A New Familial Amyotrophic Lateral Sclerosis Locus on Chromosome 16q12.1-16q12.2 (original) (raw)
Abstract
Familial amyotrophic lateral sclerosis (FALS) affects 5%–10% of cases of amyotrophic lateral sclerosis (ALS) and is inherited as an autosomal dominant condition with incomplete penetrance. One-fifth of these cases of FALS are associated with mutations in copper/zinc-dependent superoxide dismutase (SOD1), but the gene defect in the remaining 80% of familial cases is, as yet, unknown. We have carried out a preliminary genome screen, using a U.K. resource of families lacking SOD1 mutations, to identify other potential disease loci and have identified a putative locus on chromosome 16q12.1-q12.2. The region associated with disease was further refined in the major family that contributed to this result and was localized to D16S409–D16S3032, a 14.74-cM genetic interval that corresponds to a physical distance of 6.6 Mb, which coincides with a region independently identified by two further research groups in the United States and the United Kingdom.
Amyotrophic lateral sclerosis (ALS) is a late-onset, severe, debilitating condition that affects spinal cord, brain stem, and cortical motor neurons; causes progressive muscle weakness, atrophy, and paralysis; and leads finally to respiratory failure. There have been considerable advances in the understanding of disease progression from studies on the dominantly inherited familial form of ALS (familial amyotrophic lateral sclerosis [FALS]), which, although rare (accounting for 5%–10% of cases), is clinically indistinguishable from the sporadic form of the disease. Approximately 20% of FALS cases have been shown to be associated with mutations in copper/zinc-dependent superoxide dismutase (SOD1) (Rosen et al. 1993), designated “ALS1” (MIM 147450), and this has provided much insight into potential mechanisms of pathogenesis (Beckman et al. 2001; Cleveland and Rothstein 2001).
A second adult-onset locus has recently been reported in a single pedigree by Hand et al. (2002), which is designated “ALS3” (MIM 606640) and localizes to a 7.5-cM region on chromosome 18q21. A slowly progressing autosomal dominant form of ALS with juvenile onset, designated “ALS4” (MIM 602433), has been mapped to chromosome 9q34 by Chance et al. (1998) and further localized to a 500-kb interval by Blair et al. (2000). A dominant form of ALS with frontotemporal dementia (ALSFTD [MIM 105550]) has also been described, which localizes to chromosome 9q21-q22 (Hosler et al. 2000). In addition to these dominantly inherited forms of ALS, autosomal recessive forms are known. A rare autosomal recessive form of juvenile-onset ALS, “ALS2” (MIM 606352), maps to chromosome 2q33 (Hentati et al. 1994), and the gene mutated in these families has been identified as a GTPase, termed “_alsin_” (Hadano et al. 2001; Yang et al. 2001). The most prevalent recessive form of ALS with juvenile onset, FALS5 (MIM 602099) has been mapped to chromosome 15q15-q22 (Hentati et al. 1998).
However, the majority (∼80%) of adult-onset dominantly inherited FALS cases cannot be attributed to any of these loci and remain to be elucidated. With this elucidation as our major goal, we have carried out a genome screen in our 13 most extensive U.K. families that lack SOD1 mutations (de Belleroche et al. 1996; Orrell et al. 1997). Further heterogeneity is indicated from this analysis, as data have been obtained that are suggestive of several putative loci present in separate family subsets. One such subset, where heterogeneity was confirmed by use of HOMOG version 3.33, contains three families and shows a significant association between chromosome-16 markers (D16S409 and D16S3032) and disease (multipoint LOD score of 3.16 obtained from the summation of LOD scores [SIMWALK2 version 2.82] for three families at D16S3035). In this report, we describe the major family (MB2) that contributes to this LOD score, localizing the region to a 14.74-cM interval and a physical distance of 6.6 Mb (National Center for Biotechnology Information [NCBI]). Independently, two other laboratories have described three further families with FALS that map to the same region and show an overlap of 9.42 cM.
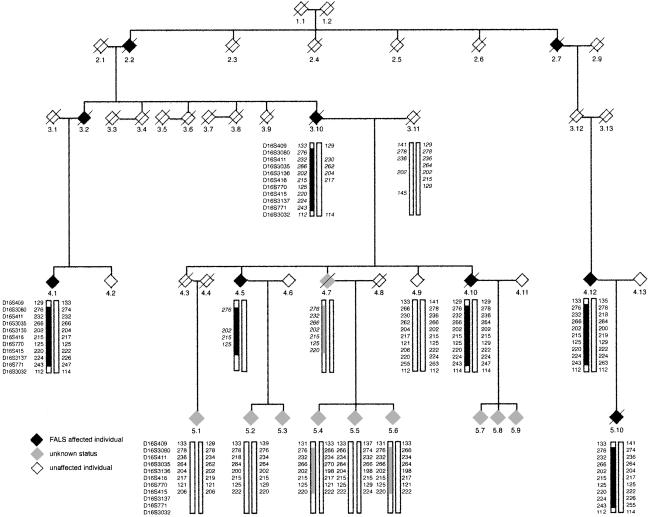
A diagnosis of ALS was confirmed clinically in the index case (fig. 1; individual 5.10), with evidence of both upper and lower motor–neuron involvement, by use of the El Escorial criteria for diagnosis of ALS (Brooks 1994). Bulbar signs were also present. The patient presented with lower limb weakness. The age at onset was 37 years, and the duration of illness was 36 mo. A less-rapid disease progression was seen in other affected family members, with disease duration lasting up to 20 years. The average age at onset for four affected individuals was 50.25 years (range 37–66 years). No SOD1 mutations were detected in the five exons of SOD1. This study has been approved by the Riverside Research Ethics Committee (Hammersmith Hospitals National Health Service Trust), with appropriate informed consent from subjects.
Figure 1.
Haplotype analysis of the chromosome 16q11.2-16q13.1 region in the family with FALS. Allele data for 11 markers in this region are shown; inferred genotypes of the reconstructed individuals 3.10, 3.11, 4.5, and 4.7 are indicated by italics. The names of the makers are listed on the far left side of each generation. Haplotypes associated with the disease are indicated by thick solid “boxing.” Crossover events are indicated by a change in the “boxing.” Both males and females are indicated by a diamond symbol, affected individuals are represented by a solid black diamond, and individuals of unconfirmed status with a light gray diamond. The index case is 5.10.
Simulation calculations, by use of SIMLINK version 4.12 (Ploughman and Boehnke 1989), were used to detect the statistical power of this family to detect genetic linkage and resulted in a theoretical two-point LOD score of 3.791 at θ=0, which demonstrated the statistical power of this family and, hence, the feasibility of using this family for further study.
DNA was extracted from whole blood or the buffy coat layer, by use of a Qiagen Blood and Tissue DNA extraction kit according to the manufacturer’s instructions, except for the elution of the DNA from the spin columns, which was performed in 200 μl distilled H2O instead of the Qiagen elution buffer. Following an initial genome screen, by use of the Human Genome Mapping Project (HGMP) linkage mapping set markers, a subset of families indicated putative linkage to chromosome 16. Further mapping of chromosome 16 was performed by use of a total of 43 microsatellite markers, fluorescently labeled with either FAM, HEX, TET, or NED fluorochromes (forward primer only), which spanned a region of 100.76 cM from D16S423 to D16S422 on chromosome 16, by use of published sequence data. The order of the microsatellite markers was determined according to the Marshfield genetic map of chromosome 16. PCRs were performed by use of Platinum Taq DNA polymerase (Invitrogen), according to the manufacturer’s instructions. PCR samples were pooled together on the basis of expected size and fluorescent label used. Electrophoresis of the pooled samples was then performed on the ABI 310 Genetic Analyzer. Information concerning peak size of products, allele assignment, and allele frequency was determined by use of the ABI software package Genotyper version 2.5.
Two-point linkage analysis of genotyping data was performed by use of the MLINK program (version 5.1) of the FASTLINK version 4.1p software package. Individuals confirmed clinically with FALS were entered as “affected status,” and unaffected individuals at risk of disease, aged ⩾80 years, were entered as “unaffected status.” Penetrance curves were not used, since no individuals of unknown status (other than obligate carriers) were entered into the analysis. The analysis was performed by use of the model that FALS was inherited in an autosomal dominant manner. The frequency of the mutant allele was taken as 0.0001. Simple counting estimates were used to determine marker allele frequencies from at least 52 individuals. Marker D16S419 has been omitted from the analysis, since its chromosomal position is unsure. Multipoint analysis on the same data was performed initially by use of Genehunter 2.1 r3 and, subsequently, by use of the SIMWALK2 version 2.82 software package. Haplotype reconstruction was initially performed manually and confirmed by use of the haplotype function of SIMWALK2 version 2.82.
Following a genome screen on 13 U.K. families with FALS, a putative locus was indicated on chromosome 16 (16q12.1-16q12.2) in a subset of families. The major family linked to this region was characterized by a dominantly inherited form of ALS that was evident in three generations. DNA was available from 10 individuals from two generations; 4 individuals were affected, and partial reconstructions of the genotypes of 4 further individuals have been performed (fig. 1). The affected individuals in this family lacked mutations in all exons of SOD1, and significant exclusion of linkage to ALS2, ALS3, and ALS4 was obtained by use of close markers for these loci. For ALS2, three markers within 7 cM of alsin were used (LOD scores and Marshfield genetic distances, where available, are shown in parentheses): D2S115 (_LOD_=-2.981; 195.65 cM), D2S309 (_LOD_=-4.367; 198.65 cM), and D2S155 (_LOD_=-3.746; 202.92 cM). For the ALS3 locus, which is flanked by D18S846 (77.36 cM) and D18S1109 (84.8 cM), two markers were used: D18S57 (_LOD_=-0.71; 62.85 cM) and D18S64 (_LOD_=-4.34; 84.8 cM). For ALS4 (9q34), the following markers, reported by Blair et al. (2000), were used: D9S159 (_LOD_=-3.782; 142.07 cM), D9S118 (_LOD_=-5.890), and D9S1847 (_LOD_=-1.112; 144.67 cM).
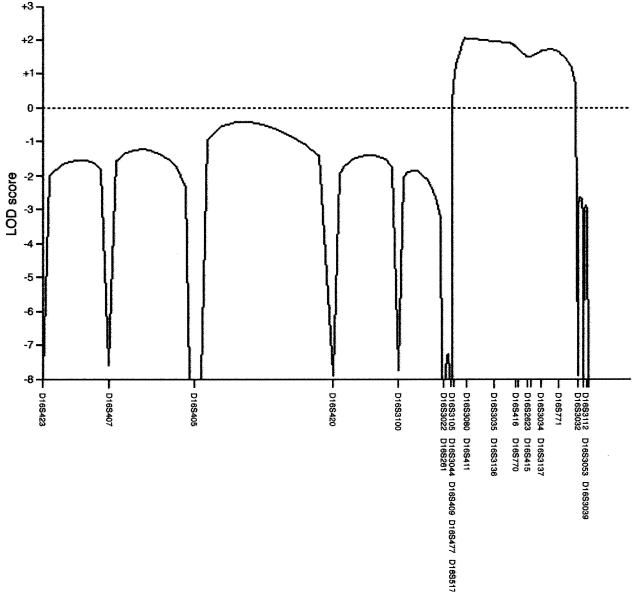
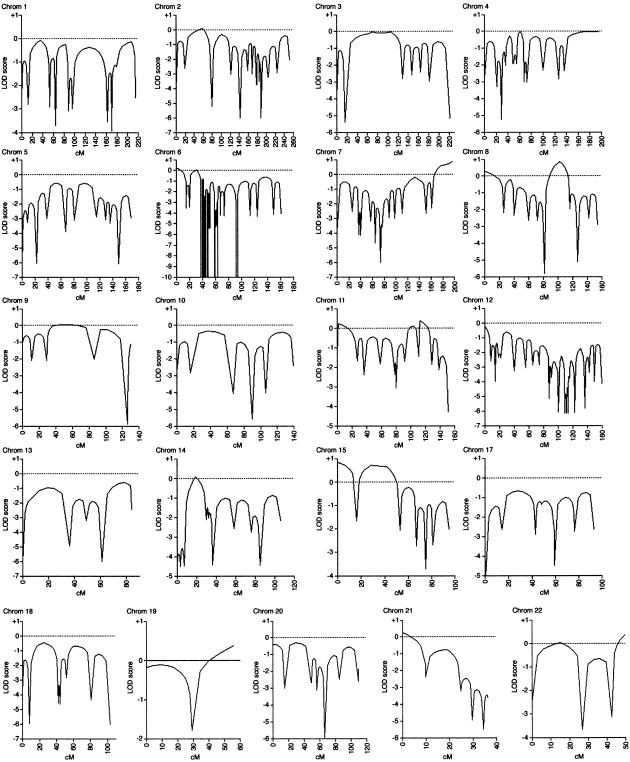
We have screened this family with 43 markers on chromosome 16 and refined the region of linkage (fig. 2). Recombinations in this family defined a region (16q12.1-16q12.2), flanked by the markers D16S409 and D16S3032, in which a common haplotype is shared by affected individuals (fig. 1). There were no positive LOD scores >0.87 obtained for this family from the genome scan of chromosomes 1–15 and 17–22 by use of 344 markers (fig. 3). The marker alleles shared in the disease haplotype (fig. 1) are D16S3080, D16S411, D16S3035, D16S3136, D16S416, D16S770, D16S415, D16S3137, and D16S771 (by use of Marshfield map distances and markers ordered at the same distance, on the basis of physical map data). The genetic distance flanked by markers D16S409 (58.46 cM) and D16S3032 (73.20 cM) is 14.74 cM, which approximates to a physical distance of 6.6 Mb (NCBI). The maximum two-point LOD score obtained by use of MLINK was 1.509 for D16S411 at θ=0 (table 1), and the multipoint analysis (fig. 2) yielded maximum multipoint LOD scores, for markers D16S3080 and D16S411, of 2.06 (P<.04).
Figure 2.
Multipoint linkage analysis of chromosome 16, generated by use of the SIMWALK2 version 2.82 software package
Figure 3.
A genome screen of family MB2 for chromosomes 1–15 and 17–22, by use of a total of 301 microsatellite markers. The multipoint linkage analysis was performed by use of Genehunter version 2.1r3 software. The numbers of microsatellite markers for each chromosome were 18, 18, 9, 18, 11, 52, 18, 17, 8, 7, 14, 33, 7, 11, 10, 12, 11, 4, 9, 9, and 5, respectively.
Table 1.
Two-Point LOD Scores for Family with FALS on Chromosome 16q12.1-q12.2[Note]
| θ= | |||||
|---|---|---|---|---|---|
| Locusa | .0 | .1 | .2 | .3 | .4 |
| D16S420 | −∞ | −.375 | −.150 | −.054 | −.012 |
| D16S3100 | .172 | .096 | .048 | .021 | .007 |
| D16S3022 | −∞ | −.713 | −.290 | −.114 | −.033 |
| D16S261 | .488 | .288 | .144 | .059 | .018 |
| D16S3105 | .626 | .425 | .252 | .121 | .039 |
| D16S3044 | .083 | .069 | .043 | .010 | −.008 |
| D16S409 | −∞ | −.096 | −.019 | −.024 | −.022 |
| D16S477 | −.067 | −.042 | −.024 | −.010 | −.002 |
| D16S517 | −.122 | −.091 | −.049 | −.018 | −.002 |
| D16S3080 | 1.192 | .902 | .619 | .362 | .148 |
| D16S411 | 1.509 | 1.079 | .664 | .315 | .091 |
| D16S3035 | .965 | .710 | .456 | .226 | .067 |
| D16S3136 | .512 | .335 | .199 | .102 | .036 |
| D16S416 | .537 | .383 | .252 | .141 | .052 |
| D16S770 | .618 | .398 | .234 | .114 | .033 |
| D16S2623 | .156 | .212 | .160 | .086 | .026 |
| D16S415 | 1.045 | .787 | .533 | .300 | .112 |
| D16S3034 | .162 | .208 | .154 | .081 | .025 |
| D16S3137 | .562 | .370 | .214 | .102 | .034 |
| D16S771 | .487 | .344 | .203 | .080 | .006 |
| D16S3032 | −∞ | −.100 | .007 | .008 | −.006 |
| D16S3112 | −.295 | −.066 | −.016 | −.007 | −.007 |
The region defined by the flanking markers contains 35 known genes, on the basis of SWISS-PROT, TrEMBL, mRNA, and ResSeq data and 138 gene predictions (UCSC Genome Browser version 17). Several of the known genes present in the region are expressed in the CNS and, in some cases, possess properties relevant to cell viability and, hence, potentially to ALS pathology: for example, trinucleotide repeat containing 9 (TNRC9) (in view of the important role of trinucleotide repeat expansions in many late-onset neurodegenerative conditions), caspase recruitment domain family (CARD15), and bromodomain-containing 7 (BRD7), which interacts with acetylated-histone peptides and affects transcriptional activation. In addition, several genes that encode transcription factors and are involved in development are also present in this region: for example, OLF-1/EBF associated zinc–finger gene (OAZ), topoisimerase-related function protein (TRF4–2), naked cuticle homolog 1 (Drosophila) (NKD1), a putative zinc finger transcription factor Sal-like 1 (Drosophila) (SALL1), retinoblastoma-like 2 (RBL2), and Iroquois homeobox proteins 5, 3, and 6 (IRX3, 5, and 6). Two further genes with a role in CNS signaling are also present: cerebellin 1 precursor (CBLN1) and adenylyl cyclase 7 (ADCY7).
Three further chromosome 16–linked families have been identified independently: an American family under investigation by Dr. R. Brown, a further U.K. family that is in collaboration with Dr. Chris Shaw, and a Dutch family that is being studied by Drs. Chris Shaw and Frank Baas. For all four families, a common region of overlap has been obtained between D16S3396 and D16S3032, which covers a region of 9.42 cM (R. H. Brown and C. Shaw, personal communications). We have initiated a collaboration to facilitate this research. Our current plan is to refine this region optimally with further markers and then to embark on sequencing the genes in the region. The small size of this region and the availability, through collaboration, of other linked families supports the feasibility of identifying the putative disease gene present at this locus. Subsequently, mutation analysis will be performed on other cohorts of case subjects with FALS, including the 190 U.K. families available at Charing Cross Hospital.
Acknowledgments
We are grateful to the American ALS Association, the Smith’s Charity, and the Motor Neurone Disease Association, for funding this research; and to the patients and their relatives involved in this research, for their willing cooperation.
Electronic-Database Information
URLs for data presented herein are as follows:
- Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ALS1, ALS2, ALS3, ALS4, ALS5, ALSFTD, and FALS5)
- National Center for Biotechnology information (NCBI), http://www.ncbi.nlm.nih.gov/
- UCSC Genome Browser version 17, http://www.genome.ucsc.edu
References
- Beckman JS, Estevez AG, Crow JP, Barbeito L (2001) Superoxide dismustase and the death of motor neurones in ALS. Trends Neurosci 24:S15–S20 [DOI] [PubMed] [Google Scholar]
- Blair IP, Bennett CL, Abel A, Rabin BA, Griffin JW, Fischbeck KH, Cornblath DR, Chance PF (2000) A gene for autosomal dominant juvenile amyotrophic lateral sclerosis (ALS4) localizes to a 500-kb interval on chromosome 9q34. Neurogenetics 3:1–6 [DOI] [PubMed] [Google Scholar]
- Brooks BR (1994) El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on motor neuron disease/amyotrophic lateral sclerosis of the World federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits on amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci 124:S96–S107 [DOI] [PubMed] [Google Scholar]
- Chance PF, Rabin BA, Ryan SG, Ding Y, Scavina M, Crain B, Griffin JW, Cornblath DR (1998) Linkage of the gene for an autosomal dominant form of juvenile amyotrophic lateral sclerosis to chromosome 9q34. Am J Hum Genet 62:633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Rothstein JD (2001) From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci 2:806–819 [DOI] [PubMed] [Google Scholar]
- de Belleroche J, Orrell RW, Virgo L (1996) Amyotrophic lateral sclerosis: recent advances in understanding disease mechanisms. J Neuropathol Exp Neurol 55:747–757 [PubMed] [Google Scholar]
- Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Deveon RS, Mivamoto N, Showguchi-Mivata J, Okada Y, Singaraja R, Figlewicz DA, Kwiatkowski T, Iiosler BA, Sagie T, Skaug J, Nasir J, Brown RII Jr, Scherer SW, Rouleau GA, Hayden MR, Ikeda JE (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29:166–173 [DOI] [PubMed] [Google Scholar]
- Hand CK, Khoris J, Salachas F, Gros-Louis F, Lopes AAS, Mayeux-Portas V, Brown RH Jr, Meininger V, Camu W, Rouleau GA (2002) A novel locus for familial amyotrophic lateral sclerosis, on chromosome 18q. Am J Hum Genet 70:251–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentati A. Bejaoui K, Pericak-Vance MA, Hentati F, Speer MC, Hung W-Y, Figlewicz DA, Haines J, Rimmler J, Ben Hamida C, Ben Hamida M, Brown RH Jr, Siddique T (1994) Linkage of recessive familial amyotrophic lateral sclerosis to chromosome 2q33-q35. Nat Genet 7:425–428 [DOI] [PubMed] [Google Scholar]
- Hentati A, Ouahchi K, Pericak-Vance MA, Nijhawan D, Ahmad A, Yang Y, Rimmler J, Hung W, Schlotter B, Ahmed A, Ben Hamida M, Hentati F, Siddique T (1998) Linkage of a commoner form of recessive amyotrophic lateral sclerosis to chromosome 15q15-q22 markers. Neurogenetics 2:55–60 [DOI] [PubMed] [Google Scholar]
- Hosler BA, Siddique, T, Sapp PC, Sailor W, Huang MC, Hossain A, Daube JR, Nance M, Fan C, Kaplan J, Hung W-Y, McKenna-Yasek D, Haines JL, Pericak-Vance MA, Horvitz HR, Brown RH Jr (2000) Linkage of familial amyotrophic lateral sclerosis with frontotemporal dementia to chromosome 9q21-q22. JAMA 284:1664–1669 [DOI] [PubMed] [Google Scholar]
- Orrell RW, Habgood J, Gardiner I, King AW, Bowe FA, Hallewell RA, Marklund SL, Greenwood J, Lane RJM, de Belleroche J (1997) Clinical and functional investigation of 10 missense mutations and a novel frameshift insertion mutation of the gene for copper-zinc superoxide dismutase in UK families with amyotrophic lateral sclerosis. Neurology 48:746–751 [DOI] [PubMed] [Google Scholar]
- Ploughman LM, Boehnke M (1989) Estimating the power of a proposed linkage study for a complex genetic trait. Am J Hum Genet 44:543–551 [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, et al (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62 [DOI] [PubMed] [Google Scholar]
- Yang Y, Hentati A, Deng H-X, Dabbagh O, Sasaki T, Hirano M, Hung W-Y, Ouahchi K, Yan J, Azim AC, Cole N, Gascon G, Yagmour A, Ben-Hamida M, Pericak-Vance M, Hentati F, Siddique T (2001) The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 29:160–165 [DOI] [PubMed] [Google Scholar]