserved N- and C-terminal domains (Figure 8). In BC1863 and NEQ395, the large-subunit module effectively occupies a position equivalent to that of a PriCT or Zn-chelating domains in other AEPs (Figure 4). These associations and the absence of other conserved C-terminal domains in members of the AEP-proper clade, which physically associate with the primase large subunit, suggest that the large subunit, PriCT domains and the β-strand-rich Zn-chelating domain of the Herpes-pox primase family have similar functions, perhaps facilitating the association of the AEP domain with DNA.
Figure 8.
Multiple alignment of the Primase Large subunit. The coloring scheme, consensus abbreviations and secondary structure representation are as in Figure 2. Short inserts are shown with a reduced font size, whereas longer inserts are represented as numbers. The secondary structure was predicted using the JPred program. The ‘a’ in the consensus abbreviations represents aromatic residues (FWY) that are shaded yellow. The cysteine residues predicted to have a role in metal-binding are shaded red. The granuloviruses appear to have lost their C-terminal cysteine cluster. Species abbreviations are as in Figure 2.
In addition to the Zn-binding domains that are fused to the AEP domains, we detected several lineage-specific Zn-clusters with characteristic patterns of cysteines and histidines, which are inserted within the AEP domain (Figure 1). These include:
- A Zn-cluster with three cysteines and a histidine between strand 1 and helix-2 in a subset of members of the prim-pol family from Magnetospirillum and Streptomyces;
- A cysteine cluster with four conserved cysteines, occurring in the same position as the previous insert, in members of the Z1568-like family, which are distinguished by a fusion to A18R-like helicases;
- A small cysteine cluster with 3–4 conserved cysteines located immediately downstream of the DxD motif in the archaeal primases;
- A cysteine cluster with at least three conserved cysteines in the loop between strands 4 and 5, preceding the conserved motif-II in tetrapod herpesviruses;
- A predicted Zn-ribbon in the terminal hairpin after the flange in the ESV ORF45 protein and its ortholog in Feldmania irregularis virus A;
- A small Zn-cluster with a histidine and three cysteines in the prim-pols of the Sulfolobus pRN-like plasmids, which occurs in the same position as the previous insert (Figure 1).
While insertions of Zn-ribbons and Zn-clusters are occasionally found in other protein domains involved in nucleic acid metabolism, the multiple, independent insertion in different positions within the same fold is an intriguing feature that is unique to the AEP superfamily. Superposition of the locations of these inserts on the structural scaffold provided by the two primase structures shows that most of the inserts are located in loops and are likely to be on the surface of the molecule (Figure 1). This suggests that the chelated Zn ions are used to stabilize particular, extended, surface-exposed loops in the AEP domain. These loops might have been recruited for recognizing specific template structures at the primer initiation sites. More specifically, there might be a selective pressure on the primases due to competing replicons to efficiently recognize cis primer initiation sites that are specific to the cognate replicon, and this recognition could be mediated by the Zn-clusters.
Conserved gene neighborhoods (predicted operons) associated with the AEP superfamily
It has been convincingly demonstrated that strongly conserved gene neighborhoods typically reflect physical interaction between the encoded proteins and/or consecutive functions in a biochemical pathway (79–81). Accordingly, analysis of gene neighborhoods throws light on the functional context of a particular gene. Certain highly characteristic mobile operons containing the AEPs of the NHEJ family have also been described previously and the predictions made on the basis of the conserved gene neighborhoods have been experimentally verified (22,23) (Figure 4). Not surprisingly, the most common conserved gene neighborhoods of the AEP superfamily involve co-occurrence with genes for the above-mentioned families of helicases and genes encoding solo versions of AEP-associated domains, such as the PriCT domains. In addition, members of the AEP superfamily were detected in conserved gene neighborhoods with genes for the single-strand-binding (SSB) protein, DNA polymerase I, PCNA, DNA polymerases of the B family and the ParA-like proteins (Figure 4). For example, the gene for the AEP protein VP2p27 of Vibriophages forms a potential operon with DNA polymerase I and SSB genes (Figure 4). Similarly, the Clostridium thermocellum pol-prim Chte02000285 is found in a potential operon with genes encoding a family B DNA polymerase (Figure 4). The primases of the plasmid-borne ColE2 Rep and RepE/S families show associations with genes for ParA-like ATPases of the MinD family and TnpR-like resolvases (Figure 4). The MinD ATPase prevents aberrant formation of the septum near the poles of the cell (82). Hence, the plasmid-borne ParA protein might cooperate with the AEP and the TnpR-like resolvase in coupling the partitioning of these plasmids with DNA replication and chromosome resolution. In Methanosarcina, Methanococcoides and crenarchaea, the AEPs are lodged in the ribosomal superoperon (80) and are usually encoded next to the ribosomal proteins L44, S27 and the translation initiation factor eIF2 (Figure 4), along with the DNA replication clamp PCNA; functional implications of this association are unclear beyond the possibility of a general, higher-order regulation of essential housekeeping processes. This gene neighborhood might be an atavistic feature in which the primase remains associated with the original core replicon of primordial cells.
The AEPs of the prim-pol proper family encoded by Low-GC Gram-positive bacteria and their plasmids and bacteriophages (e.g. Streptococcus thermophilus bacteriophage Sfi21) are encoded in a conserved gene neighborhood with an SFII helicase (in addition to the D5-like helicase) and a small, uncharacterized α+β protein (Sfi21-ORF124) (Figure 4). This protein and several of its orthologs in the Gram-positive phages contain a C-terminal region with high sequence similarity to the characteristic tail motif, which occurs C-terminal to the OB-fold in bacterial SSBs (Figure 9). It has been shown that this C-terminal tail, which is enriched in acidic residues, interacts with components of DNA repair and replication machinery, such as Exonuclease I, DNA glycosylase, the PriA helicase and the κ subunit of DNA pol III (83–86). Several residues are conserved across the Sfi21-ORF124 family; of particular note are a conserved aspartate preceding helix-1, a glutamate in strand 1 and two acidic residues after strand 4. The conservation of these acidic residues suggests the presence of a potential metal-coordination site (Figure 9). The predicted secondary structure of Sfi21-ORF124-like proteins includes a conserved core with three of the strands arranged sequentially and flanked by two helices (Figure 9). This secondary structure pattern resembles that of the RAD52-like proteins, which have a dsRNA-binding domain fold, with a core of three strands flanked by two helices (Figure 9) (87). Previous analyses have shown that members of the RAD52 superfamily of ssDNA-binding proteins can be extremely divergent in sequence and show functional equivalence to SSB in viral operons (65). Hence, these observations suggest that members of the Sfi21-ORF124 family are novel SSB proteins with a RAD52-like fold. These proteins are likely to function in conjunction with the phage-encoded AEPs by binding the ssDNA template at the primer initiation sites.
Figure 9.
Multiple alignment of the putative RAD52-like domains encoded in the same predicted operons with prim-pols. The coloring scheme, consensus abbreviations, secondary structure representation and species abbreviations are as in Figure 2. Short inserts are shown with a reduced font size, whereas longer inserts are represented by the corresponding numbers of residues.
Implications for the origins of DNA replication
The present analysis of the AEP superfamily, its evolutionary diversification and functional interactions in various cellular, viral, transposon and plasmid replication systems, and the apparent operational equivalence with the TOPRIM superfamily primases has substantial implications for the origins of DNA replication. In mechanistic terms, the monophyly of the AEP-superfamily and the RCREs is a remarkable parallel to the evolutionary history of the unrelated TOPRIM primases (6,21,88). In both cases, the primase is evolutionarily and structurally related to enzymes cleaving a DNA strand, specifically, nucleases that transfer the 5′ end of the cleaved DNA to an internal tyrosine residue in the protein (RCREs and topoisomerases, respectively) (4,89). Both the primase activity and the RCRE-/Topoisomerase-like nuclease activity, at a fundamental level, are two different solutions to the priming problem encountered by DNA polymerases. Thus, it seems plausible that, in both cases, the ancestral protein was involved in priming in a more generic fashion, whereas the descendent proteins evolved distinct, specific activities. One possibility is that the active tyrosine might have originally provided a hydroxyl group for protein priming, and subsequently, a more complex, polymerase or nuclease, activity was acquired by the priming proteins. In light of this suggestion, the OBDs of papovaviruses might be an offshoot from such an ancestor where only the origin-binding function was retained without any catalytic activity. The increase of the genome size apparently favored the primase mechanism, as opposed to the RCR mechanism; accordingly, the RCREs have been, largely, excluded from the genomes of cellular life forms. However, the topoisomerases, mainly drawn from the TOPRIM superfamily, despite catalyzing reactions similar to those catalyzed by the RCREs, were recruited for entirely different functions, such as, control of DNA superstructure, which is critical for large genomes. The prim-pol proteins have been proposed to comprise evolutionary intermediates that acted as both primases and DNA polymerases (25). Given that even the cellular AEPs have a similar DNA polymerase activity (49), this proposal appears plausible. However, if this were the case, dedicated DNA polymerases clearly displaced the primases at an early stage of evolution of larger replicons.
At a more basic level, the need for primers in DNA synthesis remains a largely unresolved problem. Almost all DNA polymerases, with extremely rare exceptions like the Mauriceville plasmid reverse transcriptase (90), require a primer for DNA synthesis. This requirement is seen across all classes of DNA polymerases that are unrelated or only distantly related to each other. In contrast, most known RNA polymerases, including unrelated ones, do not seem to require priming. DNA polymerases evolved many different solutions to the priming problem rather than acquiring a primer-independent initiation mechanism. Our analysis of the AEPs and the RCREs suggests that even the same fold was used in more than one way for solving the priming problem. These observations suggest a strong constraint against ‘invention’ of de novo initiation of DNA synthesis which, perhaps, stems from fundamental chemical differences between ribo- and deoxyribonucleotides, rather than a frozen evolutionary accident. It is tempting to speculate on the origin of priming in some more detail based on the previously reported evolutionary relationships between DNA polymerases and nucleotide cyclases (9,13,51). Even though these protein folds also include RNA polymerases, there seems to be a specific evolutionary relationship between the respective DNA polymerases and nucleotide cyclases. This suggests that the cyclases evolved from DNA polymerases independently on multiple occasions. Combining this observation with the strict requirement for a primer, one might speculate that DNA polymerases could be inefficient in initiating DNA synthesis due, at least in part, to the competing futile reaction of 3′→5′ nucleotide cyclization while using deoxyribonucleotides. Given the tendency of diverse, unrelated RNA polymerases to initiate de novo strand synthesis, it seems likely that this problem does not arise with ribonucleotides. This might explain the requirement for RNA primers for the initiation of DNA replication. This speculation is potentially testable through experimental demonstration of a cyclization reaction catalyzed by DNA polymerases.
In terms of specific evolutionary scenarios, previous reconstructions of the genome and the replication apparatus of the LUCA suggested that LUCA had the ability to synthesize DNA but certain key components of the replication apparatus, including the replicative polymerase, were missing. The apparent presence of DNA and RNAse HII, which removes primers during DNA replication, in LUCA suggests that it had a mechanism for priming DNA synthesis (18). However, the phyletic patterns of the cellular members of the AEP superfamily do not seem to indicate that this enzyme was present in LUCA. Although LUCA might have had a DnaG-like protein, the association of the archaeal version of this enzyme with the exosome makes it an unlikely candidate for the ancestral primase (20). A simple proposal made previously is that LUCA employed a reverse transcription step for DNA synthesis and might have accordingly used tRNAs as primers (18). An alternative to this scenario is that the DNA replication system of LUCA was a purely DNA-based system as in modern cells. This proposal posited that the difference in the replication systems of the bacterial and archaeo-eukaryotic branches of life arose as a result of non-orthologous displacement of certain components, such as the primases and DNA polymerases, by proteins of phage provenance (19,91). In principle, this proposal is consistent with the discovery of AEPs and DnaG-type primases in numerous viruses and, in particular, with the presence of the primase small subunit–primase large subunit–MCM combination in phages and plasmids (21,60,88). Furthermore, the functional equivalence of the two types of primases in viral replicons and evidence of multiple displacements indicate that different types of DNA replication components are extensively mixed-and-matched in the extra-chromosomal replicons. Nevertheless, there seem to be arguments against such a scenario. In the present study, we obtained indications that, both in eukaryotes and in prokaryotes, AEPs and other replication proteins were introduced into the cellular genomes from viral or mobile extrachromosomal replicons on several independent occasions. However, there is no evidence that Eukprim2 in eukaryotes or primases of the AEP superfamily in bacteria ever displaced the main replicative primase. The same observation holds true for the DNA polymerases, DNA ligases, DNA clamps and replicative helicases. Thus, the replication apparatus of cellular life forms seems to be refractory to displacement from functionally equivalent exogenous proteins, presumably due to mutual fine-tuning of individual components.
It appears plausible that there were several independent transitions to entirely DNA-based genomes via the transitional genomes involving the reverse transcription step. These parallel systems probably possessed different degrees of complexity, and certain viral and plasmid replicons might be models for the simpler systems that emerged in this period. While multiple non-orthologous displacements probably occurred in these DNA replication systems early on, after a certain level of complexity was reached in some of them, further displacements seem to have become unlikely if not altogether prohibited. Two of these systems acquired considerable complexity and became the precursors of the two principal cellular systems observed today.
General conclusions
Our study of the AEP superfamily greatly expanded the known diversity of AEPs and related proteins and revealed the unique active site shared by all members of this superfamily. Using contextual information provided by domain architectures and genome contexts of members of the AEP superfamily, we gleaned information regarding their functional associations with other conserved modules in the replication system. An interesting outcome of this investigation is the discovery of multiple, independent associations of members of the AEP superfamily with a variety of distinct ATPases that appear to function as helicases in replication initiation. Furthermore, we provide evidence for repeated in situ displacement of DnaG and AEP primase by each other in the context of the same polypeptide or operon. The discovery of the putative primases of the NCLDV class of viruses, the unification of these with the catalytic domain of the herpesvirus primases and prediction of the complete set of residues comprising the catalytic site of the latter seem to be of considerable importance. These findings might help in understanding unexplored aspects of the replication of these viruses and solving the long-standing mystery of the priming of their DNA replication. The discovery of the previously unnoticed, conserved eukaryotic AEPs related to the viral primases suggests the existence of novel DNA repair pathways. Similarly, the identification of potential primase-coding transposons points to a novel transposition mechanism. Experimental investigation of these systems has the potential of opening new vistas in the understanding of the crucial processes of DNA replication and repair.
SUPPLEMENTARY MATERIAL
A list of gis of all members of the AEP superfamily will be made available at ftp://ftp.ncbi.nih.gov/pub/aravind/.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health, USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lodish H., Berk A., Zipursky S.L., Matsudaira P., Baltimore D., Darnell J.E. Molecular Cell Biology. NY: W.H. Freeman & Co.; 1999. [Google Scholar]
- 2.Kornberg A., Baker T.A. DNA Replication, 2nd edn. NY: W.H. Freeman & Company; 1991. [Google Scholar]
- 3.Salas M. Protein-priming of DNA replication. Annu. Rev. Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 4.Noirot-Gros M.F., Ehrlich S.D. Change of a catalytic reaction carried out by a DNA replication protein. Science. 1996;274:777–780. doi: 10.1126/science.274.5288.777. [DOI] [PubMed] [Google Scholar]
- 5.Ilyina T.V., Koonin E.V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aravind L., Leipe D.D., Koonin E.V. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keck J.L., Roche D.D., Lynch A.S., Berger J.M. Structure of the RNA polymerase domain of E.coli primase. Science. 2000;287:2482–2486. doi: 10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- 8.Augustin M.A., Huber R., Kaiser J.T. Crystal structure of a DNA-dependent RNA polymerase (DNA primase) Nature Struct. Biol. 2001;8:57–61. doi: 10.1038/83060. [DOI] [PubMed] [Google Scholar]
- 9.Aravind L., Mazumder R., Vasudevan S., Koonin E.V. Trends in protein evolution inferred from sequence and structure analysis. Curr. Opin. Struct. Biol. 2002;12:392–399. doi: 10.1016/s0959-440x(02)00334-2. [DOI] [PubMed] [Google Scholar]
- 10.Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12:7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 13.Murzin A.G. How far divergent evolution goes in proteins. Curr. Opin. Struct. Biol. 1998;8:380–387. doi: 10.1016/s0959-440x(98)80073-0. [DOI] [PubMed] [Google Scholar]
- 14.Pei J., Grishin N.V. GGDEF domain is homologous to adenylyl cyclase. Proteins. 2001;42:210–216. doi: 10.1002/1097-0134(20010201)42:2<210::aid-prot80>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 15.Makarova K.S., Aravind L., Grishin N.V., Rogozin I.B., Koonin E.V. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burgers P.M., Koonin E.V., Bruford E., Blanco L., Burtis K.C., Christman M.F., Copeland W.C., Friedberg E.C., Hanaoka F., Hinkle D.C., et al. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 2001;276:43487–43490. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- 17.Braithwaite D.K., Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leipe D.D., Aravind L., Koonin E.V. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forterre P. Displacement of cellular proteins by functional analogues from plasmids or viruses could explain puzzling phylogenies of many DNA informational proteins. Mol. Microbiol. 1999;33:457–465. doi: 10.1046/j.1365-2958.1999.01497.x. [DOI] [PubMed] [Google Scholar]
- 20.Evguenieva-Hackenburg E., Walter P., Hochleitner E., Lottspeich F., Klug G. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 2003;4:889–893. doi: 10.1038/sj.embor.embor929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koonin E.V., Wolf Y.I., Kondrashov A.S., Aravind L. Bacterial homologs of the small subunit of eukaryotic DNA primase. J. Mol. Microbiol. Biotechnol. 2000;2:509–512. [PubMed] [Google Scholar]
- 22.Aravind L., Koonin E.V. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Della M., Palmbos P.L., Tseng H.-M., Tonkin L.M., Daley J.M., Topper L.M., Pitcher R.S., Tomkinson A.E., Wilson T.E., Doherty A.J. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science. 2004;306:683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- 24.Lipps G. The replication protein of the Sulfolobus islandicus plasmid pRN1. Biochem. Soc. Trans. 2004;32:240–244. doi: 10.1042/bst0320240. [DOI] [PubMed] [Google Scholar]
- 25.Lipps G., Weinzierl A.O., von Scheven G., Buchen C., Cramer P. Structure of a bifunctional DNA primase-polymerase. Nat. Struct. Mol. Biol. 2004;11:157–162. doi: 10.1038/nsmb723. [DOI] [PubMed] [Google Scholar]
- 26.Klinedinst D.K., Challberg M.D. Helicase–primase complex of herpes simplex virus type 1: a mutation in the UL52 subunit abolishes primase activity. J. Virol. 1994;68:3693–3701. doi: 10.1128/jvi.68.6.3693-3701.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dracheva S., Koonin E.V., Crute J.J. Identification of the primase active site of the herpes simplex virus type 1 helicase-primase. J. Biol. Chem. 1995;270:14148–14153. doi: 10.1074/jbc.270.23.14148. [DOI] [PubMed] [Google Scholar]
- 28.Takechi S., Matsui H., Itoh T. Primer RNA synthesis by plasmid-specified Rep protein for initiation of ColE2 DNA replication. EMBO J. 1995;14:5141–5147. doi: 10.1002/j.1460-2075.1995.tb00196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takechi S., Itoh T. Initiation of unidirectional ColE2 DNA replication by a unique priming mechanism. Nucleic Acids Res. 1995;23:4196–4201. doi: 10.1093/nar/23.20.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 32.Schaffer A.A., Aravind L., Madden T.L., Shavirin S., Spouge J.L., Wolf Y.I., Koonin E.V., Altschul S.F. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuwald A.F., Liu J.S., Lawrence C.E. Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci. 1995;4:1618–1632. doi: 10.1002/pro.5560040820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuler G.D., Altschul S.F., Lipman D.J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 35.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 37.Pei J., Sadreyev R., Grishin N.V. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics. 2003;19:427–428. doi: 10.1093/bioinformatics/btg008. [DOI] [PubMed] [Google Scholar]
- 38.Walker D.R., Koonin E.V. SEALS: a system for easy analysis of lots of sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:333–339. [PubMed] [Google Scholar]
- 39.Cuff J.A., Clamp M.E., Siddiqui A.S., Finlay M., Barton G.J. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 40.Cuff J.A., Barton G.J. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 41.Kraulis P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 42.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 43.Holm L., Sander C. The FSSP database: fold classification based on structure-structure alignment of proteins. Nucleic Acids Res. 1996;24:206–209. doi: 10.1093/nar/24.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holm L., Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 45.Gibrat J.F., Madej T., Bryant S.H. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 46.Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 47.Hasegawa M., Kishino H., Saitou N. On the maximum likelihood method in molecular phylogenetics. J. Mol. Evol. 1991;32:443–445. doi: 10.1007/BF02101285. [DOI] [PubMed] [Google Scholar]
- 48.Wolf Y.I., Rogozin I.B., Grishin N.V., Tatusov R.L., Koonin E.V. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol. Biol. 2001;1:8. doi: 10.1186/1471-2148-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lao-Sirieix S.-h., Bell S.D. The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase and 3′-terminal nucleotidyl transferase activities. J. Mol. Biol. 2004;344:1251–1263. doi: 10.1016/j.jmb.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 50.Kirk B.W., Kuchta R.D. Arg304 of human DNA primase is a key contributor to catalysis and NTP binding: primase and the family X polymerases share significant sequence homology. Biochemistry. 1999;38:7727–7736. doi: 10.1021/bi990247c. [DOI] [PubMed] [Google Scholar]
- 51.Aravind L., Koonin E.V. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steitz T.A. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 53.Campos-Olivas R., Louis J.M., Clerot D., Gronenborn B., Gronenborn A.M. The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc. Natl Acad. Sci. USA. 2002;99:10310–10315. doi: 10.1073/pnas.152342699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Enemark E.J., Chen G., Vaughn D.E., Stenlund A., Joshua-Tor L. Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus. Mol. Cell. 2000;6:149–158. [PubMed] [Google Scholar]
- 55.Luo X., Sanford D.G., Bullock P.A., Bachovchin W.W. Solution structure of the origin DNA-binding domain of SV40 T-antigen. Nature Struct. Biol. 1996;3:1034–1039. doi: 10.1038/nsb1296-1034. [DOI] [PubMed] [Google Scholar]
- 56.Hickman A.B., Ronning D.R., Kotin R.M., Dyda F. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol. Cell. 2002;10:327–337. doi: 10.1016/s1097-2765(02)00592-0. [DOI] [PubMed] [Google Scholar]
- 57.Datta S., Larkin C., Schildbach J.F. Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure (Camb) 2003;11:1369–1379. doi: 10.1016/j.str.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 58.Bradshaw E.M., Sanford D.G., Luo X., Sudmeier J.L., Gurard-Levin Z.A., Bullock P.A., Bachovchin W.W. T antigen origin-binding domain of simian virus 40: determinants of specific DNA binding. Biochemistry. 2004;43:6928–6936. doi: 10.1021/bi030228+. [DOI] [PubMed] [Google Scholar]
- 59.Hickman A.B., Ronning D.R., Perez Z.N., Kotin R.M., Dyda F. The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol. Cell. 2004;13:403–414. doi: 10.1016/s1097-2765(04)00023-1. [DOI] [PubMed] [Google Scholar]
- 60.McGeoch A.T., Bell S.D. Eukaryotic/archaeal primase and MCM proteins encoded in a bacteriophage genome. Cell. 2005;120:167–168. doi: 10.1016/j.cell.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 61.Matsui E., Nishio M., Yokoyama H., Harata K., Darnis S., Matsui I. Distinct domain functions regulating de novo DNA synthesis of thermostable DNA primase from hyperthermophile Pyrococcus horikoshii. Biochemistry. 2003;42:14968–14976. doi: 10.1021/bi035556o. [DOI] [PubMed] [Google Scholar]
- 62.Mikhailov V.S. Replication of the baculovirus genome. Mol. Biol. (Mosk) 2003;37:288–299. [PubMed] [Google Scholar]
- 63.Foiani M., Santocanale C., Plevani P., Lucchini G. A single essential gene, PRI2, encodes the large subunit of DNA primase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1989;9:3081–3087. doi: 10.1128/mcb.9.7.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans J.T., Leisy D.J., Rohrmann G.F. Characterization of the interaction between the baculovirus replication factors LEF-1 and LEF-2. J. Virol. 1997;71:3114–3119. doi: 10.1128/jvi.71.4.3114-3119.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iyer L.M., Koonin E.V., Aravind L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics. 2002;3:8. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iyer L.M., Makarova K.S., Koonin E.V., Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berg D.E., Howe M.M. Mobile DNA. Washington, DC: ASM Press; 1989. [Google Scholar]
- 68.Shinohara M., Itoh T. Specificity determinants in interaction of the initiator (Rep) proteins with the origins in the plasmids ColE2-P9 and ColE3-CA38 identified by chimera analysis. J. Mol. Biol. 1996;257:290–300. doi: 10.1006/jmbi.1996.0163. [DOI] [PubMed] [Google Scholar]
- 69.Iyer L.M., Koonin E.V., Aravind L. Extensive domain shuffling in transcription regulators of DNA viruses and implications for the origin of fungal APSES transcription factors. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-research0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iyer L.M., Leipe D.D., Koonin E.V., Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 71.Iyer L.M., Aravind L., Koonin E.V. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tye B.K. MCM proteins in DNA replication. Annu. Rev. Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 73.Leipe D.D., Aravind L., Grishin N.V., Koonin E.V. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000;10:5–16. [PubMed] [Google Scholar]
- 74.Subramanya H.S., Bird L.E., Brannigan J.A., Wigley D.B. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 75.Egelman E.H. Bacterial conjugation: running rings around DNA. Curr. Biol. 2001;11:103–105. doi: 10.1016/s0960-9822(01)00040-9. [DOI] [PubMed] [Google Scholar]
- 76.Biswas N., Weller S.K. A mutation in the C-terminal putative Zn2+ finger motif of UL52 severely affects the biochemical activities of the HSV-1 helicase–primase subcomplex. Curr. Biol. 1999;274:8068–8076. doi: 10.1074/jbc.274.12.8068. [DOI] [PubMed] [Google Scholar]
- 77.Pan H., Wigley D.B. Structure of the zinc-binding domain of Bacillus stearothermophilus DNA primase. Structure Fold Des. 2000;8:231–239. doi: 10.1016/s0969-2126(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 78.Kusakabe T., Hine A.V., Hyberts S.G., Richardson C.C. The Cys4 zinc finger of bacteriophage T7 primase in sequence-specific single-stranded DNA recognition. Proc. Natl Acad. Sci. USA. 1999;96:4295–4300. doi: 10.1073/pnas.96.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dandekar T., Snel B., Huynen M., Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 80.Wolf Y.I., Rogozin I.B., Kondrashov A.S., Koonin E.V. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 2001;11:356–372. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- 81.Huynen M., Snel B., Lathe W., III, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Margolin W. Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol. 2001;4:647–652. doi: 10.1016/s1369-5274(01)00264-8. [DOI] [PubMed] [Google Scholar]
- 83.Handa P., Acharya N., Varshney U. Chimeras between single-stranded DNA-binding proteins from Escherichia coli and Mycobacterium tuberculosis reveal that their C-terminal domains interact with uracil DNA glycosylases. J. Biol. Chem. 2001;276:16992–16997. doi: 10.1074/jbc.M100393200. [DOI] [PubMed] [Google Scholar]
- 84.Genschel J., Curth U., Urbanke C. Interaction of E.coli single-stranded DNA binding protein (SSB) with exonuclease I. The carboxy-terminus of SSB is the recognition site for the nuclease. J. Biol. Chem. 2000;381:183–192. doi: 10.1515/BC.2000.025. [DOI] [PubMed] [Google Scholar]
- 85.Kelman Z., Yuzhakov A., Andjelkovic J., O'Donnell M. Devoted to the lagging strand-the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cadman C.J., McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004;32:6378–6387. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Singleton M.R., Wentzell L.M., Liu Y., West S.C., Wigley D.B. Structure of the single-strand annealing domain of human RAD52 protein. Proc. Natl Acad. Sci. USA. 2002;99:13492–13497. doi: 10.1073/pnas.212449899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ilyina T.V., Gorbalenya A.E., Koonin E.V. Organization and evolution of bacterial and bacteriophage primase-helicase systems. J. Mol. Evol. 1992;34:351–357. doi: 10.1007/BF00160243. [DOI] [PubMed] [Google Scholar]
- 89.Wang J.C. DNA topoisomerases. Annu. Rev. Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- 90.Kuiper M.T., Lambowitz A.M. A novel reverse transcriptase activity associated with mitochondrial plasmids of Neurospora. Cell. 1988;55:693–704. doi: 10.1016/0092-8674(88)90228-0. [DOI] [PubMed] [Google Scholar]
- 91.Forterre P. The origin of DNA genomes and DNA replication proteins. Curr. Opin. Microbiol. 2002;5:525–532. doi: 10.1016/s1369-5274(02)00360-0. [DOI] [PubMed] [Google Scholar]
- 92.Ago H., Adachi T., Yoshida A., Yamamoto M., Habuka N., Yatsunami K., Miyano M. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold. Des. 1999;7:1417–1426. doi: 10.1016/s0969-2126(00)80031-3. [DOI] [PubMed] [Google Scholar]
- 93.Georgiadis M.M., Jessen S.M., Ogata C.M., Telesnitsky A., Goff S.P., Hendrickson W.A. Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase. Structure. 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 94.Hopfner K.P., Eichinger A., Engh R.A., Laue F., Ankenbauer W., Huber R., Angerer B. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc. Natl Acad. Sci. USA. 1999;96:3600–3605. doi: 10.1073/pnas.96.7.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim Y., Eom S.H., Wang J., Lee D.S., Suh S.W., Steitz T.A. Crystal structure of Thermus aquaticus DNA polymerase. Nature. 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 96.Bieger B., Essen L.O. Structural analysis of adenylate cyclases from Trypanosoma brucei in their monomeric state. EMBO J. 2001;20:433–445. doi: 10.1093/emboj/20.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]