Engineering Cytoplasmic Male Sterility via the Chloroplast Genome by Expression of β-Ketothiolase (original) (raw)
Abstract
While investigating expression of the polydroxybutyrate pathway in transgenic chloroplasts, we addressed the specific role of _β_-ketothiolase. Therefore, we expressed the _pha_A gene via the chloroplast genome. Prior attempts to express the _pha_A gene in transgenic plants were unsuccessful. We studied the effect of light regulation of the _pha_A gene using the _psb_A promoter and 5′ untranslated region, and evaluated expression under different photoperiods. Stable transgene integration into the chloroplast genome and homoplasmy were confirmed by Southern analysis. The _pha_A gene was efficiently transcribed in all tissue types examined, including leaves, flowers, and anthers. Coomassie-stained gel and western blots confirmed hyperexpression of _β_-ketothiolase in leaves and anthers, with proportionately high levels of enzyme activity. The transgenic lines were normal except for the male-sterile phenotype, lacking pollen. Scanning electron microscopy revealed a collapsed morphology of the pollen grains. Floral developmental studies revealed that transgenic lines showed an accelerated pattern of anther development, affecting their maturation, and resulted in aberrant tissue patterns. Abnormal thickening of the outer wall, enlarged endothecium, and vacuolation affected pollen grains and resulted in the irregular shape or collapsed phenotype. Reversibility of the male-sterile phenotype was observed under continuous illumination, resulting in viable pollen and copious amount of seeds. This study results in the first engineered cytoplasmic male-sterility system in plants, offers a new tool for transgene containment for both nuclear and organelle genomes, and provides an expedient mechanism for F1 hybrid seed production.
Male-sterility-inducing cytoplasms have been known for more than 100 years. Bateson and Gairdner (1921) reported that male sterility in flax (Linum usitatissimum) was inherited from the female parent. Chittenden and Pellow (1927) observed that male sterility in flax was due to an interaction between the cytoplasm and nucleus. Jones and Clarke (1943) established that male sterility in onion (Allium cepa) is conditioned by the interaction of the male-sterile cytoplasm with the homozygous recessive genotype at a single male-fertility restoration locus in the nucleus. The authors also described the technique used today to exploit cytoplasmic-genic male sterility (CMS) for the production of hybrid seed. CMS inbred lines have been widely used for hybrid production of many crops. The use of CMS in hybrid seed production has recently been reviewed by Havey (2004).
Naturally occurring CMS has been reported for maize (Zea mays; Kriete et al., 1996), oilseed rape (Brassica napus; Kriete et al., 1996), rice (Oryza sativa), and Beta beets (Kriete et al., 1996), but such a natural occurring system is not available for most other crops used in agriculture. A natural source of nuclear male sterility was identified in leek (Allium porrum; Smith and Crowther, 1995). The CMS phenotype is a maternally inherited trait caused by incompatibility between the nuclear and cytoplasmic genomes. CMS systems are used to produce commercial F1 hybrid lines, but full advantage of this system may be obtained if a nuclear restorer gene is available to suppress the male sterility in the hybrid plant. A few such nuclear fertility-restorer (Rf) genes have been isolated, including the maize _rf_2 gene that encodes an aldehyde dehydrogenase (Cui et al., 1996; Liu et al., 2001) and the petunia (Petunia hybrida) Rf gene, which encodes a pentatricopeptide repeat-containing protein (Bentolila et al., 2002). Recently, the pentatricopeptide repeat protein gene orf687 was shown to restore fertility in cytoplasmic male-sterile Kosena radish (Raphanus sativus cv Kosena; Koizuka et al., 2003). Unfortunately, introgression of Rf genes to restore fertility of rapeseed led to the deleterious effects and to the loss of genetic information (Delourme et al., 1998).
Engineered sources of nuclear male sterility have been developed in model systems (Mariani et al., 1990; Hernould et al., 1993; Perez-Prat and van Lookeren Campagne, 2002). A problem with these nuclear transformants is that they segregate for male fertility or sterility and must be over planted and rogued by hand or sprayed with herbicides to remove male-fertile plants. Male-sterility systems have been created by different mechanisms, most of these affect tapetum and pollen development (Kriete et al., 1996; Yui et al., 2003; Zheng et al., 2003). Unfortunately, additional severe phenotypic alterations that were due to interference with general metabolism and development had precluded its use in agriculture (Hernould et al., 1998; Napoli et al., 1999; Goetz et al., 2001).
In plants, three modes of inheritance of plastids and mitochondria are known: uniparental maternal, biparental, or uniparental paternal organelle inheritance. The majority of angiosperms exhibit the maternal mode of plastid inheritance, whereas in gymnosperms the paternal or the biparental modes are prevailing (Hagemann, 2004). The uniparental maternal inheritance characteristics of cytoplasmic organelles (plastid and mitochondria) are achieved through organelle exclusion from the generative cell during the first haploid pollen mitosis; organelles are distributed into the vegetative cell and not the generative cell, and, therefore, the sperm cell is free of any organelles. If the generative cell acquires a few organelles, they degenerate during maturation (Hagemann, 1992). However, rare exceptions to uniparental maternal inheritance have been reported (Hagemann, 2004).
Polyhydroxybutyrate (PHB) synthesis takes place by the consecutive metabolic action of _β_-ketothiolase (_pha_A gene), acetoacetyl-CoA reductase (_pha_B), and PHB synthase (_pha_C). Poirier et al. (1992) reported the expression of PHB in plants for the first time by expressing the _pha_B and _pha_C genes in the cytosol via nuclear transformation, taking advantage of available cytosolic acetoacetyl-CoA. This approach yielded very low levels of PHB, but severe pleiotropic effects were observed in the transgenic plants. In an attempt to increase the PHB yield in plants, Nawrath et al. (1994) introduced the _phb_A, _phb_B, and _phb_C genes in individual nuclear Arabidopsis (Arabidopsis thaliana) transgenic lines and reconstructed the entire pathway, targeting all enzymes to the plastids. This approach resulted in PHB expression up to 14% leaf dry weight, and no pleiotropic effects were evident. This suggested that the depletion of metabolites from essential metabolic pathways in the cytoplasm might have caused the pleiotropic effects, and that by targeting the enzymes to chloroplast, which is a compartment with high flux through acetyl-CoA, the adverse effects were overcome (Nawrath et al., 1994). With expression of optimized gene constructs, PHB yield increased up to 40% leaf dry weight, but this was accompanied by severe growth reduction and chlorosis (Bohmert et al., 2000), indicating that targeting the PHB pathway to the chloroplast can result in pleiotropic effects, at higher concentrations of polymer synthesis (Bohmert et al., 2000). Lossl et al. (2003) reported the expression of PHB in tobacco (Nicotiana tabacum) by expressing _pha_A, _pha_B, and _pha_C via plastid transformation. The expression of PHB resulted in severe growth reduction, and the authors concluded that in tobacco significant levels of PHB could only be achieved if a sufficient pool of acetyl-CoA precursor is generated (Lossl et al., 2003). Additionally, they observed that when the transgenic plants were grown autotrophically, PHB levels significantly decreased, which overcame the stunted phenotype, but male sterility was still observed. It was not known whether the polymer or other metabolic factors were responsible for the male-sterile phenotype (Lossl et al., 2003).
In an attempt to address the role of _pha_A expression in the pleiotropic effects observed in transgenic plants expressing PHB, Bohmert et al. (2002) expressed the _phb_A gene constitutively and under inducible promoters via the nuclear genome. Constitutive expression of the _phb_A gene led to a significant decrease in transformation efficiency, inhibiting the recovery of transgenic lines, and prevented analysis of plants expressing the _β_-ketothiolase gene (Bohmert et al., 2002). Such toxic effect exerted by _phb_A expression was speculated to be the result of PHB biosynthesis intermediates or its derivatives, the depletion of the acetyl-CoA pool, or of interaction of the _β_-ketothiolase with other proteins or substrates (Bohmert et al., 2002).
While investigating expression of the phb operon encoding the PHB pathway in transgenic chloroplasts, we proposed to address the specific role of _β_-kethiolase (_pha_A gene). Therefore, we expressed the _β_-ketothiolase gene by inserting the transgene into the chloroplast genome and studied the effect of light regulation using the _psb_A promoter and 5′ untranslated region (5′UTR), and evaluated the effects of _pha_A expression in the chloroplast transgenic plants under a specific photoperiod or continuous illumination. Fully regenerated transgenic plants facilitated evaluation of the specific effect of _β_-ketothiolase in the transgenic lines for the first time. Detailed characterization revealed that severe pleiotropic effects, such as stunted phenotype and chlorosis observed during PHB expression (Bohmert et al., 2000, 2002), were not observed during _pha_A expression via the chloroplast genome, with the exception of complete (100%) male sterility. Such elimination of pleiotropic effects was not surprising because the chloroplast genetic engineering system has been able to overcome the toxic effects (stunted growth, chlorosis, and infertility) associated with nuclear expression of the trehalose (Lee et al., 2003), xylanase (Leelavathi et al., 2003), and cholera toxin B subunit (Daniell et al., 2001a) genes producing normal plants that were healthy and fertile. In this article, we characterize _pha_A transgene site-specific integration into the chloroplast genome, transcription, translation in different tissue types, _β_-ketothiolase activity under different photoperiods, microscopic analysis of different stages of anther development, and electron microscopic analysis of pollen grains, and attempt to correlate these activities with metabolic activities. This study resulted in the first engineered CMS system in plants.
RESULTS
Chloroplast Transformation Vector
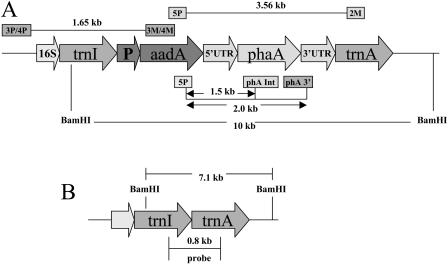
The Acinetobacter sp. (accession no. L37761) gene phaA (1,179 bp) coding for _β_-ketothiolase was amplified by PCR and cloned into the chloroplast transformation vector (pLD-ctv) to finally produce the pLDR-5′UTR-_pha_A vector (Fig. 1A). The vector contains chloroplast DNA sequences (flanking sequences), which allow site-specific integration by homologous recombination into the inverted repeat region of the chloroplast genome, in between the _trn_I (tRNA Ile) and _trn_A (tRNA Ala) genes (Daniell et al., 1998). Site-specific integration of the transgene cassette containing the _aad_A (aminoglicoside 3′-adenylyltransferase) gene conferred spectinomycin resistance. Transcription of the _aad_A and pha_A genes is driven by the constitutive 16S ribosomal RNA gene promoter (P_rrn), located upstream of the _aad_A gene, and this should produce a dicistron (_aad_A-_pha_A). In addition, the _psb_A gene promoter and 5′ regulatory sequence (5′UTR), known to enhance translation of foreign genes in the light (Eibl et al., 1999; Fernandez-San Millan et al., 2003; Daniell et al., 2004c; Dhingra et al., 2004; Watson et al., 2004), were inserted upstream of the _pha_A gene, and this should produce the _pha_A monocistron. At the 3′ end of the gene construct is the 3′ _psb_A untranslated region (3′UTR), which is known to be involved in mRNA abundance and stability (Schuster and Gruissem, 1991; Sugita and Sugiura, 1996; Monde et al., 2000).
Figure 1.
Molecular characterization of transgenic lines. A, Schematic representation of the transformed chloroplast genome and the pLDR-5′UTR-_pha_A cassette. Annealing sites for the primer pairs and expected sizes of PCR products are shown. _Bam_HI restriction sites, DNA fragment produced after restriction digestion, and the _pha_A probe used in Southern-blot analyses are shown. B, The map shows the wild-type chloroplast genome, restriction enzyme sites used for Southern-blot analysis, expected fragment sizes after digestion, and probing with the flanking sequence.
Transgene Integration into Chloroplast Genomes
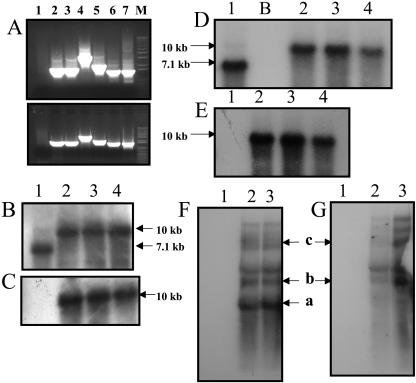
Chloroplast transgenic lines were obtained through particle bombardment following the method described previously (Daniell, 1997; Daniell et al., 2004a; Kumar and Daniell, 2004; Dhingra and Daniell, 2005). More than 10 positive independent transgenic lines were obtained. Three independent transgenic lines were characterized, confirming that independent chloroplast transgenic lines show little variation in foreign gene expression, as observed earlier (Daniell et al., 2001a). PCR-based analysis with the primer pairs 3P-3M and 4P-4M was used to test site-specific integration of the transgene cassette into the chloroplast genome (Daniell et al., 2001b). The 3P-4P primers land on the native chloroplast genome, upstream of the gene cassette, and the 3M-4M primers land on the _aad_A gene, which is located within the gene cassette (Fig. 1A). If site-specific integration had occurred, a PCR product of 1.65 kb should be obtained; this product was detected in transgenic lines (Fig. 2A, lanes 2 and 3). Untransformed plants as well as mutant plants, which had undergone spontaneous mutation of the 16S rRNA gene and acquired resistance to spectinomycin, did not show any PCR product, indicating that these plants are negative for integration (Fig. 2A, lane 1). The integration of the _aad_A and _pha_A genes was further confirmed by the use of primer pairs 5P-2M, 5P-3′phaA, and 5P-phaAinternal, which produced PCR products of sizes 3.56 kb, 2.0 kb, and 1.5 kb, respectively. These primers anneal at different locations within the transgene cassette (Fig. 1A). Results revealed expected-size PCR products in the transgenic lines (Fig. 2A, lanes 4–6), confirming the presence of the _pha_A gene.
Figure 2.
A, PCR analysis of untransformed plants and putative transformants. Top section is transgenic line 4B; bottom section is transgenic line 4A. A, Lane 1, Untransformed plant; 2, 3P-3M (1.65 kb); 3, 4P-4M (1.65 kb); 4, 5P-2M (3.56 kb); 5, 5P-3′phaA (2.0 kb); 6, 5P-_pha_A internal (1.5 kb); 7, positive control (pLD-5′UTR-_phb_A plasmid DNA) 5P-_phb_A internal (1.5 kb); M, marker. B and D, Southern-blot analysis of T0 and T1 generation transgenic lines, respectively, with the chloroplast flanking sequence probe; 10-kb fragment shows integration of transgenes; 7.1-kb fragment shows wild-type fragment. C and E, Transgenic T0 and T1 plants, respectively, and untransformed plant probed with the _pha_A gene. The 10-kb fragment is observed in transgenic lines but not in the untransformed plant. F and G, Northern-blot analysis using the _pha_A and _aad_A probes, respectively. Monocistron (a, 5′UTR-_pha_A, 1,384 nt) and polycistrons (b, _aad_A-pha_A, 2,255 nt; c, from native 16S P_rrn, 4,723 nt) containing the _pha_A gene are observed in the transgenic lines when the _pha_A probe is used. Only polycistrons are observed with the _aad_A probe. In B to G: 1, untransformed plant; 2 to 4, chloroplast transgenic lines 4A, 4B, and 4C, respectively; B, blank lane.
Total DNA from T0 and T1 generation transgenic lines as well as from untransformed tobacco plants was extracted and used for Southern-blot analysis (Fig. 2, B–E). The flanking sequence probe (Fig. 1B), which hybridizes with the _trn_I and _trn_A genes, was used to analyze heteroplasmy and investigate site-specific integration of the transgene cassette into the chloroplast genome. Additionally, the _pha_A probe was used to confirm the presence of the _pha_A gene. DNA from untransformed plants and transgenic lines was digested with _Bam_HI (Fig. 1, A and B) and probed with either a flanking sequence probe or a _pha_A probe; a 10-kb hybridizing fragment was detected in transformed chloroplast genome (Fig. 2, B–E, lanes 2–4). The detection of a 7.1-kb fragment by the flanking sequence probe in the untransformed line confirmed that these chloroplast genomes lacked foreign genes (Fig. 2, B and D, lane 1). The fact that no 7.1-kb-size fragment was observed in T0 generation (Fig. 2B, lanes 2–4) and T1 generation (Fig. 2D, lanes 2–4) transgenic samples confirmed stable integration of foreign genes within all chloroplast genomes and lack of heteroplasmy (within the limits of detection of Southern blots). The absence of any hybridizing fragment in the untransformed lines when screened with the _pha_A probe confirmed the absence of _pha_A gene (Fig. 2, C and E, lane 1). If any unexpected-size fragment were observed in the transgenic samples when probed with the transgene after prolonged exposure of Southern blots to x-ray film, nonspecific integration into the nuclear or mitochondrial genomes would be indicated. Such nonhomologous recombination was not observed.
Transcription of Transgenes Integrated into Chloroplast Genomes
Transcript abundance and stability in chloroplast transgenic lines were studied by northern-blot analysis using the _pha_A and _aad_A probes on total plant RNA (Fig. 2, F and G). The chloroplast transgenic lines were expected to transcribe a 2,255-nt dicistron (_aad_A-pha_A; Fig. 2, F and G) from the upstream 16S promoter (P_rrn), in addition to a 1,384-nt monocistron (_pha_A) transcribed from the _psb_A promoter, located upstream from the _pha_A gene (Fig. 2F, lanes 2 and 3). As expected, the monocistron was not detected with the aad_A probe, showing that polycistrons are transcribed from the engineered P_rrn promoter; less abundant polycistrons transcribed from the native 16S promoter were also detected (Fig. 2G). The monocistron was only detected with the pha_A probe. These results showed that both the monocistron and dicistron transcripts were abundant in the transgenic lines because of the efficiency of the psb_A and P_rrn promoters, which are strong chloroplast promoters. Additionally, larger-size transcripts were detected, one of them correlating with the size of a transcript initiated at the chloroplast native 16S promoter (P_rrn) and terminating at the 3′UTR _psb_A; the predicted size of this transcript is 4,723 nt (Fig. 2, F and G). Other transcripts detected may be either read-through (because 3′UTR does not terminate transcription efficiently in chloroplast) or processed products.
Translation of the _pha_A Gene Integrated into Chloroplast Genomes
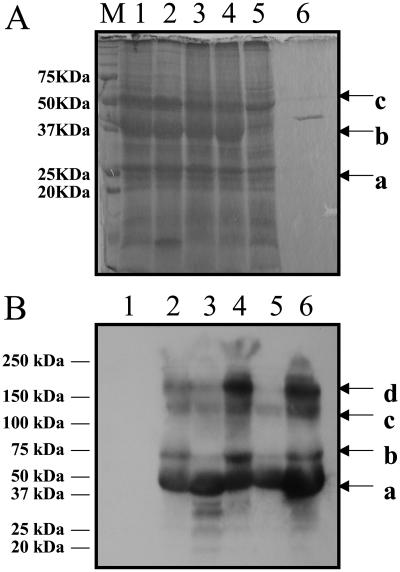
To confirm expression of _β_-ketothiolase in the chloroplast transgenic lines, untransformed and transformed plants were subjected to western-blot analysis using anti-_β_-ketothiolase antibody. Chloroplast-synthesized _β_-ketothiolase, when treated with _β_-mercaptoethanol (BME) and boiled, appeared mostly as a monomeric form (40.8 kD), or in polymeric forms, which included the homotetrameric form (163 kD; Fig. 3B, lanes 2–6). The homotetramer is the functional form of _β_-ketothiolase. No _β_-ketothiolase was detected in wild-type samples (Fig. 3B, lane 1). The appearance of a distinct band at 40.8 kD in the Coomassie-stained SDS-PAGE gel (Fig. 3A, lanes 1–4) but not in the untransformed sample (lane 5) suggests that the chloroplast transgenic plants were expressing high levels of _β_-ketothiolase.
Figure 3.
_β_-Ketothiolase characterization in transgenic lines. A, Coomassie-stained gel showing abundant _β_-ketothiolase expression in transgenic lines; 15 _μ_g of total plant protein was loaded per lane (a, D1 protein; b, _β_-ketothiolase, 40.8 kD; c, Rubisco large subunit, based on molecular range). M, Marker; 1 to 3, transgenic lines 4A, 4B, and 4C, respectively; 4, 4A T1 generation; 5, untransformed plant; 6, bacterial purified _β_-ketothiolase. B, Western-blot analysis; 10 _μ_g of total plant protein from transgenic lines and wild type was loaded per lane; anti-_β_-ketothiolase antibody was used. Bands corresponding to the 40.8-kD monomers (a), a possible modified version of the monomer (b), trimer (c), or tetramer (d) are observed in transgenic lines. 1, Untransformed plant; 2 to 4, transgenic lines 4A, 4B, and 4C, respectively; 5 and 6, 4A and 4B T1 generation, respectively.
The activity of the chloroplast-expressed _β_-ketothiolase was measured in the thiolysis direction (breaking down acetoacetyl-CoA to acetyl-CoA) spectrophotometrically at 304 nm. The chloroplast transgenic lines showed _β_-ketothiolase activities that were up to 30-fold higher than previous levels reported for nuclear transgenic plants. No endogenous _β_-ketothiolase activity was detected in untransformed tobacco plants (less than 0.0001 unit/mg plant protein; Table I). The chloroplast transgenic lines showed enzyme activity in the range of 14.08 to 14.71 unit/mg plant protein during normal photoperiod (16 h light/8 h dark; Table I). After 5 d of continuous illumination, the enzyme activity slightly increased in both transgenic lines (Table I). Thus, _β_-ketothiolase activity remained unchanged from light/dark photoperiod to continuous illumination, even though the _pha_A gene is under the control of strong psbA regulatory elements that should enhance translation in the light. The high levels of enzyme activity correlated well with the high amounts of protein detected by the Coomassie-stained gel and western-blot analyses performed on total plant extracts; these results suggested that the enzyme was in its biologically active form (homotetramer).
Table I.
β-Ketothiolase activity as a function of illumination
Enzyme activity for transgenic lines 4A and 4B and untransformed tobacco plants at the respective illumination periods are shown. Protein samples were obtained by grinding 1 g of leaf tissue in liquid nitrogen, followed by the addition of _β_-ketothiolase extraction buffer (100 mm Tris-HCl, pH 8.1, 50 mm MgCl2, 5 mm BME) and homogenization. Total plant protein concentration was determined by Bradford assay. Ten microliters of crude plant extract was used per assay. _β_-Ketothiolase activity was measured spectrophotometrically at 304 nm for 60 s in the thiolysis direction. na, Not applicable.
| Plant Line | Illumination Period Light/Dark | Enzyme Activity Units |
|---|---|---|
| h | μmol/min | |
| Untransformed | 16 h/8 h | <0.0001 |
| 4A | 16 h/8 h | 0.215 |
| 4B | 16 h/8 h | 0.259 |
| Untransformed | 5 d of light | <0.0001 |
| 4A | 5 d of light | 0.247 |
| 4B | 5 d of light | 0.239 |
| Purified bacterial _β_-ketothiolase | na | 0.017 |
Characterization of Male Sterility
Among the 10 T0 transgenic lines expressing _β_-ketothiolase, 100% of the flowers produced by transgenic plants failed to develop fruit capsules and seeds, finally senescing and falling off (Fig. 4, A–C). The male-sterile phenotype was maintained in T1 generation transgenic lines. T0 (Fig. 5A) and T1 (Fig. 5B) generation transgenic plants showed no difference in growth and development when compared to untransformed tobacco plants under the same growth conditions. Like the parental line, the T1 transgenic lines were not affected by hyperexpression of _pha_A and were phenotypically indistinguishable, with the exception of being male sterile, from untransformed control plants (Fig. 5B). Analysis of chlorophyll content of three independent T1 transgenic lines showed average chlorophyll content of 1.90 ± 0.20 mg/g fresh weight. The chlorophyll content of three wild-type plants averaged 1.92 ± 0.12 mg/g fresh weight. These results showed that the chlorophyll levels in the leaves of the transgenic lines expressing _β_-ketothiolase were similar to the levels in wild-type tobacco plants, confirming normal chloroplast biosynthetic functions and integrity of the thylakoid membranes, although _β_-ketothiolase was hyperexpressed.
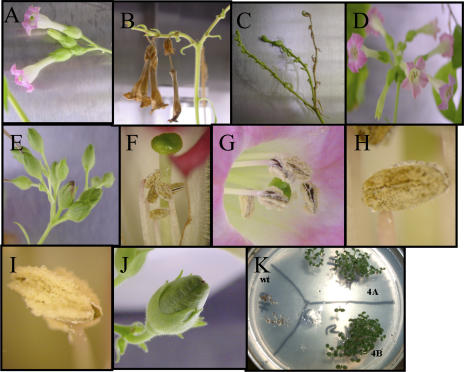
Figure 4.
Characterization of the male-sterile phenotype. A to C, Flowers from transgenic lines; note the absence of fruit capsules and fallen flowers. D and E, Untransformed tobacco flowers and fruit capsules. Comparison of stamens and stigma: note shorter stamens in transgenic lines (F) compared to untransformed (G). Comparison of mature anthers: note abundant pollen in untransformed anther (I) and the lack of pollen in transformed anther (H). J, Transgenic fruit capsule with seeds after pollination of transgenic stigma with untransformed pollen. K, Germination and growth of T1 seedlings on Murashige and Skoog medium with 500 _μ_g/mL spectinomycin. wt, Untransformed; 4A, T1 transgenic line 4A; 4B, T1 transgenic line 4B, obtained after pollination with untransformed pollen.
Figure 5.
Comparison of growth and development. A, Untransformed plant (WT) and T0 generation transgenic (T) line (4A) grown for 2 months in soil. B, Untransformed plant (WT) and T1 generation independent transgenic lines 4A, 4B, 4C, and 4D, 1.5 months after germination.
However, the chloroplast transgenic lines showed specific defects in anther development and failed to produce viable pollen. The anthers were characterized by the lack of pollen grains (Fig. 4H), or when pollen grains were formed, they were abnormal with a collapsed phenotype (Fig. 6). Additionally, the stamens were shorter (Fig. 4F) than in wild-type plants (Fig. 4G), adding a degree of severity to the male-sterile phenotype. To investigate that plants were sterile due to lack of pollen or nonviable pollen, a total of 15 emasculated wild-type flowers from three untransformed plants (five flowers from each untransformed plant) were pollinated with pollen collected from anthers from transgenic lines 4A, 4B, and 4C. Pollen from each transgenic line was used to pollinate a total of five untransformed emasculated flowers. All of these crosses failed to produce seeds. The shorter stamens may be a consequence of the failure of the cells to elongate in the central and upper parts of the anther filament. To investigate the possibility of female infertility affecting the transgenic plants, the plants were fertilized with pollen from untransformed plants. The transgenic plants developed normal fruit capsules (Fig. 4J) and normal seeds that were able to germinate and develop normally (Fig. 4K). The T1 seedlings grew well in the medium supplemented with 500 _μ_g/mL spectinomycin and were identical to the parental line (T0), and Southern-blot analysis showed the presence of the gene construct (Fig. 2, D and E).
Figure 6.
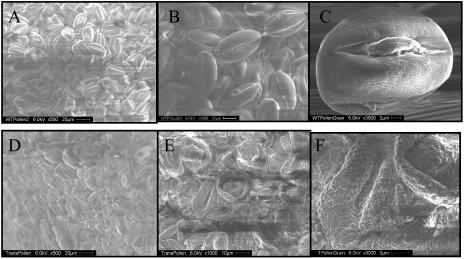
SEM pictures of pollen grains in anthers of untransformed (A–C) and transgenic (D–F) plants at different magnifications. A and D, ×500; B and E, ×1,000; C, ×3,500; F, ×3,000.
Scanning electron microscopy (SEM) was performed on transgenic anthers as well as untransformed anthers to further characterize male sterility in transgenic plants. The SEM revealed that the pollen grains in the transgenic anthers exhibited collapsed morphology and consisted of a heterogeneous population with respect to size and shape (Fig 6, D–F). Wild-type anthers showed a homogenous population of pollen grains of uniform size and shape (Fig. 6, A–C). The apparent lack of turgidity of the transgenic pollen may be produced by lack of intracellular material, resulting in the distorted and collapsed morphology. The aberrant pollen morphology observed under the SEM may account for the inability of the transgenic plant to produce seeds.
_β_-Ketothiolase Expression in Anthers
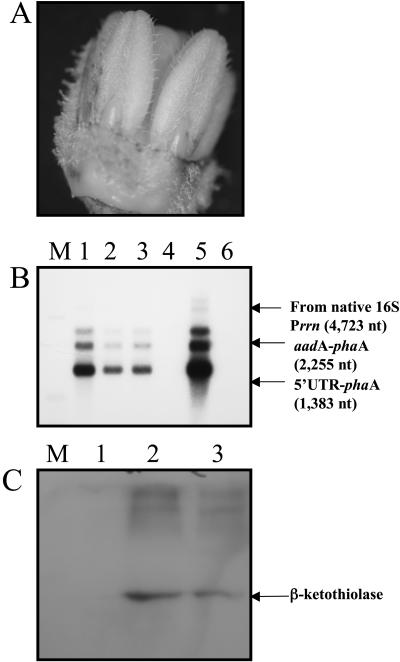
Plastids in anthers may be in low abundance when compared to the numbers in leaves, but they produce enough _β_-ketothiolase to affect pollen development in anthers. As shown in Figure 7A, flowers and specifically anthers were green during anther development (stage 1 of flower development is shown). This means that chloroplasts are present and are metabolically active. Northern-blot analysis of leaves showed that the mRNAs coding for _β_-ketothiolase are present in monocistronic and polycistronic forms, the monocistron form being the most abundant transcript (Fig. 2F). The same pattern was maintained in the _pha_A transcripts in flowers and anthers (Fig. 7B). Northern-blot analysis of transgenic flowers and anthers showed that transcription of the pha_A gene occurs in the flower and anther; this was expected because P_rrn and _psb_A promoters are constitutive promoters, allowing transcription to occur in both photosynthetically active as well as nonphotosynthetic plastids (Fig. 7B). The translation of the _pha_A monocistron is under the light-regulated _psb_A 5′UTR because the flowers as well as anthers are green, containing photosynthetically active chloroplasts; translation was quite efficient (Fig. 7C). _β_-Ketothiolase was detected by western blot in both whole flowers and anthers from transgenic plants, confirming that the _β_-ketothiolase was present during anther development and should play a significant role in the male-sterile phenotype (Fig. 7C).
Figure 7.
_β_-Ketothiolase expression in anthers. A, Pigmentation of anthers during flower development in transgenic plants. Stage 1 of flower development is shown. B, Northern-blot analysis of flowers and anthers. Three micrograms of total plant RNA was loaded per lane, and the _pha_A probe was used. M, Marker; 1, transgenic flower; 2 and 3, transgenic anthers from line 4A and 4B, respectively; 4, untransformed flower; 5, transgenic leaf; 6, untransformed leaf. C, _β_-Ketothiolase expression in transgenic flowers and anthers detected by western-blot analysis in lanes 2 and 3, respectively. RNA and protein samples used per lane were the product of the combined extraction from flowers or anthers from stages 1 and 3.
Anther Development and Male Sterility
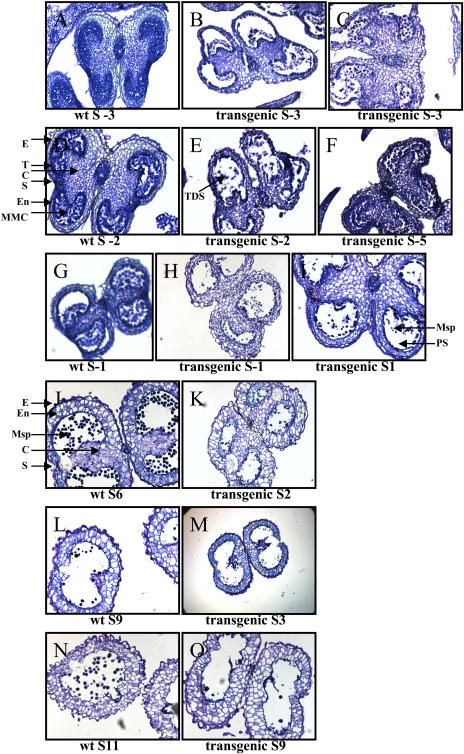
Analysis of anther development revealed that the anthers of the transgenic lines followed an accelerated pace in their development and maturation, resulting in aberrant tissue patterns (Fig. 8). Flower developmental stages were determined as described by Koltunow et al. (1990). At stage −3 of flower development, the untransformed plants showed a normal pattern of tissue development, where all major tissues were differentiated, the anther had acquired its characteristic shape, all tissues were interconnected, and there was callose deposition between microspore mother cells. This pattern was not followed in the transgenic lines, which at stage −3 showed characteristics of a more advanced stage. The transgenic anthers at stage −3 showed the microspores in tetrads, with stomium differentiation occurring, and the tapetum was shrunken and broken. This pattern presented characteristics of more advanced stages of flower development, which included stages +1 and +2. Additionally, the transgenic anthers showed abnormal development of the epidermis and endothecium, probably resulting in the aberrant shape of anthers. The anthers of the transgenic lines at stage −2 were also advanced in the aberrant phenotype with the microspores already separated, a developmental step that should have been observed at stage +2. Again the tapetal layer was broken. We noticed that the transgenic anthers at stage −5, which is a very early stage of flower development, showed great similarities with stage −2 in the untransformed plants, but aberrant pattern of tissue development could still be observed. At stage −1 in transgenic lines, the tapetum was shrunken and discontinuous, and formation of pollen grains was evident at stage 1. These morphological changes observed in stages −1 and +1 should be observed at much later stages, +3 and +4. At early stages of floral development (stages −5 to +1), transgenic lines showed accelerated anther development, which averaged +3 stages ahead from the wild-type plants. At late stages of floral development, accelerated phenotype increased even more, at an average of four to six stages ahead of untransformed plants. At stage +2 in the transgenic lines, cells adjacent to the stomium had degenerated, and only remnants of the tapetum were observed. The thickening of the outer wall was accompanied by enlarged endothecium and vacuolation. The irregular shape and collapsed phenotype of pollen grains might be a synergistic effect of the reduced locule space and the apparent lack of turgidity of the transgenic pollen produced by insufficient intracellular material. Lack of intracellular material can be attributed to the lack of tapetum in the transgenic lines. The developmental changes observed in the transgenic anthers at stage +2, although aberrant, were similar to the ones observed in untransformed at stage +6. In the untransformed anthers, abundant normal pollen grains were observed. Almost complete degradation of the connective tissue that separates the pollen sac occurred at stage +3 in transgenic anthers, while this occurred at stage +9 in untransformed plants. Finally, both untransformed and transgenic anthers were bilocular and connective tissue was absent, but the major difference was that abundant pollen was present in untransformed (stage +11) but not in transgenic anthers (transgenic, stage +9). Additionally, the pollen grains formed in transgenic anthers were collapsed.
Figure 8.
Studies on anther development. Bright-field photographs of untransformed (wt) and transgenic anthers at different developmental stages (S). Anthers at the designated stages were fixed, embedded with paraffin, and sliced into 5- and 10-_μ_m transverse sections. The fixed sections were stained with toluidine blue and visualized under the light microscope at a magnification of ×100. C, Connective tissue; E, epidermis; En, endothecium; MMC, microspore mother cells; Msp, microspores; PS, pollen sac; S, stomium; T, tapetum; TDS, tetrads.
Anther development is a very complex process involving the coordination of several genes and the specific development and maturation of several tissues and cells (Yui et al., 2003); any defect in these well-coordinated processes may lead to dysfunctional pollen. Many male-sterility systems produced by mutations or nuclear expressions of foreign proteins have shown to interfere with the function or differentiation of tapetum, indicating that this tissue is essential for the production of viable pollen (Goetz et al., 2001). Here, we observed that the tapetum of the transgenic lines was severely impaired. The tapetum is critical for the development of pollen by secreting essential substances such as proteins (Yui et al., 2003), carbohydrates (Goetz et al., 2001), and lipids (Zheng et al., 2003) into the locules. Developing microspores and the surrounding tapetal cells have been shown to be particularly active in lipid metabolism (Zheng et al., 2003). The precise differentiation and maturation of tapetum with respect to microspore development is of major importance for the successful production of pollen. Here, we observed almost total dysfunction in the anther tissue differentiation patterns, which may be caused by an alteration in chloroplast fatty acid metabolism in the transgenic lines expressing the _pha_A gene, thereby affecting the development of pollen grains.
Reversibility of Male Fertility
To test whether the acetyl-CoA pool destined for de novo fatty acid biosynthesis in chloroplast is being diverted by _β_-ketothiolase to the synthesis of acetoacetyl-CoA, resulting in the male-sterile phenotype, we explored whether continuous illumination would revert male sterility to fertility. The photoperiod experiments represent an indirect test of the proposed basis for male sterility in this system. Acetyl-CoA carboxylase (ACCase) carries the first committed step in fatty acid biosynthesis, which involves the conversion of acetyl-CoA to malonyl-CoA. Because acetyl-CoA is not imported into plastids from the cytoplasm, it should be synthesized in this organelle; the expression of _pha_A in the chloroplast should increase the competition for the same pool. Therefore, if _β_-ketothiolase and ACCase are competing for the same acetyl-CoA pool, _β_-ketothiolase will outcompete ACCase when it is hyperexpressed in transgenic chloroplasts; this could decrease the supply of acetyl-CoA for fatty acid biosynthesis. Therefore, conditions that could divert the acetyl-CoA pool back to the fatty acid biosynthesis pathway might restore the fatty acid biosynthesis. It has been shown that the intermediates of fatty acid biosynthesis change during the transition to darkness in leaves and in chloroplasts in a manner consistent with control at the level of ACCase (Post-Beittenmiller et al., 1991). The chloroplast uses photosynthesis as the mechanism to generate the reducing power and ATP for diverse metabolic reactions, including fatty acid biosynthesis. De novo fatty acid biosynthesis in the chloroplast has been shown to be regulated by the response of ACCase to the redox state of the plastid, ensuring that carbon metabolism is linked to the energy status of this organelle (Rawsthorne, 2002). Recent reports have shown light-dependent regulation of ACCase by the redox status of the plastid, whereby the enzyme is more active under the reducing conditions observed in light (Page et al., 1994; Kozaki et al., 2000).
Therefore, to test this hypothesis, two independent transgenic lines 4A and 4B were exposed to continuous light for a period of 10 d. These plants have previously been characterized and shown to be 100% male sterile, unable to produce any fruit capsules or seeds. These plants were isolated from all other plants (transgenic and wild types) in a growth chamber at the first indication of flower bud development (before any flower opened), and the flowers were covered with transparent plastic bags to avoid cross-pollination. We observed that from a total of 20 flowers produced during the 10 d of illumination by the two transgenic plants, four flowers were able to produce pollen (Fig. 9A), produce normal-length anther filaments (Fig. 9A), develop fruit capsules (Fig. 9B), and produce seeds (Fig. 9C). Transgenic line 4A produced three male-fertile flowers during the 10-d continuous-light photoperiod; all of them were produced between days 8 and 10. Transgenic line 4B produced one viable flower on day 9 and was able to produce additional viable flowers 3 d after the 10-d assay was completed. The seeds recovered (Fig. 9C) from the reverted male-fertility study were able to germinate in a medium with 500 _μ_g/mL spectinomycin (Fig. 9D), indicating that these seedlings were transgenic and contained the transgene; untransformed tobacco seedlings were bleached (Fig. 9E). These findings support the hypothesis that an increase in ACCase activity outcompetes, at least partially, the removal of acetyl-CoA by _β_-ketothiolase. Additionally, this line of evidence supports the hypothesis that male sterility is associated with _β_-ketothiolase expression in the chloroplast. Based on the lack of increase in _β_-ketothiolase activity (Table I) but increased ACCase activity during continuous illumination (see “Discussion”), de novo fatty acid biosynthesis should be enhanced in transgenic lines and this should reverse male sterility. Monitoring ACCase activity in different tissues and determining various metabolic pools would further test this hypothesis.
Figure 9.
Reversibility of male sterility after 10 d under continuous illumination. A, Transgenic flower after 9 d in continuous light; note normal length of the anther filaments and pollen grains. B, Fully developed fruit capsules containing seeds from reversed transgenic lines. C, Abundant seeds from a transgenic fruit capsule. D, Seedlings produced via the reversibility to male fertility. Transgenic seeds germinated in Murashige and Skoog medium supplied were with 500 _μ_g/mL spectinomycin. E, Bleached wild-type tobacco seedlings.
DISCUSSION
We have shown that the hyperexpression of _β_-ketothiolase via chloroplast transformation resulted in normal growth and pigmentation, even when the activity of the enzyme was very high. Successful expression of _β_-ketothiolase in transgenic plants showed that this enzyme could be safely expressed in the chloroplast and that the complete PHB pathway should be expressed to cause the stunted phenotype. Although no growth reduction was observed in the chloroplast transgenic lines expressing _β_-ketothiolase, 100% male sterility was observed. This was interesting and worthy of further investigation because of the importance of the production of male-sterile lines in gene containment and hybrid seed production (Perez-Prat and van Lookeren Campagne, 2002). Because the expression of _β_-ketothiolase did not disrupt growth and normal development, with the exception of the lack of pollen formation, the expression of _β_-ketothiolase may be used as a mechanism to generate male-sterile lines.
Here, we also demonstrate that it is possible to revert the chloroplast transgenic lines to male fertility by continuous illumination. This supports the hypothesis that _β_-ketothiolase may deplete the pool of acetyl-CoA in the chloroplast, and through increasing ACCase activity by continuous light (Post-Beittenmiller et al., 1991; Page et al., 1994; Kozaki et al., 2000), ACCase is able to compete more effectively for acetyl-CoA, thereby increasing the levels of plastidic fatty acid biosynthesis. Developing microspores and surrounding tapetal cells have been shown to be particularly active in lipid metabolism (Fatland et al., 2002; Yui et al., 2003), which is especially needed for the formation of the exine pollen wall from sporopollenin (Wang et al., 2002; Ariizumi et al., 2004).
Support for the normal growth and development observed in the chloroplast transgenic lines expressing _pha_A is provided from studies where a mutant plant with only 10% activity in acyl-CoA synthase (which catalyzes the formation of fatty acyl-CoA in chloroplast from free fatty acid, ATP, and CoA) showed normal content of lipids in leaves and normal growth (Schnurr et al., 2002), indicating that under normal growth conditions even plants severely impaired in the fatty acid biosynthesis pathway were able to grow normally. Furthermore, Nikolau et al. (2000) reported that constitutive expression of an antisense construct of acetyl-CoA synthase failed to produce any detrimental morphological phenotype or change in total plant fatty acid composition in the transgenic plant; acetyl-CoA synthase is the enzyme responsible for supplying acetyl-CoA from acetate in chloroplast. However, they observed that the use of acetate as a source for acetyl-CoA decreased in proportion to the degree of down-regulation of the enzyme in the antisense lines. Therefore, under normal developmental conditions, which do not have an excessive demand for de novo fatty acid synthesis, alternative mechanisms can compensate for the decrease in activity of key enzymes of the de novo fatty acid biosynthesis pathway. However, during high fatty acid-driven processes such as tapetum and pollen development, any such alternative mechanism might not be adequate to meet the high demand. This same scenario could apply if acetyl-CoA is in short supply and ACCase cannot synthesize enough malonyl-CoA to drive the synthesis of fatty acids.
An alternative explanation for the male-sterile phenotype might be that the formation of acetoacetyl-CoA or _β_-ketothiolase itself may be producing a detrimental effect on chloroplast metabolism. This explanation is difficult to support from our reversibility studies because under continuous light no decrease in _β_-ketothiolase level or activity was observed; this excludes a possible decrease in acetoacetyl-CoA or detrimental effect caused by _β_-ketothiolase. The results of this study clearly show that the expression of _pha_A in the chloroplast does not cause observable severe pleiotropic effects (with the exception of male sterility). This is in sharp contrast with previous reports, where attempts to insert and express the _pha_A via the nuclear genome failed to recover any transgenic plants; this effect was attributed to the putative deleterious effect of _pha_A (Bohmert et al., 2002). Compartmentalization of proteins in chloroplasts has been shown to avoid pleiotropic effects, as previously reported for cholera toxin B subunit (Daniell et al., 2001a), trehalose (Lee et al., 2003), and xylanase (Leelavathi et al., 2003).
In addition, the lack of detrimental effect can be explained by the enzymatic mechanism of _β_-ketothiolase in the chloroplast. _β_-Ketothiolase is an enzyme that can generate a carbon-carbon bond in a biological Claisen condensation (Davis et al., 1987). _β_-Ketothiolase interacts with two identical acetyl-CoA molecules differently, by using one as a carbanion and the other as an electrophile, to produce a head-to-tail condensation. Alternatively, under physiological conditions in bacteria, _β_-ketothiolase can also undergo the CoASH-mediated thiolysis of acetoacetyl-CoA (Davis et al., 1987); in chloroplast, there is no known endogenous pool of acetoacetyl-CoA. It is possible that the detrimental effects observed during the expression of the entire PHB pathway in chloroplast (Lossl et al., 2003) were due to the depletion of the chloroplast acetyl-CoA pool by the consecutive action of _pha_A, _pha_B, and _pha_C to produce PHB, which would act as a sink because PHB cannot be metabolized by plants. However, when only _pha_A is expressed in the chloroplast, acetyl-CoA will be converted to acetoacetyl-CoA, which cannot be metabolized by any known chloroplast enzyme. Accumulation of acetoacetyl-CoA will drive the kinetics of the reaction toward _β_-ketothiolase-mediated thiolysis. This ping-pong effect caused by _β_-ketothiolase could cause an imbalance between acetyl-CoA removal and production that might be enough to affect a steady supply of acetyl-CoA to the fatty acid biosynthesis pathway. The effect of such an imbalance could be more pronounced under conditions that require high de novo fatty acid biosynthesis, such as in the case of tapetum and pollen development.
Chloroplast genetic engineering approach offers a number of attractive advantages, including high-level transgene expression (De Cosa et al., 2001; Daniell et al., 2005a, 2005b), multigene engineering in a single transformation event (De Cosa et al., 2001; Daniell and Dhingra, 2002; Lossl et al., 2003; Ruiz et al., 2003), transgene containment via maternal inheritance (Daniell, 2002; Hagemann, 2004), lack of gene silencing (De Cosa et al., 2001; Lee et al., 2003; Dhingra et al., 2004), position effect due to site-specific transgene integration (Daniell et al., 2002), and lack of pleiotropic effects due to subcellular compartmentalization of transgene products (Daniell et al., 2001a; Lee et al., 2003; Leelavathi et al., 2003). Because of these advantages, the chloroplast genome has been engineered to confer several useful agronomic traits, including herbicide resistance (Daniell et al., 1998), insect resistance (McBride et al., 1995; Kota et al., 1999), disease resistance (DeGray et al., 2001), drought tolerance (Lee et al., 2003), salt tolerance (Kumar et al., 2004a), and phytoremediation (Ruiz et al., 2003). The chloroplast genome has also been used in molecular farming to express human therapeutic proteins (Guda et al., 2000; Staub et al., 2000; Fernandez-San Millan et al., 2003; Leelavathi and Reddy, 2003; Daniell et al., 2004a, 2004b, 2004c), vaccines for human (Daniell et al., 2001a, 2004c; Daniell, 2004; Watson et al., 2004; Chebolu and Daniell, 2005) or animal use (Molina et al., 2004), and biomaterials (Guda et al., 2000; Lossl et al., 2003; Vitanen et al., 2004). Genetically engineered CMS via the chloroplast genome may be used for the safe integration of foreign genes via the nuclear genome and in those rare cases where plastids genomes are paternally or biparentally transmitted (Hagemann, 2004). Recently, plastid transformation was demonstrated in carrot (Daucus carota; Kumar et al., 2004a), showing hyperexpression of the transgene in non-green plastids to levels of up to 75% the expression observed in leaf chloroplasts. Additionally, plastid transformation of recalcitrant crops such as cotton (Gossypium hirsutum; Kumar et al., 2004b) and soybean (Glycine max; Dufourmantel et al., 2004) allows the application of the cytoplasmic male-sterile system to commercially important crops.
An important consideration for hybrid development using CMS systems is the requirement (or not) for male-fertility restoration. For vegetable, fruit, or forage crops, restoration of male fertility in the hybrid is not necessary. This simplifies the production of hybrids because effort can concentrate on maintainer line development, without concern for whether the pollinator restores male fertility in the hybrid. For crops with seeds as the economically important product, such as canola, sunflower (Helianthus annuus), or maize, one or both of the hybrid's parents must bring in male-fertility restoration factors or the male-sterile hybrid seed must be blended with male-fertile hybrid seed (Havey, 2004). In currently available cytoplasmic male-sterile lines, nuclear genome controls various restoration factors, and such factors are often located at multiple loci. However, we report here restoration of male fertility by changing conditions of illumination. Also, in the case of _β_-ketothiolase-induced male sterility, fertility can be obtained by the use of regulatory or inducible elements instead of constitutive expression. Thus, this is a novel approach for creating male-sterile transgenic plants, which may help advance the field of plant biotechnology through effective transgene containment.
Havey (2004) documents the worldwide use of CMS to produce competitive hybrid cultivars. Major investments of time and resources are required to backcross a male-sterility-inducing cytoplasm into elite lines. These generations of backcrossing could be avoided by transformation of an organellar genome of the elite male-fertile inbred to produce female inbred lines for hybrid seed production. Because the male-fertile parental and male-sterile transformed lines would be developed from the same inbred, they should be highly uniform and possess the same nuclear genotype, excluding mutations and residual heterozygosity (Havey, 2004). Therefore, the male-fertile parental line becomes the maintainer line to seed-propagate the newly transformed male-sterile line (Havey, 2004). A few generations of seed increases would produce a CMS-maintainer pair for hybrid seed production. An additional advantage of organellar transformation would be the diversification of CMS sources used in commercial hybrid-seed production. Transformation of the chloroplast genome would allow breeders to introduce different male-sterility-inducing factors into superior inbred lines. Introduction of a male-sterility-inducing transgene into one of the organellar genomes of a higher plant would be a major breakthrough in the production of male-sterile inbred lines (Havey, 2004). This technique would be of great potential importance in the production of hybrid crops by avoiding generations of backcrossing, an approach especially advantageous for crop plants with longer generation times (Havey, 2004).
MATERIALS AND METHODS
Chloroplast Vector Construction
Plasmid DNA from Acinetobacter sp. coding for the _pha_A gene (pJKD 1425) was kindly provided by Metabolix (Cambridge, MA). Isolation and amplification of the _pha_A gene from the native plasmid were performed by PCR using _pha_A-specific 5′ and 3′ flanking DNA primers. All primers were designed using the QUICKPRI program of the DNASTAR software (Madison, WI). The PCR product was cloned into the vector pCR2.1-5′UTR_psb_A, which contained the functional _psb_A gene promoter and 5′ regulatory sequence, by directional cloning after _Nde_I and _Not_I restriction digestion of the PCR product and vector. The _pha_A gene and the 5′UTR_psb_A regions were sequenced and subsequently cloned into the chloroplast transformation vector pLD-ctv, by directional insertion using appropriate restriction sites.
Chloroplast Transformation and Selection of Transgenic Plants
The delivery of the pLDR-5′UTR-_pha_A-3′UTR vector to the chloroplast by particle bombardment and the subsequent selection process of the transgenic tobacco (Nicotiana tabacum var Petit Havana) lines were performed essentially as described previously (Daniell, 1997; Daniell et al., 2004a; Kumar and Daniell, 2004). Tobacco leaves were bombarded using the biolistic device PDS-1000/He (Bio-Rad, Hercules, CA). After bombardment, leaves were placed on regeneration medium of plants, supplied with 500 _μ_g mL−1 spectinomycin for two rounds of selection on plates, and subsequently moved to jars on Murashige and Skoog medium containing 500 _μ_g mL−1 spectinomycin. Finally, homoplasmic plants were transferred to high nutrient soil and grown in controlled growth chambers at 26°C in a 16-h-light/8-h-dark photoperiod.
Confirmation of Chloroplast Integration by PCR
Total plant DNA from untransformed and transgenic plants was isolated using the DNeasy plant mini kit (Qiagen, Valencia, CA) and used as the template for PCR reactions. The PCR primer pairs 3P-3M and 4P-4M were used to confirm the integration of the gene cassette into the chloroplast genome, essentially as described previously (Daniell et al., 2004a). Primer pairs 5P-2M, 5P-phaAinternal, and 5P-3′phaA were used to confirm the presence of the _pha_A gene. PCR analysis was performed using the Gene Amp PCR System 2400 (Perkin-Elmer, Chicago).
Southern-Blot Analysis
Total plant DNA was obtained from T0 and T1 transgenic plants as well as from untransformed tobacco plants using the DNeasy plant mini kit (Qiagen) and protocol. Southern-blot analyses were performed essentially as described previously (Daniell et al., 2004a). Two micrograms of plant DNA was digested with _Bam_HI and resolved on a 0.8% (w/v) agarose gel at 50 V for 2 h. The gel was soaked in 0.25 n HCl for 15 min and then was rinsed two times with water. The gel was then soaked in transfer buffer (0.4 n NaOH and 1 m NaCl) for 20 min, and the denatured DNA was transferred overnight to a nitrocellulose membrane by capillarity. The next day the membrane was rinsed twice in 2× SSC (0.3 m NaCl and 0.03 m sodium citrate), dried on Whatman paper, and then cross-linked in the GS GeneLinker (Bio-Rad) at setting C3 (150 njoules). The flanking sequence probe was obtained by _Bgl_II/_Bam_HI restriction digestion of plasmid pUC-ct, which contains the chloroplast flanking sequence (_trn_I and _trn_A genes). The _pha_A probe was obtained by _Nde_I/_Not_I restriction digestion of plasmid pCR2.1-5′UTR-_pha_A. Probes were radiolabeled with 32P dCTP by using Ready Mix and Quant G-50 microcolumns for purification (Amersham, Arlington Heights, IL). Prehybridization and hybridization were performed using the Quick-Hyb solution (Stratagene, La Jolla, CA). The membrane was washed twice for 15 min at room temperature in 2× SSC with 0.1% (w/v) SDS, followed by two additional washes at 60°C (to increase the stringency) for 15 min with 0.1× SSC with 0.1% (w/v) SDS. Radiolabeled blots were exposed to x-ray films and then developed in the Mini-Medical Series x-ray film processor (AFP Imaging, Elmsford, NY).
Northern-Blot Analysis
Total plant RNA from untransformed and chloroplast transgenic plants was isolated using the RNeasy mini kit (Qiagen) and protocol. Northern-blot analyses were performed essentially as described previously (Ruiz et al., 2003). Total RNA (2.5 _μ_g) per plant sample was resolved in a 1.2% (w/v) agarose/formaldehyde gel. The _pha_A probe generation, labeling reaction, prehybridization/hybridization, membrane washing steps, and autoradiography were performed essentially as explained above in the Southern-blot section. The _aad_A probe was amplified by PCR from the pLD-ctv-_aad_A.
Western-Blot Analysis
Protein samples were obtained from 100 mg of leaf material from untransformed plants and transgenic lines by grinding the tissue to a fine powder in liquid nitrogen, subsequent homogenization in 200 μ_L of plant protein extraction buffer (100 mm NaCl, 10 mm EDTA, 200 mm Tris-HCl, 0.05% [w/v] Tween 20, 0.1% [w/v] SDS, 14 mm BME, 400 mm Suc, and 2 mm phenylmethylsulfonyl fluoride), and a centrifugation step at 15.7_g for 1 min to remove solids. Protein concentrations were determined by Bradford assay (Bio-Rad Protein Assay) with bovine serum albumin as the protein standard. Proteins were resolved by electrophoresis in a 12% (v/v) SDS-PAGE and then transferred to a nitrocellulose membrane (Bio-Rad). The membrane was blocked for 1 h with PTM buffer: 1×PBS (phosphate-buffered solution), 0.05% (v/v) Tween 20, and 3% (w/v) nonfat dry milk. The membrane was probed for 2 h with rabbit anti-_β_-kethiolase antibody (Metabolix) in a dilution of 1:1,000, then rinsed with water twice and probed with alkaline phosphatase-conjugated secondary antibody (goat anti-rabbit; Sigma, St. Louis) for 1.5 h in a 1:20,000 dilution. Finally, the membrane was washed three times for 10 min with PT buffer (1× PBS, 0.05% [v/v] Tween 20) and one time with 1×PBS, followed by incubation in Lumi-phos WB (Pierce, Rockford, IL) reagent for the alkaline phosphatase reaction. Film exposure took place for 3 min.
_β_-Ketothiolase Activity Assay
Protein samples were obtained by grinding 1 g of leaf tissue to a fine powder in liquid nitrogen, followed by the addition of 2 mL of ice-cold β_-ketothiolase extraction buffer (100 mm Tris-HCl, pH 8.1, 50 mm MgCl2, 5 mm BME) and homogenization. The homogenates were centrifuged for 10 min at 4°C at 5,000_g, and the supernatant was passed through PD-10 columns (Amersham) containing Sephadex G-25 M for desalting, and elution was optimized for the recovery of proteins of size range 25 to 60 kD. Protein concentration was determined by a Bradford assay. _β_-Ketothiolase activity was measured spectrophotometrically at 304 nm in the thiolysis direction (breaking down acetoacetyl-CoA to acetyl-CoA) by monitoring the disappearance of acetoacetyl-CoA for 60 s, which in the presence of magnesium ion forms a magnesium enolate with _A_304; this protocol is an adaptation of the protocol by Senior and Dawes (1973). The reaction took place in a total volume of 1 mL containing 62.4 mm Tris-HCl, pH 8.1, 50 mm MgCl2, 62.5 _μ_m CoA, 62.5 _μ_m acetoacetyl-CoA (substrate is dissolved in 50 mm phosphate buffer, pH 4.7), 10 _μ_L of plant extract (_β_-ketothiolase sample), and bringing the volume with distilled water to 1 mL. The plant extract containing the _β_-ketothiolase was added at the end immediately before the sample reading. In this assay, the enzyme specific activity is given in units per milligram of total plant protein, and 1 unit is defined as the degradation of 1 _μ_mol/min of acetoacetyl-CoA under standard reaction conditions.
Scanning Electron Microscopy
SEM was performed at the AMPAC facility at the University of Central Florida. Anthers and pollen samples were gold coated on a Sputter Coater (Emitech, Houston) with a gold film thickness of 150 Å. SEM pictures were produced using the scanning electron microscope model JSM-6400F (JEOL, Peabody, MA) and the x-ray energy dispersive spectrometer (Edax, Mahwah, NY) at an acceleration voltage of 6 kV.
Histological Analysis of Anthers
Anthers at relevant developmental stages were dissected from flower buds and fixed in 3% (v/v) glutaraldehyde in phosphate buffer for 12 h at room temperature, applying a continuous vacuum for the first 3 h of incubation and degassing (by bringing the vacuum up and down slowly) for 10 min at 1, 2, and 3 h. The fixed anthers were dehydrated in an ethanol series (5%, 10% to 80% in increments of 10, 95% and 100%) for 30 min per gradient treatment. Samples were kept overnight in fresh 100% ethanol and were washed twice the next day for 1 h in 100% ethanol. Samples were then treated with a gradient (25% to 100%) of Citro Solv clearing (Fisher, Pittsburgh) reagent for 30 min per gradient treatment, and finally embedded in Paraplast Plus (Fisher). Tissue embedding was performed in molten paraffin for 3 d, changing the molten paraffin every 8 to 12 h. Paraffin-treated tissue was finally embedded into paraffin blocks by using the Leica EG 1160 paraffin embedding station (Leica, Solms, Germany). A metal blade microtome, model HM 315 (MICROM, Walldorf, Germany), was used for tissue embedded sectioning. Finally, tissue sections were put onto Superfrost/Plus microscope slides, followed by a rehydration step and tissue staining with 0.05% (w/v) toluidine blue. Tissue slides were observed under the Olympus BX60F5 light microscope and Olympus U-CMAD-2 camera (Olympus, Melville, NY). Flower developmental stages were characterized following the procedure described by Koltunow et al. (1990).
Reversibility of Male Fertility
Two independent transgenic plants were moved to a separate growth chamber after the first indication of flower bud formation and were kept away from any contact with untransformed plants and other transgenic lines; the flowers were covered with thin transparent plastic bags to inhibit any possibility of cross-pollination. Bags were only removed to take pictures. Transgenic plants were kept under continuous illumination for 10 d with a photon flux density of 11,250 _μ_E m−2 supplied throughout this period. The number of flowers developed was counted daily throughout these 10 d, while newly formed flowers, senescent flowers, and fallen flowers were recorded. Fruit capsules and seeds that developed were also counted. After the 10 d, a 16-h-light/8-h-dark photoperiod was reestablished, while the plants were kept from contact with any other plant for 20 d to allow maturation of the fruit capsules and to harvest seeds produced during continuous illumination.
Acknowledgments
We thank Metabolix, especially Dr. Kristi Snell, for kindly providing the plasmid pJKD 1425 and anti-_β_-ketothiolase antibody, help with _β_-ketothiolase assays, and Dr. Karen Bohmert for helpful discussions. Also, Drs. Karl X. Chai, Li-Mei Chen, and Jeanette Nadeau are acknowledged for their advice and help with histological analysis.
1
This work was supported in part by the National Institutes of Health (grant no. R 01 GM 63879 to H.D.), the U.S. Department of Agriculture (grant no. 3611–21000–017–00D to H.D.), and Metabolix (grants to H.D.).
References
- Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sato S, Kato T, Tabata S, Toriyama K (2004) Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J 39**:** 170–181 [DOI] [PubMed] [Google Scholar]
- Bateson W, Gairdner AE (1921) Male sterility in flax subject to two types of segregation. J Genet 11**:** 269–275 [Google Scholar]
- Bentolila S, Alfonso AA, Hanson MR (2002) A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci USA 99**:** 10887–10892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmert K, Balbo I, Kopka J, Mittendorf V, Nawrath C, Poirier Y, Tischendorf G, Trethewey RN, Willmitzer L (2000) Transgenic Arabidopsis plants can accumulate polyhydroxybutyrate to up to 4% of their fresh weight. Planta 211**:** 841–845 [DOI] [PubMed] [Google Scholar]
- Bohmert K, Balbo I, Steinbuchel A, Tischendorf G, Willmitzer L (2002) Constitutive expression of the beta-ketothiolase gene in transgenic plants. A major obstacle for obtaining polyhydroxybutyrate-producing plants. Plant Physiol 128**:** 1282–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebolu S, Daniell H (2005) Chloroplast derived vaccine antigens and biopharmaceuticals: expression, folding, assembly and functionality. Curr Trends Microbiol Immunol (in press) [DOI] [PMC free article] [PubMed]
- Chittenden RJ, Pellow CA (1927) A suggested interpretation of certain cases of anisogeny. Nature 119**:** 10–12 [Google Scholar]
- Cui X, Wise RP, Schnable PS (1996) The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 272**:** 1334–1336 [DOI] [PubMed] [Google Scholar]
- Daniell H (1997) Transformation and foreign gene expression in plants mediated by microprojectile bombardment. Methods Mol Biol 62**:** 453–488 [DOI] [PubMed] [Google Scholar]
- Daniell H (2002) Molecular strategies for gene containment in transgenic crops. Nat Biotechnol 20**:** 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H (2004) Medical molecular pharming: therapeutic recombinant antibodies, biopharmaceuticals, and edible vaccines in transgenic plants engineered via the chloroplast genome. In RM Goodman, ed, Encyclopedia of Plant and Crop Science. Marcel Decker, New York, pp 704–710
- Daniell H, Carmona-Sanchez O, Burns B (2004. b) Chloroplast derived antibodies, biopharmaceuticals and edible vaccines. In S Schillberg, ed, Molecular Farming. Wiley-VCH Verlag, Weinheim, Germany, Chapter 8, pp 113–133
- Daniell H, Chebolu S, Kumar S, Singleton M, Falconer R (2005. a) Chloroplast-derived vaccine antigens and other therapeutic proteins. Vaccine 23**:** 1779–1783 [DOI] [PubMed] [Google Scholar]
- Daniell H, Cohill PR, Kumar S, Dufourmantel N, Dubald M (2004. c) Chloroplast genetic engineering. In H Daniell, C Chase, eds, Molecular Biology and Biotechnology of Plant Organelles. Springer Publishers, Dordrecht, The Netherlands, pp 443–490
- Daniell H, Datta R, Varma S, Gray S, Lee SB (1998) Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol 16**:** 345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A (2002) Multigene engineering: dawn of an exciting new era in biotechnology. Curr Opin Biotechnol 13**:** 136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Khan MS, Allison L (2002) Milestones in chloroplast genetic engineering: an environmentally friendly era in biotechnology. Trends Plant Sci 7**:** 84–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Kumar S, Dufourmantel N (2005. b) Breakthrough in chloroplast genetic engineering of agronomically important crops. Trends Biotechnol 23**:** 238–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Panchal T, Weibe PO (2001. a) Expression of the native cholera toxin B subunit gene and assembly as functional oligomers in transgenic chloroplasts. J Mol Biol 311**:** 1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Muthukumar B, Lee SB (2001. b) Marker free transgenic plants: engineering the chloroplast genome without the use of antibiotic selection. Curr Genet 39**:** 109–116 [DOI] [PubMed] [Google Scholar]
- Daniell H, Ruiz ON, Dhingra A (2004. a) Chloroplast genetic engineering to improve agronomic traits. Methods Mol Biol 286**:** 111–137 [DOI] [PubMed] [Google Scholar]
- Davis JT, Moore RN, Imperiali B, Pratt AJ, Kobayashi K, Masamune S, Sinskey AJ, Walsh CT (1987) Biosynthetic thiolase from Zoogloea ramigera. J Biol Chem 262**:** 82–89 [PubMed] [Google Scholar]
- De Cosa B, Moar W, Lee SB, Miller M, Daniell H (2001) Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol 19**:** 71–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H (2001) Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol 127**:** 852–862 [PMC free article] [PubMed] [Google Scholar]
- Delourme R, Foisset N, Horvais R, Barret P, Champagne G, Cheung WY, Landry BS, Renard M (1998) Characterisation of the radish introgression carrying the Rfo restorer gene for _Ogu_-INRA cytoplasmic male sterility in rapeseed (Brassica napus L.). Theor Appl Genet 97**:** 129–134 [Google Scholar]
- Dhingra A, Daniell H (2005) Chloroplast genetic engineering via organogenesis or somatic embryogenesis. In J Sanchez-Serano, ed, Arabidopsis Protocols II. Humana Press, Totowa, NJ (in press) [DOI] [PubMed]
- Dhingra A, Portis AR Jr, Daniell H (2004) Enhanced translation of a chloroplast-expressed RbcS gene restores small subunit levels and photosynthesis in nuclear RbcS antisense plants. Proc Natl Acad Sci USA 101**:** 6315–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourmantel N, Pelissier B, Garçon F, Peltier JM, Tissot G (2004) Generation of fertile transplastomic soybean. Plant Mol Biol 55**:** 479–489 [DOI] [PubMed] [Google Scholar]
- Eibl C, Zou Z, Beck A, Kim M, Mullet J, Koop HU (1999) In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast transformation: tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. Plant J 19**:** 333–345 [DOI] [PubMed] [Google Scholar]
- Fatland BL, Ke J, Anderson MD, Mentzen WI, Cui LW, Allred CC, Johnston JL, Nikolau BJ, Wurtele ES (2002) Molecular characterization of a heteromeric ATP-citrate lyase that generates cytosolic acetyl-coenzyme A in Arabidopsis. Plant Physiol 130**:** 740–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-San Millan A, Mingo-Castel A, Miller M, Daniell H (2003) A chloroplast transgenic approach to hyper-express and purify human serum albumin, a protein highly susceptible to proteolytic degradation. Plant Biotechnol J 1**:** 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Godt DE, Guivarc'h A, Kahmann U, Chriqui D, Roitsch T (2001) Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc Natl Acad Sci USA 98**:** 6522–6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guda C, Lee SB, Daniell H (2000) Stable expression of biodegradable protein based polymer in tobacco chloroplasts. Plant Cell Rep 19**:** 257–262 [DOI] [PubMed] [Google Scholar]
- Hagemann R (1992) Plastid genetics in higher plants. In RG Herrmann, ed, Cell Organelles. Springer-Verlag, New York, pp 65–96
- Hagemann R (2004) The sexual inheritance of plant organelles. In H Daniell, C Chase, eds, Molecular Biology and Biotechnology of Plant Organelles. Springer Publisher, Dordrecht, The Netherlands, pp 87–108
- Havey MJ (2004) The use of cytoplasmic male sterility for hybrid seed production. In H Daniell, C Chase, eds, Molecular Biology and Biotechnology of Plant Organelles. Springer Publisher, Dordrecht, The Netherlands, pp 617–628
- Hernould M, Suharsono S, Litvak S, Araya A, Mouras A (1993) Male-sterility induction in transgenic tobacco plants with an unedited atp9 mitochondrial gene from wheat. Proc Natl Acad Sci USA 90**:** 2370–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernould M, Suharsono S, Zabaleta E, Carde JP, Litvak S, Araya A, Mouras A (1998) Impairment of tapetum and mitochondria in engineered male-sterile tobacco plants. Plant Mol Biol 36**:** 499–508 [DOI] [PubMed] [Google Scholar]
- Jones H, Clarke A (1943) Inheritance of male sterility in the onion and the production of hybrid seed. Proc Am Soc Hortic Sci 43**:** 189–194 [Google Scholar]
- Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura J (2003) Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J 34**:** 407–415 [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB (1990) Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2**:** 1201–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota M, Daniell H, Varma S, Garczynski SF, Gould F, William MJ (1999) Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in chloroplasts confers resistance to plants against susceptible and Bt-resistant insects. Proc Natl Acad Sci USA 96**:** 1840–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki A, Kamada K, Nagano Y, Iguchi H, Sasaki Y (2000) Recombinant carboxyltransferase responsive to redox of pea plastidic acetyl-CoA carboxylase. J Biol Chem 275**:** 10702–10708 [DOI] [PubMed] [Google Scholar]
- Kriete G, Niehaus K, Perlick AM, Puhler A, Broer I (1996) Male sterility in transgenic tobacco plants induced by tapetum-specific deacetylation of the externally applied non-toxic compound N-acetyl-L-phosphinothricin. Plant J 9**:** 809–818 [DOI] [PubMed] [Google Scholar]
- Kumar S, Daniell H (2004) Engineering the chloroplast genome for hyper-expression of human therapeutic proteins and vaccine antigens. Methods Mol Biol 267**:** 365–383 [DOI] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H (2004. a) Plastid-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots, and leaves confers enhanced salt tolerance. Plant Physiol 136**:** 2843–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H (2004. b) Manipulation of gene expression facilitates cotton plastid transformation by somatic embryogenesis and maternal inheritance of transgenes. Plant Mol Biol 56**:** 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Kwon HB, Kwon SJ, Park SC, Jeong MJ, Han SE, Byun MO, Daniell H (2003) Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol Breed 11**:** 1–13 [Google Scholar]
- Leelavathi S, Gupta N, Maiti S, Ghosh A, Reddy VS (2003) Overproduction of an alkali and thermostable xylanase in tobacco chloroplasts and efficient recovery of the enzyme. Mol Breed 11**:** 59–67 [Google Scholar]
- Leelavathi S, Reddy VS (2003) Chloroplast expression of His-tagged GUS-fusions: a general strategy to overproduce and purify foreign proteins using transplastomic plants as bioreactors. Mol Breed 11**:** 49–58 [Google Scholar]
- Liu F, Cui X, Horner HT, Weiner H, Schable PS (2001) Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize. Plant Cell 13**:** 1063–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossl A, Eibl C, Harloff HJ, Jung C, Koop HU (2003) Polyester synthesis in transplastomic tobacco (Nicotiana tabacum L.): significant contents of polyhydroxybutyrate are associated with growth reduction. Plant Cell Rep 21**:** 891–899 [DOI] [PubMed] [Google Scholar]
- Mariani C, De Beuckeleer M, Truettner J, Leemans J, Goldberg RB (1990) Induction of male-sterility in plants by a chimeric ribonuclease gene. Nature 347**:** 737–741 [Google Scholar]
- McBride KE, Svab Z, Schaaf DJ, Hogan PS, Stalker DM, Maliga P (1995) Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of an insecticidal protein in tobacco. Biotechnology (N Y) 13**:** 362–365 [DOI] [PubMed] [Google Scholar]
- Molina A, Herva-Stubbs S, Daniell H, Mingo-Castel AM, Veramendi J (2004) High yield expression of a viral peptide animal vaccine in transgenic tobacco chloroplasts. Plant Biotechnol 2**:** 141–153 [DOI] [PubMed] [Google Scholar]
- Monde RA, Greene JC, Stern DB (2000) The sequence and secondary structure of the 3′-UTR affect 3′-end maturation, RNA accumulation, and translation in tobacco chloroplasts. Plant Mol Biol 44**:** 529–542 [DOI] [PubMed] [Google Scholar]
- Napoli CA, Fahy D, Wang HY, Taylor LP (1999) White anther: a petunia mutant that abolishes pollen flavonol accumulation, induces male sterility, and is complemented by a chalcone synthase transgene. Plant Physiol 120**:** 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Porier Y, Somerville C (1994) Targeting of the polyhydroxybutyrate biosynthetic pathway to the plastids of Arabidopsis thaliana result in high levels of polymer accumulation. Proc Natl Acad Sci USA 91**:** 12760–12764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolau BJ, Oliver DJ, Schnable PS, Wurtele ES (2000) Molecular biology of acetyl-CoA metabolism. Biochem Soc Trans 28**:** 591–593 [PubMed] [Google Scholar]
- Page RA, Okada S, Harwood JL (1994) Acetyl-CoA carboxylase exerts strong flux control over lipid synthesis in plants. Biochim Biophys Acta 1210**:** 369–372 [DOI] [PubMed] [Google Scholar]
- Perez-Prat E, van Lookeren Campagne MM (2002) Hybrid seed production and the challenge of propagating male sterile plants. Trends Plant Sci 7**:** 199–203 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Dennis DE, Klomparens K, Somerville C (1992) Polyhydroxybutyrate, a biodegradable thermoplastic in transgenic plants. Science 256**:** 520–523 [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller D, Jaworski JG, Ohlrogge JB (1991) In vivo pools of free and acylated acyl carrier proteins in spinach. Evidence for sites of regulation of fatty acid biosynthesis. J Biol Chem 266**:** 1858–1865 [PubMed] [Google Scholar]
- Rawsthorne S (2002) Carbon flux and fatty acid synthesis in plants. Prog Lipid Res 41**:** 182–196 [DOI] [PubMed] [Google Scholar]
- Ruiz ON, Hussein H, Terry N, Daniell H (2003) Phytoremediation of organomercurial compounds via chloroplast genetic engineering. Plant Physiol 132**:** 1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurr JA, Shockey JM, de Boer GJ, Browse JA (2002) Fatty acid export from the chloroplast. Molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis. Plant Physiol 129**:** 1700–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G, Gruissem W (1991) Chloroplast mRNA 3′ end processing requires a nuclear-encoded RNA-binding protein. EMBO J 10**:** 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior PJ, Dawes EA (1973) The regulation of poly-beta-hydroxybutyrate metabolism in Azotobacter beijerinckii. Biochem J 134**:** 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Crowther T (1995) Inbreeding depression and single cross hybrids in leeks (Allium ampeloprasum ssp. porrum). Euphytica 86**:** 87–94 [Google Scholar]
- Staub JM, Garcia B, Graves J, Hajdukiewicz PTJ, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll JA, Spatola L, et al (2000) High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat Biotechnol 18**:** 333–338 [DOI] [PubMed] [Google Scholar]
- Sugita M, Sugiura M (1996) Regulation of gene expression in chloroplasts of higher plants. Plant Mol Biol 32**:** 315–326 [DOI] [PubMed] [Google Scholar]
- Vitanen PV, Devine AL, Kahn S, Deuel DL, Van Dyk DE, Daniell H (2004) Metabolic engineering of the chloroplast genome using the Escherichia coli ubiC gene reveals that chorismate is a readily abundant precursor for p-hydroxybenzoic acid synthesis. Plant Physiol 136**:** 4048–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Xia Q, Xie W, Dumonceaux T, Zou J, Datla R, Selvaraj G (2002) Male gametophyte development in bread wheat (Triticum aestivum L.): molecular, cellular, and biochemical analyses of a sporophytic contribution to pollen wall ontogeny. Plant J 30**:** 613–623 [DOI] [PubMed] [Google Scholar]
- Watson J, Koya V, Leppla SH, Daniell H (2004) Expression of Bacillus anthracis protective antigen in transgenic tobacco chloroplasts: development of an improved anthrax vaccine in a non-food/feed crop. Vaccine 22**:** 4374–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui R, Iketani S, Mikami T, Kubo T (2003) Antisense inhibition of mitochondrial pyruvate dehydrogenase E1alpha subunit in anther tapetum causes male sterility. Plant J 34**:** 57–66 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Xia Q, Dauk M, Shen W, Selvaraj G, Zou J (2003) Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15**:** 1872–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]