Comprehensive analysis of CpG islands in human chromosomes 21 and 22 (original) (raw)
Abstract
CpG islands are useful markers for genes in organisms containing 5-methylcytosine in their genomes. In addition, CpG islands located in the promoter regions of genes can play important roles in gene silencing during processes such as X-chromosome inactivation, imprinting, and silencing of intragenomic parasites. The generally accepted definition of what constitutes a CpG island was proposed in 1987 by Gardiner-Garden and Frommer [Gardiner-Garden, M. & Frommer, M. (1987) J. Mol. Biol. 196, 261–282] as being a 200-bp stretch of DNA with a C+G content of 50% and an observed CpG/expected CpG in excess of 0.6. Any definition of a CpG island is somewhat arbitrary, and this one, which was derived before the sequencing of mammalian genomes, will include many sequences that are not necessarily associated with controlling regions of genes but rather are associated with intragenomic parasites. We have therefore used the complete genomic sequences of human chromosomes 21 and 22 to examine the properties of CpG islands in different sequence classes by using a search algorithm that we have developed. Regions of DNA of greater than 500 bp with a G+C equal to or greater than 55% and observed CpG/expected CpG of 0.65 were more likely to be associated with the 5′ regions of genes and this definition excluded most _Alu_-repetitive elements. We also used genome sequences to show strong CpG suppression in the human genome and slight suppression in Drosophila melanogaster and Saccharomyces cerevisiae. This finding is compatible with the recent detection of 5-methylcytosine in Drosophila, and might suggest that S. cerevisiae has, or once had, CpG methylation.
Dinucleotide clusters of CpGs or “CpG islands” (1) are present in the promoter and exonic regions of approximately 40% of mammalian genes (2). By contrast, other regions of the mammalian genome contain few CpG dinucleotides and these are largely methylated (2). The decreased occurrence of CpGs is best explained by the fact that methylated cytosines are mutational hotspots (3) leading to CpG depletion during evolution. A large number of experiments have shown that methylation of promoter CpG islands plays an important role in gene silencing (4), genomic imprinting (5), X-chromosome inactivation (6), the silencing of intragenomic parasites (7), and carcinogenesis (8, 9).
The first large-scale computational analysis of CpG islands using vertebrate sequences in GenBank was performed by Gardiner-Garden and Frommer (1), who defined a CpG island as being a 200-bp region of DNA with a high G+C content (greater than 50%) and observed CpC/expected CpG ratio(ObsCpG/ExpCpG) of greater or equal to 0.6. The exact definition of what constitutes a CpG island is somewhat arbitrary because the cutoffs for the parameters used to describe them can make significant differences to what sequences are included within the definition. For example, the human _Alu_s, which are highly repetitive short interspersed elements, have an approximately 280-bp consensus sequence, and some of these have relative high %GC and ObsCpG/ExpCpG (10). This composition makes it difficult to distinguish bona fide CpG islands from the nearly 1,000,000 Alu copies per haploid genome. Here we have analyzed the complete sequences of human chromosomes 21 (11) and 22 (12), which make up ≈2% of the total human genome (11) and therefore contain approximately 750 genes (11). The use of whole chromosome sequences results in less bias being introduced to define these regions than that introduced in the earlier studies using gene exon databases. We designed an algorithm to search for and describe CpG islands, and we suggest a modification of the original criteria of Gardiner-Garden and Frommer (1), which now excludes _Alu_s and many CpG islands not located within the promoters of genes. This more rigorous description of a CpG island might be used to better define an island for studies on the potential role of methylation in promoter silencing. Also, our description reduced the number of CpG islands located on these chromosomes from 14,062 to 1,101, which is more consistent with the expected number of genes (≈750) located on these two chromosomes.
The recent sequencing of the complete genomes of Escherichia coli (13), Saccharomyces cerevisiae (14), Drosophila melanogaster (15), Caenorhabditis elegans (16), and Arabidopsis thaliana (17) also allowed us to conduct comparative studies on the frequency of occurrence of the dinucleotide CpG within these genomes and compare that to the human genome. We also used genome sequences to show strong CpG suppression in the human genome and slight suppression in D. melanogaster and S. cerevisiae.
Materials and Methods
All sequence data were obtained from the GenBank Database. We used the contigs, NT_011511–15 (chromosome 21), and NT_011516, NT_011517, NT_011519, NT_011520, NT_011521, NT_011522, NT_011523, NT_011534, NT_011525, NT_019197, and NT_011526 (chromosome 22). When we analyzed the chromosomes, approximately 350 genes were mapped on both chromosomes. CpG islands were extracted from these contigs with the following algorithm, consisting of several steps (Fig. 1). To exclude “mathematical CpG islands” (for example, a 300-bp sequence containing one G, 150 Cs, and only one CpG, which would meet the criteria of a CpG island), we added one more condition: that there are at least seven CpGs in these 200 bp. This number was selected on the basis that there would be 200/16 (i.e., 12.5) CpGs in a random DNA fragment containing no suppression of CpG. Because Gardiner-Garden and Frommer's criterion (1) of ObsCpG/ExpCpG of 0.6 would accommodate (0.6 × 12.5) CpGs (i.e., 7.5), we selected seven CpGs as being a reasonable cutoff for the initial analysis.
Figure 1.
Schematics for the algorithms for CpG island extraction from human genome sequences. (A) Set a 200-base window in the beginning of a contig, compute %GC and ObsCpG/ExpCpG. Shift the window 1 bp after evaluation until the window meets the criteria of a CpG island. (B) If the window meets the criteria, shift the window 200 bp and then evaluate again. (C and D) Repeat these 200-bp shifts until the window does not meet the criteria. (E) Shift the last window 1 bp toward the 5′ end until it meets the criteria. (G) Evaluate total %GC and ObsCpG/ExpCpG. (H) If this large CpG island does not meet the criteria, trim 1 bp from each side until it meets the criteria. (I) Two individual CpG islands were connected if they were separated by less than 100 bp. (J) Values for ObsCpG/ExpCpG and %GC were recalculated to remain within the criteria.
Alu repetitive elements (_Alu_s) were detected by the repeatmasker mail server (University of Washington Genome Center, Seattle, http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). We also found which CpG islands contain the first coding exon or other exons according to mapping information of the contigs from GenBank. CpG islands were categorized into four categories in this order: “5′ region” included at least the first coding exon of a known gene and might or might not include downstream introns and exons and Alu_s. An “Exon” CpG island did not include a known first coding exon and possibly included intronic and Alu sequences. An “_Alu ” did not include a known exonic sequence. “Unknown” sequences did not satisfy any of the above criteria.
We first extracted 14,062 CpG islands on the basis of the original criteria of Gardiner-Garden and Frommer (1) and analyzed the change of proportions of the categories of 5′ region, Exon, Alu, and unknown CpG island. We then reanalyzed these by applying modified criteria on all 14,062 CpG islands that had been identified by Gardiner-Garden and Frommer's criteria (1). On this analysis, we analyzed the variables for a 50% and 55% %GC, 0.60 and 0.65 ObsCpG/ExpCpG, and 200- and 500-bp length.
The algorithm developed to identify CpG islands in genomes with strong CpG suppression was not suitable for the analysis of other genomes not so suppressed. Therefore, to determine the distribution of %GC and ObsCpG/ExpCpG throughout the sequenced genomes of various organisms, these parameters were calculated in consecutive nonoverlapping 500-bp windows starting at one end of a contig and progressing to the other. A random sample of 5,000 sequences was then picked for each organism and 5,000 data points were displayed on each plot. Nearest neighbor base sequence analysis was performed by using a shifting 2-bp window for each possible dinucleotide and the frequencies calculated.
For these analyses we used all contigs of human chromosomes 21 and 22, NC_000862 (Arabidopsis thaliana, chromosome 4), NC_001133–48 (S. cerevisiae, chromosomes 1–16), AE002566, AE002593, AE002611, AE002620, AE002629 (sequencing scaffolds of D. melanogaster, chromosome 1), NC_000965 (C. elegans, chromosome 1), and NC_000913 (E. coli K-12 strain). All of these analyses were performed with perl script coded by D.T. with perl compiler (ActiveState, Vancouver, http://www.activestate.com/).
Results and Discussion
CpG Islands in Human Chromosomes 21 and 22 and Their Nature.
We set the criteria for CpG islands as being the original ones defined by Gardiner-Garden and Frommer (1) (length ≥ 200 bp, ObsCpG/ExpCpG ≥ 0.6, and %GC ≥ 50%) and analyzed the entire lengths of chromosomes 21 and 22. The algorithm used to extract these regions is indicated in Fig. 1 and has the advantage over existing search programs that it reduces the cycle of calculations required and results in the extraction of symmetrical CpG islands from both the 5′ and the 3′ ends. With this algorithm, we extracted 5,039 CpG islands from chromosome 21 and 9,023 from chromosome 22 (Table 1). Although the two chromosomes are similar in size, chromosome 22 had almost twice the number of CpG islands as chromosome 21, probably because of the existence of gene-poor regions constituting a third of chromosome 21 (11). However, because 40% of genes are thought to have CpG islands associated with them (2), the 14,062 CpG islands extracted by these criteria vastly exceeded the number expected to be associated with the approximately 750 genes located on the two chromosomes. This 50-fold excess suggested that the criteria might be too lenient, as has been noticed (11).
Table 1.
Number of CpG islands in chromosomes 21 and 22
| Category | 21 | 22 | 21 + 22 |
|---|---|---|---|
| 5′ region | 57 | 138 | 195 |
| Exon | 334 | 423 | 757 |
| Alu repeats | 2,520 | 5,131 | 7,651 |
| Unknown | 2,128 | 3,331 | 5,459 |
| Total | 5,039 | 9,023 | 14,062 |
The data obtained from the combination of both chromosomes were analyzed with respect to whether the CpG islands occurred in the 5′ region of a gene, within an exonic region, or within _Alu_s. The mean values and distributions of these analyses with respect to %GC, ObsCpG/ExpCpG, and length are shown in Fig. 2. The data showed that, not unexpectedly, the majority of CpG islands extracted by the criteria of Gardiner-Garden and Frommer (1) corresponded to _Alu_s (Fig. 2 A_–_C). However, a large number of unknown sequences were also identified. The majority of these two categories of sequences had properties that placed them at the lower limits of the criteria currently used to extract CpG islands. For example, the majority had %GC <59%, ObsCpG/ExpCpG of <0.72, and a length <600 bp. This result suggested that altering the stringency by which CpG islands were defined would markedly reduce the occurrence of these sequences within the data set.
Figure 2.
Distributions of %GC, ObsCpG/ExpCpG and length of CpG islands in human chromosomes 21 and 22. Mean value and SD are also indicated in each histogram. (A–C) Distribution of %GC, ObsCpG/ExpCpG, and length of all categories. In these histograms, CpG islands containing both the 5′ of gene and an Alu are included in the 5′ region category, and CpG islands containing both exons and _Alu_s are categorized as exon. (D–F) Distribution of %GC, ObsCpG/ExpCpG, and length of CpG island containing the 5′ region. The occurrence of _Alu_s within sequences defined as 5′ regions is also indicated by horizontal hatching. (G–I) Distribution of %GC, ObsCpG/ExpCpG, and length of CpG islands containing exons. In these three histograms, CpG islands containing both an exon and Alu are represented as _Alu_s. The occurrence of _Alu_s within sequences defined as exon are also indicated by horizontal hatching. (J–L) Distribution of %GC, ObsCpG/ExpCpG, and length of CpG island containing Alu. (M–O) Distribution of %GC, ObsCpG/ExpCpG, and length of CpG islands containing unknown sequences. Both the exponential and Gaussian curves are shown in D, F, G, and I. In E and H, both the exponential curve and the minus second-order curves are shown.
The CpG islands associated with the 5′ regions of genes (Fig. 2 D–F) showed a markedly different distribution when compared with the _Alu_s (Fig. 2 J–L). These 5′ elements had a mean %GC of 65%, and showed a biphasic distribution for the occurrence of ObsCpG/ExpCpG. There was also a biphasic distribution with respect to length, with a significant proportion of CpG islands being in the small region of 200–400 bp and an average length of 1,300 bp for all 5′ regions analyzed. As has been pointed out earlier (2), CpG islands can also occur within the coding regions of genes and this was again found to be the case in our analysis (Fig. 2 G–I); however, they tended to have a lower %GC on average than the 5′ CpG islands, tended to have a slightly decreased mean for the occurrence of ObsCpG/ExpCpG, and tended to be shorter.
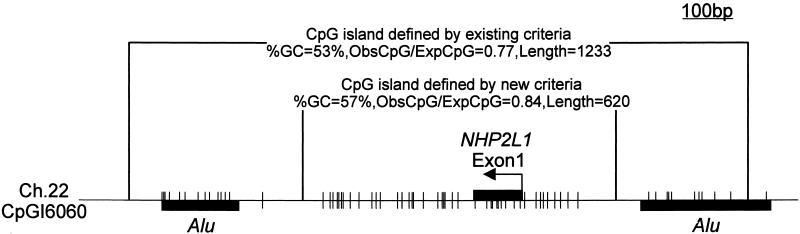
Table 2 shows the change of proportions of the four categories depending on the three parameters used to define a CpG island in an attempt to develop more rigid criteria that would exclude the _Alu_s and small unknown islands from the definition and increase the proportion of CpG islands located in the 5′ regions of genes. This table shows that modifying the criteria to a %GC ≥ 55% and a length ≥ 500 bp with ObsCpG/ExpCpG ≥ 0.65 resulted in the exclusion of the vast majority of _Alu_s and unknown sequences, while only slightly decreasing the number of CpG islands that occur within the 5′ regions of genes. The increased stringency also substantially reduced the number of exonic CpG islands. The biological functions of these islands are not well understood, but CpG islands located in nonpromoter regions can play significant roles in gene regulation (18); they also seem to be frequent targets for de novo methylation in cancer and aging (19). Therefore, although the increased stringency preferentially locates CpG islands in the 5′ regions of genes, it may also result in the loss of smaller regions of DNA from the data set that may be functionally important in gene control. The modified criteria also helped remove Alu sequences previously identified as part of 5′ CpG islands (Fig. 3). In this example of the NHP2L1 gene, the entire 1,233-bp fragment originally extracted by the algorithm included two Alu sequences with some CpG suppression. The modified stringent criteria reduced the size of the island to 620 bp and excluded the Alu sequences.
Table 2.
Effect of modifying criteria on CpG island distribution
| Length | 200 | 200 | 200 | 200 | 500* | 500* | 500* | 500* |
|---|---|---|---|---|---|---|---|---|
| %GC | 50 | 55* | 50 | 55* | 50 | 55* | 50 | 55* |
| ObsCpG/ExpCpG | 0.6 | 0.6 | 0.65* | 0.65* | 0.6 | 0.6 | 0.65* | 0.65* |
| 5′ region | 195 | 188 | 173 | 172 | 166 | 164 | 163 | 161 |
| Exon | 757 | 620 | 529 | 460 | 143 | 133 | 126 | 120 |
| Alu repeats | 7,651 | 871 | 1,026 | 138 | 506 | 168 | 310 | 122 |
| Unknown | 5,459 | 7,804 | 7,955 | 6,511 | 669 | 711 | 767 | 698 |
| Total | 14,062 | 9,483 | 9,683 | 7,281 | 1,484 | 1,176 | 1,366 | 1,101 |
Figure 3.
The modified criteria also helped remove Alu sequences previously identified as part of 5′ region CpG islands. In this example, a 1,233-bp fragment originally extracted by the algorithm included two Alu sequences with some CpG suppression associated with the nonhistone chromosome protein 2 like 1 (NHP2L1). The modified stringent criteria reduced the size of the island to 620 bp and excluded the Alu sequences.
CpG Distribution in Other Species.
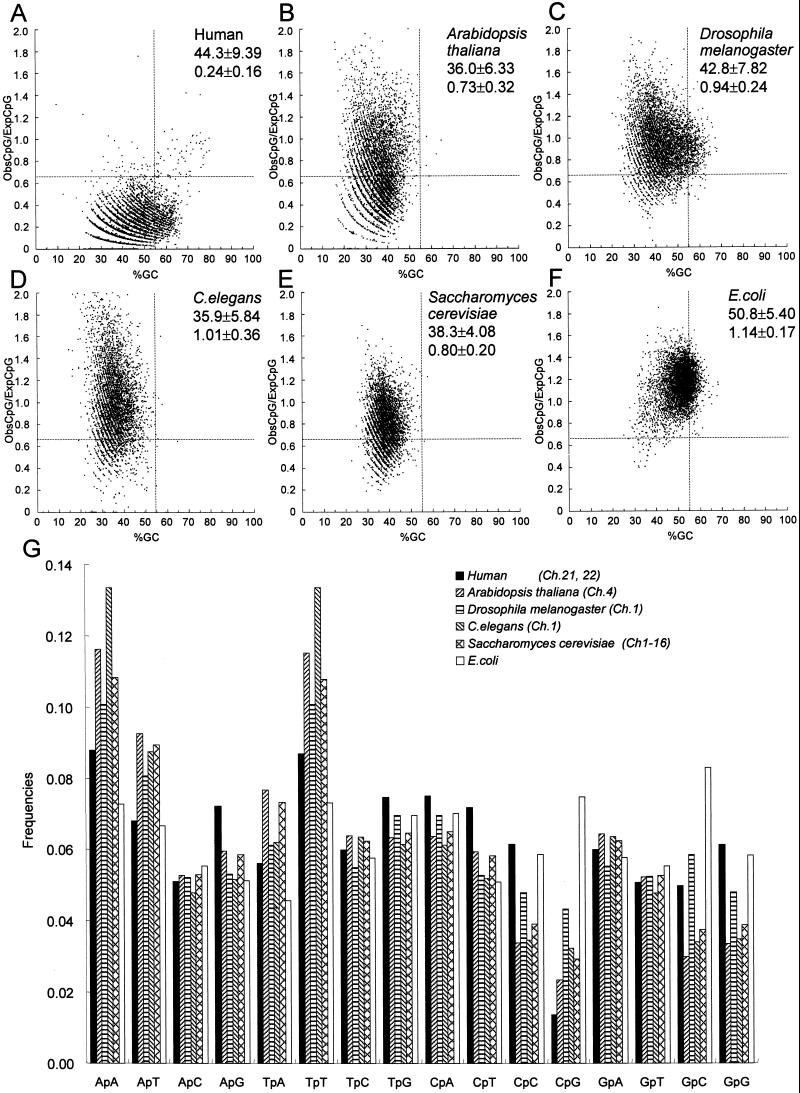
The recent cloning and sequencing of the genomes of several model organisms allowed us to analyze of those genomes and compare them with human chromosomes 21 and 22. Consecutive 500-bp windows of human chromosome 21 and 22 compared with these other species with respect to ObsCpG/ExpCpG and the %GC (Fig. 4 A–F). The strong suppression of CpG in human chromosomes 21 and 22 was analyzed and was clearly visible (Fig. 4A), and the CpG islands are indicated by using the criteria established in this paper. However, it should be noted that there is no clear demarcation between regions that are called CpG islands and those that are not. Rather, there is a continuum of 500-bp regions of DNA that move between this bulk DNA and the properties of a CpG island. The human genome showed the strongest suppression of CpG. Several sequences plotted in the lower left field of the plot of %GC vs. ObsCpG/ExpCpG of the human genome (Fig. 4A) turned out to be simple repetitive sequences such as (TA)n and (TTTAA)n (data not shown). CpG suppression in the human genome is caused not only by CpG depletion through evolution but also by the high content of simple repetitive sequences and a low rate of sequence utilization for genes. A. thaliana contains 5-methylcytosine, and its genome shows a wide distribution of the occurrence for CpG (Fig. 4B). However, because of the low GC content in this organism, few fragments fulfilling our criteria for a CpG island are visible in the A. thaliana genome. In this respect, the A. thaliana genome and that of C. elegans (Fig. 4D) are quite similar and not as tightly clustered with respect to %GC and ObsCpG/ExpCpG as those of S. cerevisiae (Fig. 4E) and E. coli (Fig. 4F). The genome of E. coli showed a distribution around the middle of the plot, which is consistent with the fact that E. coli does not have a recognizable sequence for a CpG methyltransferase in its genome and therefore probably does not have CpG methylation. The D. melanogaster genome is not suppressed for the occurrence of CpG and contains a large number of fragments that would fulfill the criteria of a CpG island that we have defined (Fig. 4C).
Figure 4.
(A–F) %GC vs. ObsCpG/ExpCpG plot of a randomly selected 5,000 set of 500-bp-long sequences. Mean value and SD are presented on the plot, and new criteria (%GC ≥ 55%, ObsCpG/ExpCpG ≥ 0.65) are shown as dashed lines. (G) Nearest-neighbor sequence analysis of human chromosomes 21 and 22 and other model organisms.
Nearest-neighbor sequence analysis of these model organisms (Fig. 4G) also shows that the frequency of occurrence of the CpG sequence is suppressed in several organisms, including those that are not known to have DNA methylation. Thus, with the exception of E. coli, the other five organisms examined all show that the CpG dinucleotide is the most infrequent dinucleotide within their genomes. In D. melanogaster and S. cerevisiae, the genome showed slight suppression of CpG. Previously, no methylated cytosine had been detected in the genome of either organism; however, recently 5-methylcytosine was detected in D. melanogaster (20, 21). Thus, S. cerevisiae might also have, or once had, methylcytosine considering that S. cerevisiae showed much more suppression than D. melanogaster in both of the plots of %GC vs. ObsCpG/ExpCpG and in the nearest-neighbor analysis.
Acknowledgments
We are grateful to Dr. Takako Takai-Igarashi for programming suggestions. D.T. and P.A.J. are supported by National Cancer Institute Grants R01 CA82422 and R01 CA83867.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gardiner-Garden M, Frommer M. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 2.Larsen F, Gundersen G, Lopez R, Prydz H. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 3.Coulondre C, Miller J H, Farabaugh P J, Gilbert W. Nature (London) 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- 4.Bird A. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 5.Feil R, Khosla S. Trends Genet. 1999;15:431–435. doi: 10.1016/s0168-9525(99)01822-3. [DOI] [PubMed] [Google Scholar]
- 6.Panning B, Jaenisch R. Cell. 1998;93:305–308. doi: 10.1016/s0092-8674(00)81155-1. [DOI] [PubMed] [Google Scholar]
- 7.Yoder J A, Walsh C P, Bestor T H. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 8.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J P. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 9.Jones P A, Laird P W. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 10.Schmid C W. Nucleic Acids Res. 1998;26:4541–4550. doi: 10.1093/nar/26.20.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hattori M, Fujiyama A, Taylor T D, Watanabe H, Yada T, Park H S, Toyoda A, Ishii K, Totoki Y, Choi D K, et al. Nature (London) 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- 12.Dunham I, Shimizu N, Roe B A, Chissoe S, Hunt A R, Collins J E, Bruskiewich R, Beare D M, Clamp M, Smink L J, et al. Nature (London) 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- 13.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 14.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. , 563–547. [DOI] [PubMed] [Google Scholar]
- 15.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 16.The C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 17.Mayer K, Schuller C, Wambutt R, Murphy G, Volckaert G, Pohl T, Dusterhoft A, Stiekema W, Entian K D, Terryn N, et al. Nature (London) 1999;402:769–777. [Google Scholar]
- 18.Jones P A, Takai D. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen C, Liang G, Nguyen T T, Tsao-Wei D, Groshen S, Lubbert M, Zhou J H, Benedict W F, Jones P A. J Natl Cancer Inst. 2001;93:1465–1472. doi: 10.1093/jnci/93.19.1465. [DOI] [PubMed] [Google Scholar]
- 20.Lyko F, Ramsahoye B H, Jaenisch R. Nature (London) 2000;408:538–540. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- 21.Gowher H, Leismann O, Jeltsch A. EMBO J. 2000;19:6918–6923. doi: 10.1093/emboj/19.24.6918. [DOI] [PMC free article] [PubMed] [Google Scholar]