Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans (original) (raw)
Abstract
In Caenorhabditis elegans, numerous ‘synMuv' (synthetic multivulval) genes encode for chromatin-associated proteins involved in transcriptional repression, including an orthologue of Rb and components of the NuRD histone deacetylase complex. These genes antagonize Ras signalling to prevent erroneous adoption of vulval fate. To identify new components of this mechanism, we performed a genome-wide RNA interference (RNAi) screen. After RNAi of 16 757 genes, we found nine new synMuv genes. Based on predicted functions and genetic epistasis experiments, we propose that at least four post-translational modifications converge to inhibit Ras-stimulated vulval development: sumoylation, histone tail deacetylation, methylation, and acetylation. In addition, we demonstrate a novel role for sumoylation in inhibiting LIN-12/Notch signalling in the vulva. We further show that many of the synMuv genes are involved in gene regulation outside the vulva, negatively regulating the expression of the Delta homologue lag-2. As most of the genes identified in this screen are conserved in humans, we suggest that similar interactions may be relevant in mammals for control of Ras and Notch signalling, crosstalk between these pathways, and cell proliferation.
Keywords: C. elegans, chromatin remodelling, Ras signalling, RNAi, vulva
Introduction
In the past decade, a wealth of studies has shown the importance of chromatin modifications and chromatin remodelling in gene regulation (reviewed in Jenuwein and Allis, 2001). These include modification of histone tails by acetylation, deacetylation, and methylation, binding of factors to modified histones, and nucleosome remodelling. A large number of different complexes are known to have activities in modifying chromatin, but it is poorly understood how they function in a developmental context.
The Caenorhabditis elegans vulva represents a simple developmental system in which to study the function and mechanism of chromatin regulation. The so-called synthetic multivulval (synMuv) genes, many of which encode chromatin-associated proteins (see below), inhibit vulval development by antagonizing Ras signalling through an unknown mechanism (Fay and Han, 2000). Identification of the factors involved in this regulation and study of their functions should lead to an understanding of how these work together to bring about correct cellular development.
The vulva develops from three of six multipotent vulval precursor cells (VPCs; P(3–8).p) positioned along the ventral midline. In wild-type animals, P(5–7).p form the vulva (Sulston and Horvitz, 1977). An EGF signal (LIN-3) emanating from an overlying gonad cell (the anchor cell, AC) (Kimble, 1981; Hill and Sternberg, 1992) activates the Ras signalling cascade in the closest VPCs through the EGF receptor LET-23 (Figure 1A) (Aroian et al, 1990; Beitel et al, 1990). The VPC closest to the AC, P6.p, receives the highest level of inductive LIN-3 EGF signal and becomes a 1° vulval cell (Sternberg and Horvitz, 1986). It responds by producing Notch ligands (LAG-2, APX-1, and DSL-1) and downregulating LIN-12/Notch in a Ras-dependent manner (Shaye and Greenwald, 2002; Chen and Greenwald, 2004). The ligands activate the Notch signalling pathway in the flanking P5.p and P7.p cells (Sternberg, 1988), which in turn activate the transcription of negative modulators of Ras signalling (Yoo et al, 2004). This lowers the level of Ras signalling in P5.p and P7.p and triggers adoption of the 2° vulval fate (Figure 1B) (Sternberg, 1988; Sundaram, 2004; Yoo et al, 2004). This crosstalk between Ras and Notch is critical for specification of vulval fates. The remaining three VPCs (P(3, 4, 8).p) have a non-vulval 3o fate, dividing once and fusing with the syncytial hypoderm, hyp7 (Sulston and Horvitz, 1977). It is thought that hyp7, which is in contact with the VPCs, counteracts Ras signalling by producing an as yet undefined signal (Herman and Hedgecock, 1990; Hedgecock and Herman, 1995; Myers and Greenwald, 2005).
Figure 1.
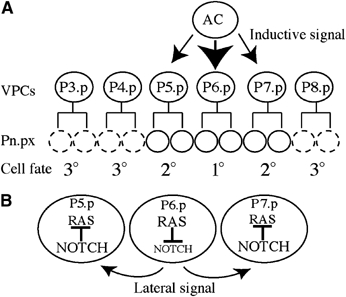
Vulval cell fate specification by the inductive and lateral signals. (A) The inductive signal emanates from the AC. The AC secretes LIN-3 EGF inducing vulval fates in P(5–7).p, which will produce 22 vulval cells. The remaining VPCs have the 3° cell fate, dividing once (dashed circles) and fusing to the hypoderm hyp7. (B) Upon activation of the Ras signalling pathway in P6.p, LIN-12/NOTCH is downregulated in a Ras-dependant manner and its ligands (LAG-2, APX-1, and DSL-1) are expressed, mediating the lateral signal essential for expression of the 2° fate in P5.p and P7.p. In these cells, Notch signalling is activated and negative regulators of Ras are upregulated, reducing Ras signalling.
Ras signalling in the VPCs is antagonized by two genetically redundant groups of genes, the class A and class B synMuv genes (Ferguson and Horvitz, 1989). Animals mutant for a gene of either class have a normal vulva, but in animals doubly mutant in both a class A and a class B gene, one or more of the VPCs that should have a 3° fate are instead induced to have a vulval fate, leading to the multivulval (Muv) phenotype. Recently, a third class of synMuv mutants (class C) was identified, which are Muv at low frequency, but show a high-penetrance Muv phenotype when combined with either a class A or a class B synMuv mutant (Ceol and Horvitz, 2004). The Muv phenotype of synMuv mutants requires the Ras signalling cascade, but is independent of the ligand LIN-3, suggesting that the synMuv genes prevent a basal level of Ras signalling from inducing VPCs to the vulval fate. At least some of the synMuv genes (e.g., LIN-35 Rb, see below) function in the hyp7 hypodermal syncytium, suggesting that they may be part of the antagonistic signalling from the hypodermis (Herman and Hedgecock, 1990; Hedgecock and Herman, 1995; Myers and Greenwald, 2005).
The molecular nature of some of the synMuv proteins suggests that they function via transcriptional repression. For example, in the synMuv B class, lin-35 is a C. elegans homologue of the tumour suppressor Rb (Lu and Horvitz, 1998), which can regulate the G1/S-phase transition via repression of transcription (reviewed in Frolov and Dyson, 2004). Rb and Rb-related factors can function in a complex with the heterodimer DP/E2F and HDAC1 (histone deacetylase) to prevent expression of S-phase-promoting genes by deacetylation of histone tails (Brehm et al, 1998; Morrison et al, 2002). Two recent publications described related Drosophila Rb/E2F complexes that contain homologues of multiple other synMuv B proteins; these complexes, termed dREAM and Myb–MuvB, have transcriptional repressor activity (Korenjak et al, 2004; Lewis et al, 2004; Lipsick, 2004). Other synMuv proteins have also been linked to repression of transcription. For example, the NuRD complex is a chromatin modifier complex that represses transcription through nucleosomal remodelling and deacetylase activities. Most members of this complex have been shown to display synMuv activities (Lu and Horvitz, 1998; Solari and Ahringer, 2000; von Zelewsky et al, 2000). In mammalian cells, deregulation of NuRD has been linked to metastatic growth (Feng and Zhang, 2003; Kumar et al, 2003). Recently, several components of the TRRAP-containing TIP60 histone acetlytransferase complex were shown to have synMuv activity (Ceol and Horvitz, 2004); the TIP60 complex has links to both transcriptional activation and repression (McMahon et al, 1998; Bouchard et al, 2001; Lang et al, 2001; Ard et al, 2002; Cai et al, 2003; Frank et al, 2003; Xiao et al, 2003). Many of the remaining synMuv genes are nuclear proteins of unknown function (Clark et al, 1994; Huang et al, 1994; Hsieh et al, 1999; Thomas and Horvitz, 1999; Thomas et al, 2003). It is not yet known in which tissue(s) most of the synMuv proteins act. However, LIN-35/Rb functions in tissue adjacent to the vulva (the syncytial hypodermis hyp7) and not in the vulva itself, so Ras-regulated vulval genes are probably not its direct targets (Myers and Greenwald, 2005).
To further understand this process, we sought to identify the majority of synMuv genes in the genome through genome-wide RNA interference (RNAi) screens in C. elegans. Our results suggest that multiple chromatin regulatory complexes cooperate to repress Ras-induced vulval development. In addition, through epistasis studies, we find that sumoylation regulates synMuv function and Notch signalling. This systematic reverse genetic approach also identified novel synMuv genes with human homologues that might potentially be involved in uncontrolled cell growth.
Results
Genome-wide screens for synMuv genes
We previously constructed a library of 16 757 bacterial strains for RNAi of 86% of C. elegans genes (Fraser et al, 2000; Kamath et al, 2003). Each bacterial strain is designed to produce dsRNA corresponding to a single predicted gene. Through feeding upon a given bacterial strain, worms ingest the dsRNA, inducing a knockdown of gene function via RNAi (Fire et al, 1998; Timmons and Fire, 1998).
To identify systematically synMuv genes, we carried out two genome-wide RNAi screens, starting with a class A synMuv mutant, lin-15A(n767), or a class B synMuv mutant, lin-15B(n744) (Clark et al, 1994; Huang et al, 1994). Animals singly mutant for lin-15A or lin-15B have a normal vulva, but in double mutants of lin-15A and any class B synMuv gene or of lin-15B and any class A synMuv gene, the vulva is hyperinduced, causing multiple vulval protrusions, the Muv phenotype (Ferguson and Horvitz, 1989) (Figure 2). To identify new synMuv genes, we fed lin-15A or lin-15B mutants each bacterial strain in the feeding library and screened for those that induced a multiple protrusion phenotype. We screened candidates using vulval GFP markers to find those with extra vulval cells (not shown) and then directly counted the number of induced vulval cells by observation using DIC microscopy (see below).
Figure 2.
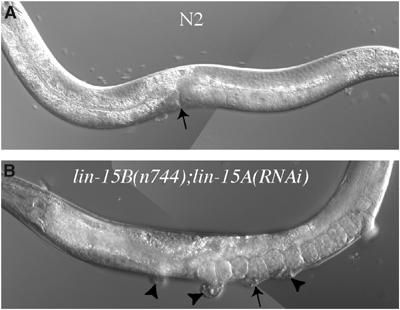
The Muv phenotype. (A) Wild-type N2 adult with normal vulva (arrow). (B) lin-15B(n744); lin-15A(RNAi) adult with multiple vulval protrusions. Arrowheads point to extra vulvae and the arrow to the normal vulva.
We identified a total of 18 synMuv genes in the screens (Table I); one other was identified by subsequent candidate testing (see below). Of these, nine are new synMuv genes, all showing ectopic vulval fate induction in a class A or class B background, or in both backgrounds (Table II). The 10 previously studied loci represent 83% of the synMuv genes in the RNAi library (Table I). All of the new synMuv genes have a human homologue (Table I), compared to around 60% of all C. elegans genes (Harris et al, 2004). In addition, based on sequence similarities, seven new synMuv genes have links to transcription regulation (Table I and see below).
Table 1.
New and previously studied synMuv genes
| Predicted gene | Locus | Human homologue | Brief description | Found in screen | Essential gene | Previously known |
|---|---|---|---|---|---|---|
| C03B8.4 | lin-13/lin-51 | ZFP28 (3.4E-18) | Zinc finger protein | ND | No | Yes |
| C32F10.2 | lin-35 | RBL2 (1E-46) | Retinoblastoma-like | B | No | Yes |
| C47D12.1 | trr-1 | TRRAP (1.7E-266) | TRRAP protein | B | Yes | Yes |
| C53A5.3 | hda-1/gon-10 | HDAC1 (1.7E-170) | Histone deacetylase 1 | B | Yes | Yes |
| F26F12.7 | let-418 | Mi-2 beta (0.0) | Chromodomain helicase protein | ND* | Yes | Yes |
| F26G5.9 | tam-1 | TRIM9 (7.9E-14) | RING finger/B-Box protein | No | No | Yes |
| F44B9.6 | lin-36 | None | None | B | No | Yes |
| JC8.6 | — | MTL5 (3.1E-42) | Tesmin | B | No | Yes |
| K01G5.2 | hpl-2 | CBX1 (1.6E-21) | Chromobox protein | No | Yes | Yes |
| K07A1.12 | lin-53/rba-2 | RBBP4 (1.8E-171) | Rb associated protein 48 | B | Yes | Yes |
| M04B2.1 | mep-1/gei-2 | ZNF406 (7E-04) | Zinc finger protein | B | Yes | Yes |
| T23G7.1 | dpl-1 | TFDP1 (4.1E-64) | Transcription factor DP-1 | ND* | Yes | Yes |
| T27C4.4 | egr-1 | MTA1L1 (4E-66) | Metastasis associated protein | A | Yes | Yes |
| VC5.4 | mys-1 | HTATIP (3.5E-121) | TIP6O histone acetyltransferase | No | Yes | Yes |
| Y1O2A5C.18 | efl-1 | E2F4 (9.2E-41) | Transcription factor E2F4 | B | Yes | Yes |
| ZK632.13 | lin-52 | L0C91750 (2.3E-06) | Novel protein | B | No | Yes |
| ZK637.7 | lin-9 | TGS (1.3E-67) | Tudor domain | B | No | Yes |
| ZK662.4 | lin-15b | None | None | ND | No | Yes |
| ZK678.1 | lin-15a | None | None | ND | No | Yes |
| E01A2.4 | — | NKAP (8.3E 50) | Nuclear NF-κB activ. protein | B | Yes | No |
| F29B9.6 | ubc-9 | UBE2I (2.3E-70) | Ubc9/SUMO-1 conj. enzyme | No | Yes | No |
| K12C11.2 | smo-1 | UBL1 (2.5E-25) | SUMO | A, B | Yes | No |
| R05D3.11 | met-2 | SETDB1 (5.3E-44) | Histone H3 lysine-9 methyltransferase | B | No | No |
| R06C7.7 | rls-1/lin-61 | L3MBTL2 (2E-42) | Lethal (3) malignant brain tumor | B | Yes | No |
| W01G7.3 | — | POLR2J (1.6E-37) | RNA pol II (13.3 kDa) | B | Yes | No |
| W02A11.4 | uba-2 | UBE1 (2.4E-107) | Uba2/SUMO activating enzyme subunit 2 | A, B | Yes | No |
| W07B3.2 | gei-4 | CALD1 (1.2E-10) | Unknown | B | Yes | No |
| Y71G12B.9 | — | ACRC (2E-13) | Unknown | B | No | No |
| Shown are predicted gene name, locus, closest human homologue with BlastP _E_-value in parentheses, a brief description of the protein, whether the gene was identified in the lin-15A screen (designated ‘B'), the lin-15B screen (designated ‘A'), or not screened (designated ‘ND' for clones not in the library or ‘ND*' for incorrect RNAi clones), if the gene is essential (defined as embryonic lethal, larval lethal, or sterile) and if it is a previously known synMuv gene. |
Table 2.
Induction of vulval fate
| Predicted gene | Locus | VPCs induced (% Muv; n) | VPCs induced (% Muv; n) | VPCs induced (% Muv; n) |
|---|---|---|---|---|
| WT | lin-15B | lin-15A | ||
| K12C11.2 | smo-1 | 3.2 (7; 15) | 5.07 (86; 7) | 3.70 (80;5) |
| W02A11.4 | uba-2 | 3.0 (0; 28) | 3.85 (62; 13) | 3.21 (33; 12) |
| F29B9.6 | ubc-9 | 3.0 (0; 20) | 3.24 (24; 17) | 5.13 (100; 8) |
| W01G7.3 | — | 3.0 (0; 18) | ND, let | 3.50 (60; 12) |
| W07B3.2 | gei-4 | 3.0 (0; 20) | ND, let | 3.25 (29; 14) |
| E01A2.4 | — | 3.0 (0; 15) | 3.03 (3; 31) | 3.94 (66; 9) |
| Y71G12B.9 | — | 3.0 (0; 20) | 3.00 (0; 30) | 3.27 (22; 32) |
| R05D3.11 | met-2 | 3.0 (0; 10) | 3.00 (0; 24) | 3.38 (38; 8) |
| R06C7.7 | rls-1/lin-61 | 3.0 (0; 8) | 3.00 (0; 20) | 3.34 (32; 25) |
| Indicated are the predicted gene, the locus, and VPC induction after RNAi in WT, lin-15B(n744), or lin-15A(n767) backgrounds (assessed by cell lineage at the L4 stage). Given is the average number of VPCs induced and in parentheses the percentage of hyperinduced animals and the number of animals analysed (% Muv; n). | ||||
| ND, let: not done because of lethality. |
Most of the new synMuv genes inhibit a ligand-independent activity of the Ras pathway
The classical synMuv phenotype depends on the LET-60 Ras signalling cascade, but is independent of the ligand LIN-3 EGF, indicating a function in inhibiting ligand-independent signalling through the RTK pathway (Lu and Horvitz, 1998). To investigate whether this is also true for the new synMuv genes, we tested whether let-60(n2021), a loss-of-function allele of let-60 Ras, suppresses the synMuv phenotype (Beitel et al, 1990). After RNAi of the synMuv genes in either let-60(n2021); lin-15A or let-60(n2021); lin-15B backgrounds, nearly no Muv animals are produced (Table III and data not shown). Therefore, the ectopic vulval development induced by loss of synMuv activity for the new synMuv genes depends on let-60 Ras activity. We also tested if these synMuv phenotypes are ligand independent using lin-3(n378), which was previously used for that assay (Ceol and Horvitz, 2004). Except for one gene (W01G7.3), the synMuv phenotype is expressed independently of the LIN-3 EGF ligand (Table III). Therefore, similar to previously studied synMuv genes, all but one of the new synMuv genes identified act to antogonize a ligand-independent activity of the Ras pathway.
Table 3.
Ras epistasis with new synMuv candidates
| | lin-15A | let-60; lin-15A | lin-3; lin-15A | | | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | ----------------- | ---------------- | -------------- | | No RNAi | 0% (23); 3.00 | 0% (25); 3.00 | 0% (27); 0.98 | | lin-15B(RNAi) | 94% (17); ND | 17% (30); ND | 100% (20); ND | | smo-1(RNAi) | 42% (19); 3.34 | 0% (40); 3.0 | 37% (19); 2.65 | | met-2(RNAi) | 59% (17); 3.94 | 0% (25); 3.00 | 19% (25); 2.83 | | gei-4 (RNAi) | 91% (11); 4.40 | 0% (15); 3.00 | 33% (21); 3.17 | | E01A2.4(RNAi) | 18% (28); 3.25 | 0% (15); 3.00 | 12% (26); 2.42 | | rls-1/lin-61(RNAi) | 93% (15); 5.30 | 3% (22); 3.27 | 52% (21); 3.29 | | W01G7.3(RNAi) | 60% (10); 3.70 | 0% (28); 3.00 | 0% (23); 1.17 | | Y71G12B.9(RNAi) | 20% (20); 3.23 | 0% (31); 3.00 | 21% (19); 2.84 | | The first column includes gene targeted by RNAi. The next three columns include the background in which the RNAi has been performed: lin-15A(n767), let-60(n2021);lin-15A(n767), and lin-3(n378);lin-15A(n767). Values represent the percentage of Muv animals, the number of animals analysed, and the average number of VPCs induced. No RNAi of let-60(n2021) or lin-3(n378) are, respectively, 0% (18); 2.75 and 0% (20); 0.48. The synMuv A activity of smo-1(RNAi) was analysed in lin-15B(n744), let-60(n2021);lin-15B(n744), and lin-3(n378);lin-15B(n744) producing, respectively, 92% (12); 4.75, 7% (29); 3.16, and 44% (20); 3.00. No RNAi for these three backgrounds are 0% (20); 3.00, 0% (18); 2.75, and 0% (20); 2.30, respectively. | | | | | ND: not determined. | | | |
The finding that W01G7.3 is ligand dependent suggests that it may act at the level of the AC, for example in downregulating lin-3 expression. W01G7.3 encodes a 13.3 kDa subunit of RNA polymerase II that might have a specific function in vulval development.
SUMO pathway genes interact with synMuv A, B, and C
The RNAi screens identified two components of the sumoylation pathway as synMuv genes: smo-1, encoding the small ubiquitin-related modifier protein SUMO, and uba-2, encoding a SUMO-activating enzyme. RNAi of either gene causes vulval hyperinduction in both class A and class B synMuv backgrounds (Table II). In the case of smo-1 RNAi, there is also a weak Muv phenotype (Table II; Broday et al, 2004; Leight et al, 2005). Post-translational modification of proteins by SUMO can affect nuclear targeting, the formation of subcellular structures, the control of protein stability, and the modulation of transcription factor activities (reviewed in Gill, 2004). Similar to ubiquitination, sumoylation requires E1, E2, and E3 enzyme activities. The E1 activity (the SUMO-activating enzyme) is composed of a heterodimeric complex consisting of Aos1 and Uba2. The SUMO-conjugating enzyme E2 is provided by Ubc9 (Wilson and Rangasamy, 2001). Finally, SUMO protein ligases (E3s) are numerous and are thought to be responsible for substrate specificity (Seeler and Dejean, 2003). To investigate further the requirement for sumoylation in vulval development, we used RNAi to test whether the C. elegans homologue of the E2 enzyme Ubc9 (F29B9.6 ubc-9) has synMuv activity. Similar to our findings with smo-1 SUMO and uba-2, we found that ubc-9 displays synMuv activity in both class A and class B backgrounds (Table II).
We then tested if smo-1 or ubc-9 synthetically interacted with the novel synMuv C class. We found that RNAi of smo-1 or ubc-9 in the class C background mys-1(n4075) did produce a synMuv phenotype (45% Muv, 3.50 VPC induced, _n_=20 and 53% Muv, 3.60 VPC induced, _n_=15, respectively, compared to 3% for mys-1(n4075) alone). The synMuv phenotype in class A, B, or C backgrounds places the SUMO pathway into a separate synMuv class.
The finding that RNAi of smo-1, uba-2, or ubc-9 produces a synMuv phenotype suggests that sumoylation might regulate one or more synMuv protein activities. The number of substrates for sumoylation is steadily growing. The human homologue of hda-1, HDAC1, is sumoylated and mutation of the lysines targeted by SUMO reduces HDAC1-mediated transcriptional repression (Kirsh et al, 2002). To test whether the HDAC HDA-1, a synMuv gene, is sumoylated in C. elegans, we performed a Western blot for this protein in extracts from wild-type and smo-1 knockout strains. We observed two bands in N2 worm extracts, a fast-migrating one at about 65 kDa, and another one at 75 kDa, the expected size increase for a SUMO modified HDA-1 (Figure 3). In a smo-1 mutant extract, the slow-migrating band is absent. Post-transcriptional modification of HDA-1 thus depends on SMO-1, suggesting that HDA-1 is sumoylated (Figure 3).
Figure 3.
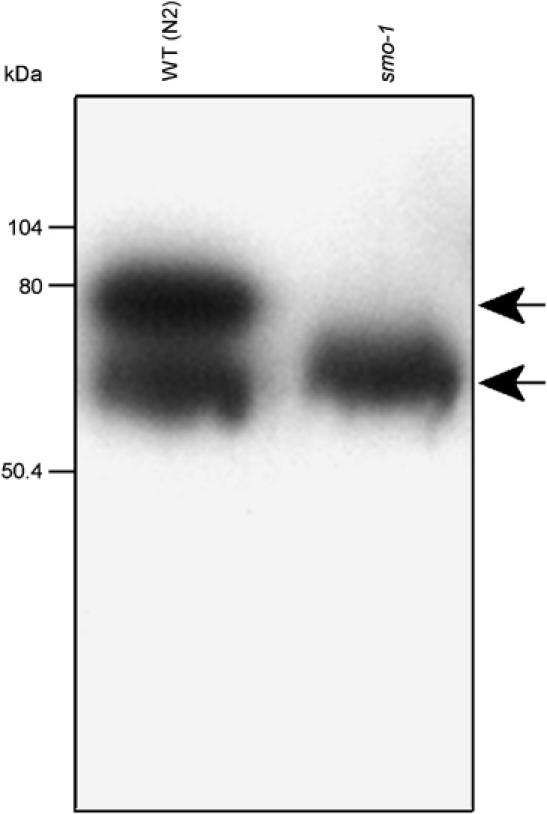
HDA-1 is sumoylated. Western blot of wild-type (lane 1) or smo-1 SUMO mutant (lane 2) total worm extract, probed with an anti-HDA-1 antibody. The N2 (WT) extract shows a fast-migrating band at about 65 kDa and a slow-migrating band at about 75 kDa. The slow-migrating band is absent from smo-1 SUMO mutant extract.
Sumoylation inhibits LIN-12/Notch signalling in the vulva
RNAi of smo-1 on its own induces a low percentage of Muv animals (Table II; Broday et al, 2004; Leight et al, 2005). We tested if this phenotype also depends on let-60 Ras using the reduction-of-function allele let-60(n2021). To our surprise, rather than a reduced Muv frequency, smo-1(RNAi); let-60(n2021) animals show a striking increase in the percentage of Muv animals compared to smo-1(RNAi) in N2 (56 versus 7%; Table IV). We observed a similar increase in Muv phenotype when two other sumoylation pathway components, uba-2 or ubc-9, are knocked down in let-60(n2021) animals (Table IV). Further, smo-1(RNAi) also induces a high percentage of Muv animals in mutants of the ligand lin-3, another background that should reduce Ras signalling (Table IV). In contrast, no Muv animals were detected after knockdown of six other tested synMuv genes in the let-60(n2021) background: _trr-1/_TRRAP, lin-35 Rb, met-2, rls-1/lin-61, lin-15A, or lin-15B (data not shown). The increase in Muv frequency does not appear to be due to an increase in RNAi sensitivity of let-60(n2021), as it is not seen in the RNAi supersensitive strain rrf-3(pk1426) (Simmer et al, 2002) (Table IV). In addition, the embryonic lethality observed by smo-1(RNAi) is not rescued by let-60(n2021), therefore an increase in survival cannot explain our observation (data not shown). Taken together, these results indicate that sumoylation affects vulval development differently from other synMuv genes.
Table 4.
Genetic interactions observed between smo-1 and let-60 Ras signalling
| Genotype | % Muv (n) | VPCs induced |
|---|---|---|
| smo-1(RNAi) | 7 (15) | 3.20 |
| smo-1 (RNAi);let-60(n2021) | 56 (50) | 3.66 |
| ubc-9(RNAi);let-60(n2021) | 50 (12) | 3.42 |
| uba-2(RNAi);let-60(n2021) | 50 (12) | 3.67 |
| smo-1(RNAi);lin-3(n378) | 50 (10) | 2.30 |
| smo-1 (RNAi);let-23(sy1) | 0 (29) | 0.94 |
| smo-1 (RNAi);rrf-3 (pk1426) | 10 (21) | 3.07 |
| let-60(n2021) | 0 (18) | 2.83 |
| lin-3(n378) | 0 (20) | 0.48 |
| let-23(sy1) | 0 (17) | 0.21 |
| rrf-3(pk1426) | 0 (18) | 3.00 |
| Indicated are the genotype, the percentage of Muv animals and the number of animals analysed, and the average number of VPCs induced. |
Since the let-60(n2021) weak loss-of-function mutant still allows significant Ras signalling, we used a stronger inhibition of the pathway to assess whether the Muv phenotype of smo-1(RNAi) animals requires Ras signalling. The let-23(sy1) loss-of-function mutant of the RTK receptor reduces VPC induction to 0.21; in this background, smo-1(RNAi) does not produce Muv animals (Table IV). This indicates that the Muv phenotype induced by smo-1(RNAi) requires a minimal level of Ras signalling that is provided by let-60(n2021), but not by let-23(sy1). If sumoylation inhibited signalling through the Ras pathway directly, we would expect RNAi of smo-1 in a wild-type background to cause a higher level of Muv animals than RNAi in a background where Ras signalling is reduced. However, the opposite is seen: a higher percentage of Muv animals is seen when Ras signalling is reduced (as in let-60(n2021 mutants) compared to wild type. These results suggest that sumoylation negatively modulates vulval development via another pathway that is dependent on Ras and which is more active when Ras signalling is reduced.
The LIN-12/Notch pathway is a good candidate for mediating this sumo-dependent inhibition of vulval development. LIN-12 is necessary and sufficient for the 2° vulval fate, and the LIN-12/Notch signalling pathway crosstalks with the Ras signalling cascade: a high level of Ras signalling leads to production of LIN-12/Notch ligands and downregulation of the LIN-12 receptor; subsequent activation of the LIN-12/Notch receptor in adjacent cells causes a reduction of Ras signalling (Shaye and Greenwald, 2002; Chen and Greenwald, 2004; Sundaram, 2004; Yoo et al, 2004). It has also been shown that hyperactivation of the LIN-12/Notch signalling pathway leads to a Muv phenotype (Greenwald et al, 1983).
If LIN-12/Notch signalling is responsible for the Muv phenotype of let-60(n2021); smo-1(RNAi) animals, then this phenotype should be suppressed by mutation of lin-12. Indeed, we found that neither smo-1(RNAi); let-60(n2021); lin-12(n941)/+ nor smo-1(RNAi); let-60(n2021); lin-12(n676n930) animals display a high percentage of Muv phenotype (Table V). This suggests that sumoylation negatively regulates one or more components of the LIN-12/Notch signalling pathway.
Table 5.
Notch signalling is required for the smo-1(RNAi);let-60(n2021) Muv phenotype
| Genotype | % Muv (n) | VPCs induced |
|---|---|---|
| smo-1(RNAi);let-60n2021 | 60 (15) | 3.87 |
| smo-1(RNAi);lin-12(n941/+);let-60(n2021) | 0 (22) | 3.00 |
| lin-12(n941/+);let-60(n2021) | 0 (29) | 3.00 |
| smo-1(RNAi);lin-12(n676n930);let-60(n2021) | 4 (24) | 3.02 |
| lin-12(n676n930);let-60(n2021) | 0 (25) | 2.90 |
| Indicated are the genotype, the percentage of Muv animal and the number of animals analysed, and the average number of VPCs induced. |
Two new synMuv B genes are linked to transcription repression
We identified five additional genes with synMuv B activity, two of which (R06C7.7 and R05D3.11) have links to transcription repression (Table II). R06C7.7 (rls-1) is similar to the Drosophila and human lethal (3) malignant brain tumor (L(3)MBT)) proteins containing MBT repeats (Table II) and was also previously identified in a screen for genes involved in genome stability (Pothof et al, 2003). R05D3.11 met-2 encodes a homologue of SETDB1, a histone H3 lysine 9 methyltransferase of the Suv39 family (Schultz et al, 2001). The remaining three new synMuv genes (E01A2.4, W07B3.2 gei-4, and Y71G12B.9) have undefined biochemical function, but a clear role in antagonizing Ras-induced vulval development (Tables II and III).
SynMuv proteins transcriptionally repress a lag-2 reporter gene
As a step towards understanding how the synMuv proteins function, we sought to identify a transcriptional target. Although functions of synMuv genes have been studied primarily in vulval development, many encode widely expressed nuclear proteins that have clear roles in other tissues (e.g., hda-1; Dufourcq et al, 2002). Furthermore, LIN-35 Rb inhibits vulval development by acting in the hypodermal syncytium hyp7 and not in vulval cells (Myers and Greenwald, 2005). Previously, it was shown that inhibition of hda-1 results in ectopic expression of a lag-2 reporter gene in gut and epidermal cells, indicating that repression of lag-2 outside the vulva requires HDA-1 (Dufourcq et al, 2002). Because HDA-1 cooperates with other synMuv proteins in the antagonism of vulval development, we decided to test whether other synMuv proteins might repress lag-2 in gut cells. We used RNAi to inhibit new and previously identified synMuv genes and scored for ectopic lag-2_∷_gfp expression.
As previously reported, we found that RNAi of hda-1 causes ectopic lag-2_∷_gfp expression in the gut (Table III). Remarkably, RNAi of many other synMuv genes results in similar ectopic reporter expression (Table VI and Figure 4). These include the NuRD components let-418 Mi-2, rba-2/lin-53 RbAp48 , the let-418 Mi-2-interacting mep-1, and a homologue of the NuRD component p66 (called dcp-66) (Table VI). This suggests that hda-1 may act within the NuRD complex for repression of lag-2. Similarly, RNAi of the sumoylation pathway genes smo-1, ubc-9, or uba-2 leads to ectopic lag-2 expression (Table VI and Figure 4). No misexpression was seen for RNAi of lin-15A or several class B synMuv genes (Table VI). In addition, lag-2_∷_gfp was not induced in a synthetic fashion by RNAi of any of three class B genes in a lin-15A mutant background (Table VI). However, surprisingly, single RNAi of some class B synMuv genes including lin-35 Rb, lin-15B, lin-9, JC8.6 (Owen et al, 2003) and the novel synMuv genes W07B3.2 gei-4 and W01G7.3 did induce ectopic lag-2 expression (Table VI). This indicates that at least some class B synMuv genes have nonredundant functions in transcriptional repression. Of interest, RNAi of efl-1 and R06C7.7 caused ectopic expression in tissues outside the gut and epidermis (data not shown).
Table 6.
Ectopic expression of lag-2_∷_gfp after RNAi of synMuv and related genes
| Gene inhibited by RNAi | Genetic background | % ectopic expression (n) |
|---|---|---|
| Sumoylation pathway | ||
| K12C11.2 smo-1 | WT | 75 (20) |
| W02A11.4 uba-2 | WT | 34.8 (69) |
| F29B9.6 ubc-9 | WT | 72.7 (11) |
| NuRD components | ||
| F26F12.7 let-418 | WT | 90 (80) |
| KO7A1.12 rba-2 | WT | 100 (11) |
| C53A5.3 hda-1 | WT | 94 (50) |
| C26C6.5 dcp-66 | WT | 93 (71) |
| M04B2.1 mep-1 | WT | 80 (20) |
| synMuv B genes involved in lag-2 repression | ||
| W07B3.2 gei-4 | WT | 100 (10) |
| C32F10.2 lin-35 | WT | 40 (20) |
| ZK662.4 lin-15B | WT | 95 (20) |
| ZK637.7a lin-9 | WT | 75 (20) |
| JC8.6 | WT | 76 (25) |
| W01G7.3 | WT | 55 (20) |
| TIP60 complex | ||
| C47D12.1 trr-1 | WT | 61.5 (78) |
| VC5.4 mys-1 | WT | 31 (62) |
| Y111B2A.23 ssl-1 | WT | 25 (24) |
| Y111B2A.11 epc-1 | WT | 100 (11) |
| C27H6.2 ruvb-1 | WT | 65 (17) |
| T22D1.10 ruvb-2 | WT | 75 (20) |
| No ectopic expression | ||
| ZK678.1 lin15A | WT | 0 (154) |
| T27C4.4a egr-1 | WT | 0 (40) |
| K07A1.11 rba-1 | WT | 0 (16) |
| T14G8.1 chd-3 | WT | 0 (40) |
| R05D3.11 met-2 | WT | 0 (40) |
| EO1A2.4 | WT | 0 (60) |
| Y71G12B.9 | WT | 0 (40) |
| Y47G6A.6 pcaf-1 | WT | 0 (60) |
| F32A5.1 ada-2 | WT | 0 (45) |
| F44B9.6 lin-36 | WT | 0 (70) |
| F44B9.6 lin-36 | lin-15A(n767) | 0 (30) |
| ZK418.4 lin-37 | WT | 0 (50) |
| ZK418.4 lin-37 | lin-15A(n767) | 0 (25) |
| ZK632.13 lin-52 | WT | 0 (50) |
| ZK632.13 lin-52 | lin-15A(n767) | 0 (25) |
| Empty vec | WT | 0 (128) |
| The predicted gene, locus, the genetic background (WT or lin-15A(n767)), and the percentage of animals with ectopic gut expression with the number of animals analysed (n) are indicated. The results have been divided into five groups: the sumoylation pathway, NuRD components, synMuv B genes involved in lag-2_∷_gfp expression, the TIP60 complex, and no ectopic gut expression. |
Figure 4.
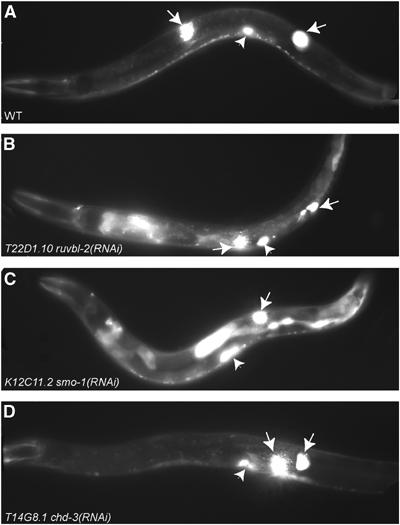
The lag-2_∷_gfp reporter assay. (A) Wild-type mid-L4 stage worm carrying the lag-2_∷_gfp reporter shows strong expression only in the distal tip cells (white arrows) and the vulva (white arrowheads). RNAi of T22D1.10 ruvb-2 helicase (B) or K12C11.2 smo-1 (C) induces strong ectopic expression of lag-2_∷_gfp in the intestine. Only one DTC is visible in the focal plane shown in (C). (D) RNAi of T14G8.1 chd-3 does not induce ectopic lag-2_∷_gfp expression.
Finally, we tested six components of the TRRAP/TIP60 acetyltransferase complex, including the ATP-dependent DNA helicases homologous to human RUVB1 and RUVB2. We found that RNAi of each caused derepression of the lag-2 reporter (Table VI and Figure 4). In contrast, RNAi of pcaf-1 or ada-2, two presumed components of the SAGA complex, which is also expected to contain TRRAP (reviewed in Carrozza et al, 2003), had no effect on reporter expression (Table VI). Taken together, these results show that in contrast to the redundant roles of synMuv genes in vulval development, many synMuv genes have a nonredundant role in the repression of lag-2 in the gut.
Discussion
We took a systematic RNAi screening approach to identify new synMuv genes involved in antagonizing Ras signalling in the vulva. The nine new genes we identified all have human homologues and many have links to transcriptional control. Three of the new genes are components of the sumoylation pathway, which we showed can positively regulate synMuv activity and inhibit Notch signalling in the vulva. In addition, we demonstrate that many of the new and previously studied synMuv proteins function in nonredundant transcriptional repression outside the vulva, suggesting that they may act together in multiple developmental contexts.
Classification of synMuv genes
synMuv genes are grouped into classes defined by their genetic interactions. Classically, class A gene mutations cause a synMuv phenotype in class B mutant backgrounds and vice versa, but not in mutant backgrounds of the same class. Similarly, class C mutations induce a synMuv phenotype in either class A or class B mutant backgrounds, but not in class C backgrounds. Recent data have linked molecular complexes to some of these classes. Class B gene products appear to belong to the recently described Myb–MuvB or dREAM complex (Korenjak et al, 2004; Lewis et al, 2004; see below), whereas class C genes encode components of a TIP60 histone acetyltransferase (HAT) complex (Ceol and Horvitz, 2004). NuRD complex components have a different behaviour, showing synthetic phenotypes in class A or class B backgrounds (Solari and Ahringer, 2000; von Zelewsky et al, 2000; Chen and Han, 2001; Unhavaithaya et al, 2002). Here, we showed that the sumoylation pathway defines a separate synMuv class, as its inhibition causes a synMuv phenotype in class A, B, or C backgrounds. It is still unclear what molecular activities are encoded by class A genes.
No new class A genes were found in our screen, raising the possibility that most or all of the class A genes have already been identified. However, we did discover genes where knockdown caused highly penetrant synthetic lethality in a class B background, which precluded scoring of the vulva. In addition, the screen identified genes where RNAi caused synthetic morphogenetic vulval defects in a class B background, rather than hyperinduction (G Poulin and J Ahringer, unpublished). Therefore, there may be class A genes remaining to be discovered that have wider developmental roles than those identified to date.
It is difficult to estimate how many additional synMuv genes exist, although it is likely that most of the as yet undiscovered synMuv genes are either essential genes or are members of gene families with partially redundant functions. In contrast to the genes identified by classical forward genetics, most of the new genes identified by these RNAi screens are essential (78 versus 50%), suggesting that the classical screens have probably saturated for synMuv genes with homozygous viable phenotypes.
The synMuv genes, chromatin complexes, and transcription regulation
Most of the studied synMuv proteins are found in the nucleus, and homologues of some are components of characterized chromatin regulating complexes (e.g., the NuRD complex). One of the new synMuv genes we identified is R06C7.7; it is similar to the Drosophila and human L(3)MBT proteins and encodes MBT repeats. Recently, L(3)MBT was identified as a component of the Myb–MuvB complex that acts in transcriptional repression, gene amplification, and G1/S-phase transition (Korenjak et al, 2004; Lewis et al, 2004); except for Myb, which has not been found in C. elegans, all the components of this complex are synMuv proteins (Lipsick, 2004; R06C7.7/L(3)MBT, LIN-9/Mip130, LIN-53/RbAp48, JC8.6/Mip120, LIN-37/Mip40, LIN-35 RB/RBF1,2, EFL-1/E2F2, DPL-1/DP, HDA-1/RPD3, and LIN-52/dLIN-52). Human L(3)MBT also has transcriptional repressor activity (Boccuni et al, 2003). In C. elegans, it is likely that R06C7.7 rls-1/lin-61 has a role in transcription repression as part of a similar Myb–MuvB complex.
Another new synMuv gene we identified that has links to transcription repression is met-2, a conserved histone methyltransferase related to mammalian SETDB1. In mammalian systems, it has been shown that after deacetylation of histone H3 tails by HDAC1, lysine 9 is methylated and then bound by HP1, a chromodomain protein, to maintain the repressed chromatin state (Boggs et al, 2002; Peters et al, 2002). The C. elegans chromodomain protein HPL-2, similar to HP1, was previously shown to have synMuv activity (Couteau et al, 2002). Because of their function in chromatin remodelling and their role in vulval development, we suggest that HDA-1, MET-2, and HPL-2 form a cassette for repression of vulval target genes: HDA-1 could deacetylate H3 lysine 9 at target genes as part of a NuRD or Myb–MuvB complex, MET-2 would methylate these residues, and HPL-2 would subsequently bind the methylated sites.
Role for synMuv proteins outside the vulva
We found that lag-2 expression in the gut is negatively regulated by many of the synMuv genes. Based on sequence homology, some of these synMuv proteins are predicted to be members of at least three different chromatin-remodelling complexes: the NuRD nucleosome-remodelling and HDAC complex, the Myb–MuvB HDAC complex, and the TIP60 HAT complex. Most of the predicted NuRD components negatively regulate lag-2 expression, including the conserved dcp-66 (Table VI). However, knockdown of only five out of nine of the Myb–MuvB complex genes tested (lin-35 Rb, hda-1, rba-2/lin-53, lin-9, and JC8.6) caused ectopic gut expression of lag-2 (Table VI). In contrast, all nine have synMuv activity. This difference suggests the possibility that there may be several different complexes containing Myb–MuvB components. Alternatively, for some genes, the RNAi assay for synMuv activity might be more sensitive than for ectopic lag-2_∷_gfp expression in the gut.
All of the tested homologues of TIP60 complex components are also needed for lag-2 repression. The TIP60 complex (similar to the NuA4 complex in yeast) contains the MYST family HAT TIP60, which has been linked with transcriptional activation through acetylation of histone H4 (Grant et al, 1998; Allard et al, 1999; Sterner and Berger, 2000; Vignali et al, 2000; Cai et al, 2003). It is intriguing that a HAT complex linked to transcription activation acts in the same process as HDAC complexes linked to transcription repression. One possibility is that the TIP60 HAT and NuRD and/or Myb–MuvB HDAC complexes have different target genes both in vulval development and in lag-2 repression, where TIP60 could be required for activation of repressors. A simpler alternative is that acetylation of histone H4 (a preferred substrate for TIP60) or of a non-histone factor has a direct role in transcriptional repression. Indeed, a recent study showed transcription repression activity for TIP60 (Xiao et al, 2003). Future work will be required to determine whether TIP60 and the HDAC complexes function on the same or different target genes.
Sumoylation and repression of transcription
Several recent reports have linked sumoylation with transcriptional repression. For example, histone H4 is sumoylated and SUMO-H4 can be co-immunoprecipitated from chromatin along with HDAC1 and HP1 (Shiio and Eisenman, 2003). SUMO modification is also required for the Polycomb group protein SOP-2 to repress Hox genes (Zhang et al, 2004). In C. elegans, it was recently shown that the ETS domain transcription factor LIN-1 is sumoylated (Leight et al, 2005). LIN-1 represses vulval fates and is negatively regulated by RTK signalling (Beitel et al, 1995; Tan et al, 1998). Sumoylation of LIN-1 promotes transcriptional repression and mediates an interaction with MEP-1, also a NuRD interacting protein (Leight et al, 2005). Sumoylation might antagonize Ras-induced vulval development by modulating the activity of multiple repressors of transcription, such as HDA-1, LIN-1, histone H4, and/or other synMuv genes. However, it is unlikely that the synMuv phenotype caused by inhibition of sumoylation is due to loss of lin-1 activity. The synMuv phenotype is dependent on Ras signalling, whereas the Muv phenotype of lin-1 mutants is not. It is intriguing that the sumoylation pathway, which could produce SUMO-H4, and the TIP60 HAT complex, which could acetylate histone H4, might function together in transcription repression. It will be of interest to investigate whether these two modifications together mark chromatin for a repressed state.
Sumoylation and signalling
Three components of the SUMO pathway, _smo-1/_SUMO, uba-2, and ubc-9, are synMuv genes. As with other synMuv genes, we found that the synMuv phenotype requires Ras signalling, but is independent of the ligand LIN-3. Surprisingly, although inhibition of sumoylation in wild-type or strong let-60/Ras mutant backgrounds has little effect on vulval development, under conditions of reduced Ras function (e.g., in the let-60(n2021) background), a high percentage Muv phenotype is induced (Table VI). We found that this latter phenotype requires LIN-12/Notch signalling.
It may seem counterintuitive that inhibiting sumoylation causes a Muv phenotype where Ras function is reduced, but not where it is wild type or strongly inhibited. This might be explained by considering the role of the Ras pathway in regulating LIN-12 signalling. Normally, Ras signalling in the 1o cell activates the LIN-12/Notch pathway (and the 2o cell fate) in neighbouring cells by inducing production of LIN-12 ligands (Chen and Greenwald, 2004; Yoo et al, 2004). In the absence of Ras signalling (e.g., in the let-23(sy1) background), no LIN-12 ligands should be produced, consequently LIN-12 signalling should not be activated even if sumoylation is inhibited, and therefore it would be unable to induce a Muv phenotype. Ras signalling also inhibits LIN-12/Notch signalling in the 1o cell by promoting LIN-12 endocytosis and degradation (Sternberg, 1988; Shaye and Greenwald, 2002; Chen and Greenwald, 2004; reviewed in Sundaram, 2004). Inhibiting sumoylation in a wild-type background might cause a higher than normal level of Ras signalling in all vulval cells due to partial abrogation of the synMuv activity, which might generally decrease LIN-12/Notch levels, preventing induction of the Muv phenotype. In the case of compromising sumoylation where there is a reduced level of Ras activity (e.g., in the let-60(n2021) background), there might be sufficient Ras signalling to produce LIN-12 ligands, but not enough to promote LIN-12 receptor downregulation, leading to a Muv phenotype dependent upon LIN-12 signalling (Table V). If, in this background, a synMuv A or a synMuv B mutation is also present, this may be sufficient to elevate Ras signalling to the point where it could trigger LIN-12 downregulation and thus prevent the occurrence of the Muv phenotype. The next goal is to investigate these models directly to learn at what level sumoylation acts to inhibit Notch signalling.
In summary, sumoylation appears to have at least two functions in vulval development: inhibition of ligand-independent RTK/Ras activity through positively regulating one or more synMuv protein activities (e.g., HDA-1) and inhibition of Notch signalling. This latter activity might be part of the crosstalk mechanism between Ras and Notch signalling pathways.
Concluding remarks
Through this systematic approach and previous studies, most of the synMuv genes in the genome are likely to have been identified. Based on the biochemical functions of new and previously known synMuv genes, it appears that sumoylation and multiple chromatin-remodelling activities (acetylation, deacetylation, and methylation) converge to antagonize Ras-induced vulval development through multiple complexes (Myb–MuvB, NuRD, and TIP60). Sumoylation positively modulates synMuv activity and inhibits Notch signalling, and these activities are likely to be conserved in higher organisms. The key goal for the future is to identify the target genes regulated and to uncover the mechanisms through which these proteins and complexes act together.
Materials and methods
RNAi screening
RNAi using the feeding library of 16 757 clones (Fraser et al, 2000; Kamath et al, 2003) was performed as described previously (Kamath et al, 2001), with minor modifications. Briefly, six-well plates containing NGM agar, 1 mm IPTG, and 25 μg/ml carbenicillin were inoculated with bacterial cultures grown for 6–10 h at 37°C in LB medium supplemented with 100 μg/ml ampicillin. Three to eight synchronized lin-15A (n767) or lin-15B (n744) L3–L4 stage worms were placed in individual wells containing one bacterial strain and the plates maintained at 21°C. Scoring for the Muv phenotype was performed using a dissecting microscope after 72 h or when F1 progeny reached adulthood. Bacterial strains inducing multiple protrusions in lin-15A or lin-15B mutants were rescreened in both backgrounds; only those strains that induced greater than 5% mutant animals were analysed further. All positive RNAi clones in this study were sequence verified.
RNAi constructs
Constructs used for RNAi are described by Fraser et al (2000) and Kamath et al (2003) except for lin-15A and lin-15B, which were made using the primers below to amplify fragments from genomic DNA: lin-15A primers: AGGTGGTGAGGTTGGACTTG and CACAGAACTTTAGTGGCGCA; lin-15B primers: CGTTCAGCAGTCGTGGTAGA and CAGGAACGGTGGATGAAAAT.
Vulval induction assays
VPC (P3p–P8p) cell fates were followed by counting the number of descendants produced by each VPC, as described in by Sternberg and Horvitz (1986) and Moghal and Sternberg (2003). Briefly, a score of 1.0 is assigned to a VPC when the vulval fate has been adopted by both direct descendants of any of the VPCs (P3p–8p) and a score of 0.5 if only one direct descendant has a vulval fate. Descendants from VPCs that have fused with the hypodermis (nonvulval fate) have a score of 0. In wild type, three VPCs (P5.p, P6.p, and P7.p) fully adopt the vulval fate and three VPCs fuse, giving a score of 3.0. A score of more than 3.0 indicates VPC hyperinduction. RNAi was carried out on six-well plates by feeding three L3/L4 worms for 42 h, then transferring these worms to a new well with the same bacteria for 24 h and then transferring again. Feeding was performed at 21°C and scoring was usually performed using F1 progeny from the second well.
lag-2∷gfp ectopic expression assay
The lag-2_∷_fp strain used (JK2868, qIs56) and assay conditions are as described by Dufourcq et al (2002). Mid-L4 worms were mounted on 3% agar pads in tetramisole (5 mM) and individually scored for ectopic expression in the gut using 40 × optics on a Zeiss Axioplan microscope.
Western blot
Whole-worm extracts were prepared using RIPA buffer followed by sonication. A total of 25 N2 worms and 50 smo-1 mutant worms were used for extract preparation. The latter were selected on the basis of a protruding vulva observed on escapers. The samples were run on SDS–PAGE 10% gel and an anti-HDA-1 antibody (Santa Cruz) was used for probing. Detection was performed according to the manufacturer's instructions (Amersham Biosciences, ECL plus). The smo-1 strain (VC186: smo-1(ok359)/szT1[lon-2(e678)]; +/szT1 X) was generated by the C. elegans knock-out consortium and obtained from the CGC.
Strains used in this study
N2, lin-15A(n767), lin-15B(n744), let-60(n2021), lin-3(n378) dpy-20(e1282), gap-1(ga133), let-23(sy1), rrf-3(pk1426), ncl-1(e1865)unc-36(e251)lin-12(n941)III/hT2[qIs48](I;III), lin-12(n676n930), lin-12(n676n930);let-60(n2021), ncl-1(e1865)unc-36(e251)lin-12(n941)III/hT2[qIs48](I;III);let060(n2021), lin-3(n378)dpy-20(e1282);lin-15A(n767), lin-3(n378) dpy-20(e1282);lin-15B(n744), let-60(n2021);lin-15A(n767), let-60(n2021);lin-15B(n744), and mys-1(n4075) V/nT1[qIs51] (IV;V) were used.
Acknowledgments
We are thankful to Verena Wolfram for her valuable help with the RNAi screen and the media facility for technical support. In addition, we are very grateful to Joe Hayward and Carly Dix for help in the early stages of the lag-2_∷_gfp work. We thank David Welchman for help with strain construction and Muv scoring, Ramkumar Nandakumar for bioinformatics help, and James Durbin for help with statistical analyses. We extend special thanks to Nathalie LeBot, Robert Andrews, and Welchy for comments on the manuscript. We are grateful to the C. elegans knock-out consortium and the CGC for providing strains. GP was supported by a fellowship from the Canadian Institutes for Health Research and from the Wellcome Trust. YD and AGF were supported by the Wellcome Trust. NAH is a Wellcome Trust Research Career Development Fellow (064988). JA is a Wellcome Trust Senior Research Fellow (054523).
References
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 18: 5108–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ard PG, Chatterjee C, Kunjibettu S, Adside LR, Gralinski LE, McMahon SB (2002) Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes. Mol Cell Biol 22: 5650–5661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroian RV, Koga M, Mendel JE, Ohshima Y, Sternberg PW (1990) The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature 348: 693–699 [DOI] [PubMed] [Google Scholar]
- Beitel GJ, Clark SG, Horvitz HR (1990) Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature 348: 503–509 [DOI] [PubMed] [Google Scholar]
- Beitel GJ, Tuck S, Greenwald I, Horvitz HR (1995) The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes Dev 9: 3149–3162 [DOI] [PubMed] [Google Scholar]
- Boccuni P, MacGrogan D, Scandura JM, Nimer SD (2003) The human L(3)MBT polycomb group protein is a transcriptional repressor and interacts physically and functionally with TEL (ETV6). J Biol Chem 278: 15412–15420 [DOI] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD (2002) Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet 30: 73–76 [DOI] [PubMed] [Google Scholar]
- Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Luscher B (2001) Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev 15: 2042–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391: 597–601 [DOI] [PubMed] [Google Scholar]
- Broday L, Kolotuev I, Didier C, Bhoumik A, Gupta BP, Sternberg PW, Podbilewicz B, Ronai Z (2004) The small ubiquitin-like modifier (SUMO) is required for gonadal and uterine-vulval morphogenesis in Caenorhabditis elegans. Genes Dev 18: 2380–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW (2003) Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem 278: 42733–42736 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Utley RT, Workman JL, Cote J (2003) The diverse functions of histone acetyltransferase complexes. Trends Genet 19: 321–329 [DOI] [PubMed] [Google Scholar]
- Ceol CJ, Horvitz HR (2004) A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev Cell 6: 563–576 [DOI] [PubMed] [Google Scholar]
- Chen N, Greenwald I (2004) The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev Cell 6: 183–192 [DOI] [PubMed] [Google Scholar]
- Chen Z, Han M (2001) Role of C. elegans lin-40 MTA in vulval fate specification and morphogenesis. Development 128: 4911–4921 [DOI] [PubMed] [Google Scholar]
- Clark SG, Lu X, Horvitz HR (1994) The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics 137: 987–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F, Guerry F, Muller F, Palladino F (2002) A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep 3: 235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourcq P, Victor M, Gay F, Calvo D, Hodgkin J, Shi Y (2002) Functional requirement for histone deacetylase 1 in Caenorhabditis elegans gonadogenesis. Mol Cell Biol 22: 3024–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Han M (2000) The synthetic multivulval genes of C. elegans: functional redundancy, Ras-antagonism, and cell fate determination. Genesis 26: 279–284 [DOI] [PubMed] [Google Scholar]
- Feng Q, Zhang Y (2003) The NuRD complex: linking histone modification to nucleosome remodeling. Curr Top Microbiol Immunol 274: 269–290 [DOI] [PubMed] [Google Scholar]
- Ferguson EL, Horvitz HR (1989) The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123: 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B (2003) MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep 4: 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330 [DOI] [PubMed] [Google Scholar]
- Frolov MV, Dyson NJ (2004) Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci 117: 2173–2181 [DOI] [PubMed] [Google Scholar]
- Gill G (2004) SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev 18: 2046–2059 [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Yates JR III, Workman JL (1998) The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell 2: 863–867 [DOI] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, Horvitz HR (1983) The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34: 435–444 [DOI] [PubMed] [Google Scholar]
- Harris TW, Chen N, Cunningham F, Tello-Ruiz M, Antoshechkin I, Bastiani C, Bieri T, Blasiar D, Bradnam K, Chan J, Chen CK, Chen WJ, Davis P, Kenny E, Kishore R, Lawson D, Lee R, Muller HM, Nakamura C, Ozersky P, Petcherski A, Rogers A, Sabo A, Schwarz EM, Van Auken K, Wang Q, Durbin R, Spieth J, Sternberg PW, Stein LD (2004) WormBase: a multi-species resource for nematode biology and genomics. Nucleic Acids Res 32 (Database issue): D411–D417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, Herman RK (1995) The ncl-1 gene and genetic mosaics of Caenorhabditis elegans. Genetics 141: 989–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman RK, Hedgecock EM (1990) Limitation of the size of the vulval primordium of Caenorhabditis elegans by lin-15 expression in surrounding hypodermis. Nature 348: 169–171 [DOI] [PubMed] [Google Scholar]
- Hill RJ, Sternberg PW (1992) The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature 358: 470–476 [DOI] [PubMed] [Google Scholar]
- Hsieh J, Liu J, Kostas SA, Chang C, Sternberg PW, Fire A (1999) The RING finger/B-box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans. Genes Dev 13: 2958–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LS, Tzou P, Sternberg PW (1994) The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol Biol Cell 5: 395–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J (2001) Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2, RESEARCH0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J (1981) Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev Biol 87: 286–300 [DOI] [PubMed] [Google Scholar]
- Kirsh O, Seeler JS, Pichler A, Gast A, Muller S, Miska E, Mathieu M, Harel-Bellan A, Kouzarides T, Melchior F, Dejean A (2002) The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J 21: 2682–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenjak M, Taylor-Harding B, Binne UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A (2004) Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119: 181–193 [DOI] [PubMed] [Google Scholar]
- Kumar R, Wang RA, Bagheri-Yarmand R (2003) Emerging roles of MTA family members in human cancers. Semin Oncol 30: 30–37 [DOI] [PubMed] [Google Scholar]
- Lang SE, McMahon SB, Cole MD, Hearing P (2001) E2F transcriptional activation requires TRRAP and GCN5 cofactors. J Biol Chem 276: 32627–32634 [DOI] [PubMed] [Google Scholar]
- Leight ER, Glossip D, Kornfeld K (2005) Sumoylation of LIN-1 promotes transcriptional repression and inhibition of vulval cell fates. Development 132: 1047–1056 [DOI] [PubMed] [Google Scholar]
- Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR (2004) Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev 18: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsick JS (2004) synMuv verite—Myb comes into focus. Genes Dev 18: 2837–2844 [DOI] [PubMed] [Google Scholar]
- Lu X, Horvitz HR (1998) lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 95: 981–991 [DOI] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94: 363–374 [DOI] [PubMed] [Google Scholar]
- Moghal N, Sternberg PW (2003) A component of the transcriptional mediator complex inhibits RAS-dependent vulval fate specification in C. elegans. Development 130: 57–69 [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Sardet C, Herrera RE (2002) Retinoblastoma protein transcriptional repression through histone deacetylation of a single nucleosome. Mol Cell Biol 22: 856–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers TR, Greenwald I (2005) lin-35 Rb acts in the major hypodermis to oppose Ras-mediated vulval induction in C. elegans. Dev Cell 8: 117–123 [DOI] [PubMed] [Google Scholar]
- Owen AB, Stuart J, Mach K, Villeneuve AM, Kim S (2003) A gene recommender algorithm to identify coexpressed genes in C. elegans. Genome Res 13: 1828–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Mermoud JE, O'Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T (2002) Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet 30: 77–80 [DOI] [PubMed] [Google Scholar]
- Pothof J, van Haaften G, Thijssen K, Kamath RS, Fraser AG, Ahringer J, Plasterk RH, Tijsterman M. (2003) Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev 17: 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ III (2001) Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev 15: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Dejean A (2003) Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol 4: 690–699 [DOI] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I (2002) Endocytosis-mediated downregulation of LIN-12/Notch upon Ras activation in Caenorhabditis elegans. Nature 420: 686–690 [DOI] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN (2003) Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA 100: 13225–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH (2002) Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol 12: 1317–1319 [DOI] [PubMed] [Google Scholar]
- Solari F, Ahringer J (2000) NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr Biol 10: 223–226 [DOI] [PubMed] [Google Scholar]
- Sternberg PW (1988) Lateral inhibition during vulval induction in Caenorhabditis elegans. Nature 335: 551–554 [DOI] [PubMed] [Google Scholar]
- Sternberg PW, Horvitz HR (1986) Pattern formation during vulval development in C. elegans. Cell 44: 761–772 [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL (2000) Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 64: 435–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56: 110–156 [DOI] [PubMed] [Google Scholar]
- Sundaram MV (2004) Vulval development: the battle between Ras and Notch. Curr Biol 14: R311–R313 [DOI] [PubMed] [Google Scholar]
- Tan PB, Lackner MR, Kim SK (1998) MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell 93: 569–580 [DOI] [PubMed] [Google Scholar]
- Thomas JH, Ceol CJ, Schwartz HT, Horvitz HR (2003) New genes that interact with lin-35 Rb to negatively regulate the let-60 ras pathway in Caenorhabditis elegans. Genetics 164: 135–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Horvitz HR (1999) The C. elegans gene lin-36 acts cell autonomously in the lin-35 Rb pathway. Development 126: 3449–3459 [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A (1998) Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC (2002) MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline–soma distinctions in C. elegans. Cell 111: 991–1002 [DOI] [PubMed] [Google Scholar]
- Vignali M, Steger DJ, Neely KE, Workman JL (2000) Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J 19: 2629–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zelewsky T, Palladino F, Brunschwig K, Tobler H, Hajnal A, Muller F (2000) The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development 127: 5277–5284 [DOI] [PubMed] [Google Scholar]
- Wilson VG, Rangasamy D (2001) Intracellular targeting of proteins by sumoylation. Exp Cell Res 271: 57–65 [DOI] [PubMed] [Google Scholar]
- Xiao H, Chung J, Kao HY, Yang YC (2003) Tip60 is a co-repressor for STAT3. J Biol Chem 278: 11197–11204 [DOI] [PubMed] [Google Scholar]
- Yoo AS, Bais C, Greenwald I (2004) Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science 303: 663–666 [DOI] [PubMed] [Google Scholar]
- Zhang H, Smolen GA, Palmer R, Christoforou A, van den Heuvel S, Haber DA (2004) SUMO modification is required for in vivo Hox gene regulation by the Caenorhabditis elegans Polycomb group protein SOP-2. Nat Genet 36: 507–511 [DOI] [PubMed] [Google Scholar]