Lipid mediator-induced expression of bactericidal/ permeability-increasing protein (BPI) in human mucosal epithelia (original) (raw)
Abstract
Epithelial cells which line mucosal surfaces are the first line of defense against bacterial invasion and infection. Recent studies have also indicated that epithelial cells contribute significantly to the orchestration of ongoing inflammatory processes. Here, we demonstrate that human epithelial cells express bactericidal/permeability-increasing protein (BPI), an antibacterial and endotoxin-neutralizing molecule previously associated with neutrophils. Moreover, we demonstrate that such BPI expression is transcriptionally regulated by analogs of endogenously occurring anti-inflammatory eicosanoids (aspirin-triggered lipoxins, ATLa). Initial studies to verify microarray analysis revealed that epithelial cells of wide origin (oral, pulmonary, and gastrointestinal mucosa) express BPI and each is similarly regulated by aspirin-triggered lipoxins. Studies aimed at localization of BPI revealed that such expression occurs on the cell surface of cultured epithelial cell lines and dominantly localizes to epithelia in human mucosal tissue. Functional studies employing a BPI-neutralizing anti-serum revealed that surface BPI blocks endotoxin-mediated signaling in epithelia and kills Salmonella typhimurium. These studies identify a previously unappreciated “molecular shield” for protection of mucosal surfaces against Gram-negative bacteria and their endotoxin.
Keywords: mucosa‖infection‖inflammation‖eicosanoid
During both acute and chronic inflammatory processes, epithelial cells coordinate mucosal responses to infection. For this reason, much recent attention has been paid to understanding innate, anti-inflammatory pathways used by mucosal epithelial cells. Of particular interest are a group of lipid mediators termed the lipoxins (1). Lipoxins are bioactive eicosanoids derived from membrane arachidonic acid by the combined action of 5-lipoxygenase (LO) and 12-LO or 15-LO (2). A number of recent in vitro and in vivo studies have revealed that lipoxins, and specifically lipoxin A4 (LXA4), function as innate “stop signals,” serving to control local inflammatory processes (3). Synthetic lipoxin analogs exhibit greater potency for these actions than the native compound, likely because of decreased metabolism (4). Recently, synthetic analogs [e.g., 15-epi-16-(_para_fluoro)-phenoxy-LXA4 (aspirin-triggered lipoxins, ATLa; ref. 5)] have been modeled on 15-epi-LXA4, a native lipoxin generated in vivo in the presence of aspirin via cyclooxygenase-2 acetylation (5).
Among the innate anti-infective defense molecules of humans is BPI, a 55- to 60-kDa protein found in neutrophil azuorphilic granules, on the neutrophil cell surface, and to a lesser extent, in specific granules of eosinophils (6–8). BPI selectively exerts multiple anti-infective actions against Gram-negative bacteria, including cytotoxicity through damage to bacterial inner/outer membranes, neutralization of bacterial lipopolysaccharide (endotoxin), as well as serving as an opsonin for phagocytosis of Gram-negative bacteria by neutrophils (6, 9). Structural characterization of BPI reveals a symmetrical bipartite molecule containing a cationic N-terminal region for antibacterial and endotoxin neutralization and a C-terminal motif necessary for bacterial opsonization (10).
Experiments aimed at identifying new anti-inflammatory molecules on mucosal surfaces revealed that epithelial cells express surface BPI and that BPI expression was regulated by epithelial exposure to stable analogs of aspirin-triggered lipoxins. Epithelial BPI was found to promote bacterial killing and to diminish endotoxin activation of epithelia. These results identify a new pathway by which anti-inflammatory eicosanoids enhance anti-microbial and endotoxin-neutralizing activity through transcriptional activation of BPI, heretofore solely associated with phagocytes.
Materials and Methods
Epithelial Cell Culture.
T84 and Caco2 intestinal epithelial cells were grown and maintained as confluent monolayers on collagen-coated permeable supports as previously described in detail (11), and used 6–12 days after plating. The oral epithelial line (KB cells) were grown as described (12). Immortalized keratinocytes of oral mucosa origin, generated by ectopic expression of the catalytic subunit of telomerase (hTERT), were a kind gift from Jim Rheinwald, Harvard Medical School and were plated and cultured as described (13, 14).
Transcriptional Analysis.
The transcriptional profile of epithelial cells (KB cells) exposed to LXA4 analog (0, 4, or 8 h exposure to 1 μM 15-epi-16-(_para_-fluoro)-phenoxy-LXA4, ATLa, prepared by total organic synthesis with Nicos Petasis, University of Southern California, under Program Project DE13499) (15) was assessed in RNA by using quantitative genechip microarrays (Affymetrix, Santa Clara, CA) (16). Reverse transcription–PCR (RT-PCR) analysis of mRNA levels was performed by using DNase-treated total RNA as previously described (17), by using the following primer sets; human BPI sense (5′-GCA CCT GTT CCT GAT GGG-3′) and antisense primer (5′-AGC ACA AAT GGA AAT TTC TTG-3′, 255-bp product); human ICAM-1 sense (5′-CAC AGT CAC CTA TGG CAA CG-3′) and antisense primer (5′-TTC TTG ATC TTC CGC TGG C-3′); and human β-actin sense (5′-TGA CGG GGT CAC CCA CAC TGT GCC CAT CTA-3′, 750-bp product) and antisense primer (5′-CTA GAA GCA TTT GCG GTG GAC GAT GGA GGG-3′, 661-bp product).
Confocal Laser-Scanning Microscopy.
OKF6 or Caco2 cells were grown to confluence on acid-washed 12-mm glass coverslips. Monolayers were exposed to indicated experimental conditions washed once in PBS and fixed for 10 min at room temperature in 1% paraformaldehyde in cacodylate buffer (0.1 M sodium cacodylate, pH 7.4/0.72% sucrose). After washing twice with PBS, the cells were incubated for 1 h with rabbit polyclonal BPI antisera (1:300 dilution) or control sera (equivalent dilution depleted of specific Ab through three consecutive adsorptions with Sepharose beads covalently linked to rBPI via cyanogen bromide coupling, kit from Pierce). After washing, the monolayers were incubated with goat anti-rabbit Oregon Green (1 μg/ml, Molecular Probes). Cells were imaged on a BioRad MRC-600 confocal fluorescence microscope.
Immunoprecipitation and Western Blotting.
Epithelial cells were grown to confluence on 45-cm2 permeable supports and exposed to ATLa or vehicle (0.01% ethanol), as indicated. Cells were washed extensively in HBSS and cooled to 4°C, and extracellular proteins were biotinylated [1 mM NHS-biotin (Pierce) in HBSS] as described (18). Plasma membranes were isolated by using nitrogen cavitation (200 psi, 8 min, 4°C) as described (18). Recombinant human BPI (100 ng/ml, a kind gift from Stephen Carroll, Xoma) was directly biotinylated, and excess biotin was removed by multiple washes on a 5-kDa cut-off membrane filter (Amicon). Fractions were precleared with 50 μl of preequilibrated protein-G Sepharose (Amersham Pharmacia). Immunoprecipitation of BPI was performed with goat polyclonal anti-BPI (a kind gift from Peter Elsbach and Jerrold Weiss) followed by addition of 50 μl of preequilibrated protein-G Sepharose and overnight incubation. Washed immunoprecipitates were boiled in nonreducing sample buffer, resolved by nonreducing SDS/PAGE, transferred to nitrocellulose, labeled with streptavidin-peroxidase, and visualized by enhanced chemiluminescence as described (18).
Cell-Surface Immunoassay.
Epithelial cells were grown and assayed for Ab binding after exposure to indicated concentrations of LPS (from Salmonella minnesota Re595, List Biological Laboratories, Campbell, CA) in the presence of 5% heat-inactivated normal human serum. After such exposure, ICAM-1 cell surface expression was quantified by using a cell surface ELISA, as described (19). In subsets of experiments, a polyclonal anti-BPI antisera with demonstrated BPI-neutralizing activity (21) (goat anti-human, used at 1:300 in HBSS) or normal goat serum (Invitrogen, 1:300 in HBSS), as indicated, were added before incubation with endotoxin. Data are presented as the mean ± SE OD at 405 nm (background subtracted).
Bacterial Killing Assays.
Salmonella typhimurium (strain 14028 from American Type Culture Collection) was cultured and grown in LB as described (22). In subsets of experiments, Enterococcus faecalis (strain PCI 1326 from American Type Culture Collection) was cultured as described (23). Caco2 epithelial cells were grown to confluence of 60-mm Petri dishes and exposed to indicated experimental conditions. Cells were washed once with HBSS, and washed bacteria were added to epithelial monolayers at a ratio of 50 bacteria per adherent epithelial cell. Incubations were allowed to proceed for 90 min or as indicated, on a rotating platform. After incubation, supernatants were collected and epithelial cells were hypotonically lysed with 1-ml ice-cold water. Bacteria were pelleted, dilutions of both pellets and supernatants plated and incubated overnight at 37°C, and colony counts performed. In subsets of experiments, anti-BPI antisera (1:300 in HBSS) or anti-BPI antisera preadsorbed with rBPI (1:300 in HBSS), as indicated, were added 30 min before incubation with bacteria. Data are presented as the mean ± SE colony-forming unit (CFU).
Localization of BPI in Human Tissue.
Normal human esophageal or colonic specimens were obtained under an approved human institutional review board protocol. Sections were fixed in 10% buffered formalin, paraffin embedded, and sectioned by using standard methods. Antigen retrieval was performed in a pressure cooker with EDTA Decloaker solution, pH 8.0 (Zymed) according to manufacturer's recommendations. Sections were stained with rabbit polyclonal BPI antisera (1:100) and peroxidase-coupled secondary Abs (1 μg/ml, Zymed) and visualized by peroxidase method according to manufacturers recommendations (Vectastain, Vector Laboratories). Control sections were incubated with BPI-preadsorbed Ab (1:100 dilution), as indicated. Sections were visualized with a Nikon E600 microscope at X200 magnification.
Data Analysis.
BPI bioactivity results were compared by two-factor ANOVA or by Student's t test, where appropriate. Values are expressed as the mean and SEM of n monolayers from at least three separate experiments.
Results
Epithelial Cells Express BPI and Such Expression Is Regulated by Lipoxins.
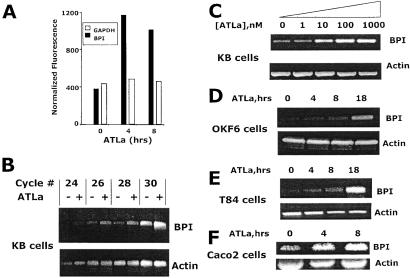
Lipoxins possess potent anti-inflammatory properties for mucosal inflammation (3). Epithelial cells of diverse origin express functional receptors for lipoxins; however, little is known about downstream transcriptional pathways elicited through ligation of the lipoxin receptor. Thus, a transcriptional profiling approach (16) was used to identify potential lipoxin-regulated gene expression in model epithelia (KB cells). This analysis revealed that 97 of 7,129 genes screen (1.4%) were induced by greater than 3-fold by ATLa and 36 of 7,129 screened (0.5%) were decreased by exposure to ATLa. This analysis identified the expression and up-regulation of BPI by ATLa in epithelial cells, providing the interesting possibility that this molecule may provide anti-infective qualities for the epithelium. Indeed, basal expression of the BPI mRNA comparable to glyceraldehyde phosphate dehydrogenase and a dominant induction of BPI by ATLa (3.2- and 2.9-fold increase over control at 4 and 8 h exposure to ATLa, respectively, Fig. 1A). RT-PCR analysis was used to verify these microarray results at the RNA level. As shown in Fig. 1B, semiquantitative RT-PCR revealed that, relative to β-actin, ATLa (1 μM exposure, 8 h) induced a prominent induction of BPI compared to vehicle control (maximal increase of 4.3-fold increase by densitometry). Dose-response analysis revealed an approximate EC50 of 50 nM (Fig. 1C). Epithelial exposure to similar concentrations of 15-deoxy-LXA4, a lipoxin analog lacking demonstrable bioactivity (4), resulted in no induction of BPI at the mRNA level (data not shown).
Figure 1.
BPI induction by ATLa. Confluent epithelial monolayers were exposed to indicated concentrations or time periods of ATLa (1 μM). (A) Quantitative microarray data for BPI in epithelial cells exposed to indicated conditions. (B) Total RNA was isolated from ATLa-exposed KB cells (1 μM, 8 h) and examined for BPI transcript by semiquantitative RT-PCR (increasing numbers of PCR cycles). β actin transcript was examined under similar conditions as an internal standard. (C) KB cells were exposed to indicated concentrations of ATLa for 8 h and examined for BPI transcript by using 28 cycles of PCR. (D_–_F) Indicated epithelial cell lines were exposed to indicated periods of ATLa, and BPI transcript was determined by 26 cycles of RT-PCR. β actin transcript was used as an internal standard.
As shown in Fig. 1 D_–_F, similar analysis by using RNA (26 cycles of PCR) derived from epithelial cells other than KB cells (OKF6, T84, and Caco2 cells) revealed a prominent pattern of time-dependent BPI induction by ATLa (1 μM) relative to β-actin. Similar results were observed with pulmonary (A549) epithelial cells (data not shown). Importantly, such results were not universal for all cell types because no detectable BPI transcript was evident with as many as 35 cycles of PCR in RNA derived from human dermal microvascular endothelial cells (data not shown), suggesting that these findings may be relatively specific for epithelia. Moreover, consistent with previous studies indicating that lipoxin signaling is a G protein-coupled event (24), Caco2 preexposure to pertussis toxin (1 μM, 30 min) resulted in a 65% decrease in BPI induction by ATLa (1 μM, 18 h, data not shown). Taken together, these findings indicate specific transcriptional activation of BPI by ATLa.
Localization of BPI Protein to the Epithelial Cell Surface.
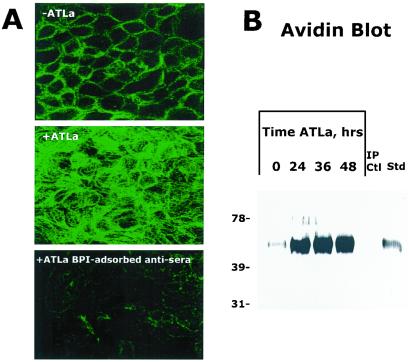
Previous studies have indicated that BPI can exist as a granule-bound protein or as a surface-associated protein on neutrophils (25). Initial attempts to detect soluble BPI by using a sulfuric acid extraction process known to release granule-associated BPI from neutrophils (26) revealed undetectable levels of BPI by BPI-specific ELISA and Western blot (sensitivity <100 pg/ml, data not shown). Thus, in an attempt to localize expression patterns of epithelial-expressed BPI, confocal microscopy was used on nonpermeabilized epithelia. As shown in Fig. 2A, BPI was expressed in a surface-bound form on Caco2 cells, with increased expression associated with ATLa exposure (1 μM for 24 h, see Fig. 2A). As a control for specificity, parallel samples exposed to ATLa (1 μM for 24 h) were incubated with BPI-preadsorbed antisera and revealed a nearly complete loss of surface staining.
Figure 2.
Localization of BPI to the cell surface. (A) BPI was localized by confocal microscopy in nonpermeabilized Caco2 cells exposed to vehicle (Top) or ATLa (1 μM, 24 h, Middle and Bottom). BPI adsorbed antisera was used as a control (Bottom). Shown here are confocal sections through the mid-zone, subjunctional portion of epithelial monolayers. Representative experiment from n = 2. (B) T84 cells were preexposed to ATLa (1 μM) for indicated periods of time. Cell surface proteins were nonspecifically labeled with biotin, BPI was immunoprecipitated from cell lysates, resolved by SDS/PAGE, and Western blots were probed with avidin peroxidase. Also shown is the immunoprecipitation control (omission of primary Ab) as well as a biotinylated BPI standard. Representative experiment from n = 3.
As biochemical verification of these observations, and to examine whether ATLa (1 μM) induces BPI at the protein level, immunoprecipitation of biotinylated plasma membrane protein followed by avidin blot was used. This approach allows for detection of surface membrane proteins derived from intact epithelial cells (27). As depicted in Fig. 2B, a time course of ATLa (1 μM) exposure was performed and revealed a dominant induction (maximal 12.5-fold increase at 36 h by densitometry) of a ≈55-kDa surface protein consistent with BPI (biotinylated recombinant BPI is shown for comparative purposes). Controls incorporating a polyclonal Ab directed against a soluble epithelial protein (IL-8) revealed no detectable protein at this level. Such analysis indicates the likelihood that BPI exists predominantly as a membrane-bound protein on the surface of epithelial cells and that such expression is regulated by ATLa.
Endotoxin Neutralization by Epithelial BPI.
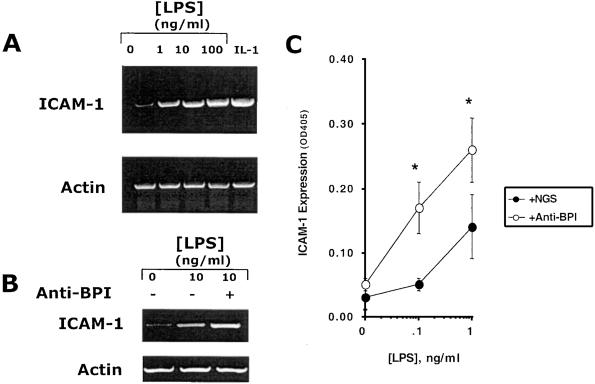
We next extended these studies to examine the functional activity of BPI on the epithelial surface. Previous studies have indicated that BPI possesses not only bactericidal, but also endotoxin-neutralizing activity (6). Because epithelial cells have been previously shown to transcriptionally respond to endotoxin in the presence of serum (28), we first determined whether endotoxin might induce ICAM-1, an endotoxin-responsive marker, which functions as a leukocyte adhesion molecule (29). As shown in Fig. 3A, addition of endotoxin to KB cells in the presence of 5% heat-inactivated normal human serum induced ICAM-1 mRNA, comparable to a known ICAM-1 agonist (IL-1, 10 ng/ml).
Figure 3.
BPI functionally regulates epithelial endotoxin responses. (A) Transcriptional induction of ICAM-1 was examined in KB cells in response to indicated concentrations of endotoxin (LPS) in the presence of NHS (5% vol/vol) or IL-1 (10 ng/ml) for 8 h. Total RNA was isolated and used to assess ICAM-1 transcripts by RT-PCR. β actin transcript was used as an internal standard. (B) Endotoxin-induced ICAM-1 expression was examined in the presence polyclonal anti-BPI (+) or in the presence of NGS (−). β actin transcript was used as an internal standard. (C) KB cells were preexposed to ATLa (1 μM, 8 h), and cell surface ELISA was used to screen ICAM-1 protein induction by endotoxin/5% NHS (additional 24 h) in the presence of anti-BPI or control NGS. (*, significantly different from NGS; P < 0.025)
Having shown that epithelial cells respond to endotoxin, we next determined whether inhibition of basally expressed BPI (i.e., in the absence of LXA4) might function to enhance endotoxin-mediated induction of ICAM-1 surface protein. To this end, epithelial cells were preexposed to anti-BPI or control normal goat serum (NGS) and subsequently activated with endotoxin (concentration range 0–1 ng/ml) in the presence of 5% normal human serum. Transcriptional analysis of the ICAM-1 response to endotoxin is shown in Fig. 3B. The addition of anti-BPI serum significantly increased endotoxin-induced ICAM-1 transcript (2.3 ± 0.45-fold, n = 3, P < 0.01 compared to NGS), suggesting that BPI provides an endotoxin-neutralizing function for epithelial cells. Similar results were found by using Caco2 cells (2.0 ± 0.61-fold increase with anti-BPI compared to NGS, P < 0.05). Moreover, as shown in Fig. 3C, anti-BPI shifted the endotoxin dose-response curve for ICAM-1 induction to the left compared to control NGS, with significant differences evident at 0.1 and 1 ng/ml (both P < 0.025 by ANOVA). When higher concentrations of endotoxin were used, the influence of anti-BPI was less apparent (data not shown). Such data indicate that surface expressed BPI might normally function to dampen epithelial endotoxin responses.
Role of Surface BPI in Bacterial Killing.
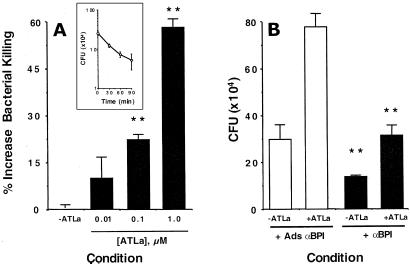
We next determined whether intact, adherent epithelial cells kill a BPI-sensitive bacteria. Confluent Caco2 epithelial cells were exposed to S. typhimurium and examined for bacterial killing in standard CFU analysis by using adherent epithelial cells. Such analysis revealed a nearly 1-log order reduction in CFU over a 90-min period (83 ± 11% killing, P < 0.025 by ANOVA, see Fig. 4A Inset ). We extended these studies to determine whether epithelial exposure to ATLa might functionally enhance killing of S. typhimurium. As shown in Fig. 4A, epithelial preexposure to ATLa increased bacterial killing in a concentration-dependent fashion (P < 0.025 by ANOVA), with a nearly 60% increase in killing at 1 μM ATLa. As shown in Fig. 4B, a significant component of increased bacterial killing was attributable to induction of BPI because the addition of anti-BPI inhibited such bacterial killing (P < 0.01 compared to anti-BPI adsorbed with rBPI). These results were not explained by bactericidal activity or nonspecific agglutination by either ATLa or anti-BPI (based on direct incubation of ATLa or anti-BPI with bacteria and colony counts, data not shown). Parallel experiments assessing epithelial killing of a Gram-positive bacterium (Enterococcus faecalis) that is not sensitive to BPI indicated a smaller degree of killing (0.3 ± 0.05-log order reduction in CFU over 90 min) but no influence of anti-BPI on such killing (5.5 ± 2.1% decrease in killing, P = not significant). Such data indicate that BPI contributes to bacterial killing by epithelial cells and that ATLa-induced BPI enhances this functional response.
Figure 4.
ATLa enhances BPI-dependent bacterial killing. Adherent Caco2 cells were incubated with vehicle or indicated concentrations of ATLa (24 h) and the role of BPI in Caco2 killing of S. typhimurium over a 60-min period was assessed. (A) Concentration-dependent increases in bacterial killing with previous exposure to ATLa. (Inset) A killing curve in the absence of ATLa. (B) Relative contribution of BPI was examined by incubation anti-BPI or control antisera preadsorbed with rBPI. Shown here are pooled results from three experiments. (**, indicates significantly different from control condition; P < 0.01)
Localization of BPI in Native Mucosal Tissue.
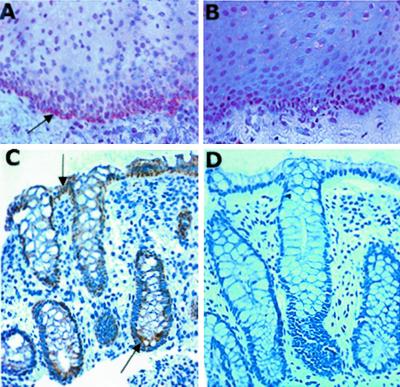
We extended these findings to examine whether native tissues express BPI and whether such expression localizes to the epithelium. Because our findings above (Fig. 1) suggest that both columnar (e.g., T84 and Caco2 cells) and squamous epithelia (e.g., KB and OKF6 cells) express BPI, we examined squamous and columnar epithelial-bearing tissues (esophagus and colon, respectively). As shown in Fig. 5, analysis of normal human esophagus (A) and colon (C) sections revealed dominant localization of BPI to the epithelium. In the case of esophageal tissue, BPI was most strongly expressed at the transition zone between epithelia and the lamina propria, with graded decreasing expression toward surface epithelia. In the colon, BPI was expressed dominantly in crypt and villus epithelia, with less expression along the crypt-villus axis. In both the esophagus and colon, localization with preadsorbed anti-BPI revealed no specific signal (B and D, respectively). These findings in native human tissue demonstrate that BPI is expressed in vivo.
Figure 5.
Localization of epithelial BPI in human mucosal tissues. Normal human esophageal (A and B) or colon (C and D) specimens were obtained, fixed in 10% buffered formalin, paraffin embedded, and sectioned. After antigen retrieval, sections were stained with rabbit polyclonal BPI antisera (A and C) or control sera (BPI preadsorbed Ab, B and D) followed by peroxidase-coupled secondary Ab and visualized by peroxidase method. Sections were visualized at ×200 magnification. Arrows indicate areas of dominant BPI localization.
Discussion
Mucosal epithelial cells provide a first line of defense against bacterial invasion and infection. Although much is known regarding innate mechanisms of bacterial clearance by leukocytes, it is only recently appreciated that epithelial cells might also function in a similar capacity (7, 30). In these studies, we explored the broad regulation of epithelial genes by ATLa, and in the course of these experiments, identified previously unappreciated expression of functional epithelial BPI. Such expression was localized to the membrane surface in cell lines and in diverse mucosal tissues in situ, and data are provided that epithelial-associated BPI serves to both inhibit endotoxin signaling as well as provide a pathway to dampen bacterial infection.
Mucosal epithelial cells harbor a number of antimicrobial factors that form a biochemical barrier to microbial colonization (7, 30, 31). Numerous studies have indicated that these antimicrobial factors are critical to maintenance of host-microbe homeostasis at the mucosal surface (7, 30). Initial studies in the present work identified the expression of BPI mRNA in epithelial cells, and extensions of these findings revealed broad expression on epithelia of diverse origin. Without exception, BPI expression has been described only in cells of myeloid lineage (7). Two conceptual points exemplify the potential importance of BPI expression on mucosal epithelia. First, epithelia provide the initiation point for host–microbial interactions. Although microbial flora are necessary and beneficial to the host, some degree of selectivity is also prerequisite for homeostasis. Epithelial-expressed BPI could play such a role. BPI is remarkable for its potent (nanomolar) and selective bioactivity against Gram-negative bacterial species (8). Moreover, the finding that functional BPI is expressed on the epithelial surface, and not in the soluble milieu, could provide an additional degree of selectivity for invasive/host-interactive pathogens. Second, a basic feature of many mucosal surfaces is the presence of high concentrations of endotoxin. Previous work has indicated that under appropriate conditions, epithelial cells can respond to endotoxin (28), and recent studies have clearly defined the existence of LPS receptors (e.g., CD14 and TLR4) on epithelial surfaces (32), the latter of which may be differentially regulated in selective mucosal diseases (33). For this reason, endogenous mechanism(s) likely exist to diminish aberrant activation of epithelial cells. Our present findings of BPI expression in epithelial cell lines and in native epithelia could provide an innate dampening mechanism against endotoxin by effectively competing for the binding of endotoxin, and as such, preventing endotoxin binding to such pro-inflammatory receptors. Indeed, endotoxin activation of epithelia (i.e., ICAM-1 induction) was significantly enhanced by the addition of functionally inhibitory anti-BPI sera, suggesting a protective role for BPI in mucosal endotoxin homeostasis. Of note, at higher concentrations of endotoxin (e.g., >50 ng/ml), the influence of epithelial expressed BPI was less obvious, suggesting that the relative concentration of BPI and/or the LPS affinity of BPI compared with LPS receptors (CD14/Toll-like receptors) may favor activation at high LPS concentrations. Taken together, epithelial BPI may contribute to the innate biochemical barrier characteristic of mucosal surfaces but also may provide a degree of selectivity necessary for effective host responses.
Lipoxins have been implicated in a number of anti-inflammatory pathways. Here, we show that ATLa, a stable analog of aspirin-triggered lipoxin (15), potently induces transcriptional activation of BPI. At present, we have not detailed the molecular pathway(s) by which ATLa induces BPI expression. Little is known about transcriptional pathways of BPI induction, and to date, the BPI promoter has not been characterized. Lipoxins have been widely studied as anti-inflammatory agents and have been demonstrated to inhibit polymorphonuclear leukocyte transmigration across both endothelia and epithelia (4, 24), block polymorphonuclear leukocyte diapedesis within the microcirculation (34) and may initiate the resolution phase of ongoing inflammation (35). Noteworthy is the finding that lipoxins are potent inhibitors of bacterial-induced inflammation in the murine air pouch model (36). In this model, lipoxins inhibited expression of COX-2, an endotoxin-stimulated gene product (37). Thus, it is possible that lipoxins might also dampen inflammatory processes via control of bacterial overgrowth and/or inhibit endotoxin activation via transcriptional induction of BPI.
Our findings have importance to a number of mucosal disease processes. Infectious agents have been implicated as etiologic agents in diseases ranging from periodontal disease (38) to inflammatory bowel disease (39). Attempts to attribute individual diseases to single specific bacterial strains have failed, and thus, attention has turned toward understanding bacterial–host interactions. As such, BPI has become an important expression marker for a number of diseases. For example, high levels of neutrophil-associated BPI are found in the colonic mucosa of patients with ulcerative colitis (40, 41), and autoAbs directed against BPI are proposed seromarkers for the inflammatory bowel diseases (42). Moreover, BPI congeners are currently being evaluated as novel therapies for diseases in which endotoxin is thought to play a role (43), including Crohn's disease (44). Our study provides further support for the notion that BPI plays important anti-infective roles in the gastrointestinal tract, particularly as a molecular shield that dampens the inflammatory influence of endotoxin.
In summary, these results contribute to the present knowledge of mucosal defense mechanisms, and define a previously unappreciated expression of BPI on the surface of alimentary tract epithelia, including those derived from the oral cavity, esophagus and intestine. Moreover, regulated expression of BPI by ATLa provides additional clues to the potent nature of these anti-inflammatory agents and provides for the possible therapeutic induction of BPI in the treatment of mucosal infections.
Acknowledgments
We thank Dr. James Rheinwald for OKF6 cells, Dr. Stephen Carroll for recombinant BPI and anti-BPI Abs, and Drs. Peter Elsbach and Jerrold Weiss for BPI antisera and critical review of the manuscript. This work was supported by National Institutes of Health Grants DK50189, GM38765, and DE13499, by a grant from the Crohn's and Colitis Foundation of America (CCFA), by an unrestricted grant from Xoma U.S. (to L.L.C.), and by a fellowship from the Health Research Board of Ireland.
Abbreviations
LPS
lipopolysaccharide
RT-PCR
reverse transcription–PCR
CFU
colony-forming unit
ATLa
aspirin-triggered lipoxins
LXA
lipoxin A
NGS
normal goat serum
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 3357.
References
- 1.Serhan C N, Haeggstrom J Z, Leslie C C. FASEB J. 1996;10:1147–1158. doi: 10.1096/fasebj.10.10.8751717. [DOI] [PubMed] [Google Scholar]
- 2.Serhan C N. Biochim Biophys Acta. 1994;1212:1–25. doi: 10.1016/0005-2760(94)90185-6. [DOI] [PubMed] [Google Scholar]
- 3.Serhan C N, Oliw E. J Clin Invest. 2001;107:1481–1489. doi: 10.1172/JCI13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serhan C N, Maddox J F, Petasis N, Papayianni A, Brady H R, Colgan S P, Madara J L. Biochemistry. 1995;34:14609–14615. doi: 10.1021/bi00044a041. [DOI] [PubMed] [Google Scholar]
- 5.Claria J, Lee M H, Serhan C N. Mol Med. 1996;2:583–596. [PMC free article] [PubMed] [Google Scholar]
- 6.Elsbach P, Weiss J. Curr Opin Immunol. 1998;10:45–49. doi: 10.1016/s0952-7915(98)80030-7. [DOI] [PubMed] [Google Scholar]
- 7.Ganz T, Weiss J. Semin Hematol. 1997;34:343–354. [PubMed] [Google Scholar]
- 8.Levy O. Antimicrob Agents Chemother. 2000;44:2925–2931. doi: 10.1128/aac.44.11.2925-2931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy O. Blood. 2000;96:2664–2672. [PubMed] [Google Scholar]
- 10.Beamer L J, Carroll S F, Eisenberg D. Science. 1997;276:1861–1864. doi: 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- 11.Dharmsathaphorn K, Madara J L. Methods Enzymol. 1990;192:354–389. doi: 10.1016/0076-6879(90)92082-o. [DOI] [PubMed] [Google Scholar]
- 12.Madianos P N, Papapanou P N, Sandros J. Infect Immun. 1997;65:3983–3990. doi: 10.1128/iai.65.10.3983-3990.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lennon P F, Taylor C T, Stahl G L, Colgan S P. J Exp Med. 1998;188:1433–1443. doi: 10.1084/jem.188.8.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickson M A, Hahn W C, Ino Y, Ronfard V, Wu J Y, Weinberg R A, Louis D N, Li F P, Rheinwald J G. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clish C B, O'Brien J A, Gronert K, Stahl G L, Petasis N A, Serhan C N. Proc Natl Acad Sci USA. 1999;96:8247–8252. doi: 10.1073/pnas.96.14.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 17.Taylor C T, Fueki N, Agah A, Hershberg R M, Colgan S P. J Biol Chem. 1999;274:19447–19450. doi: 10.1074/jbc.274.27.19447. [DOI] [PubMed] [Google Scholar]
- 18.Parkos C A, Colgan S P, Liang A, Nusrat A, Bacarra A E, Carnes D K, Madara J L. J Cell Biol. 1996;132:437–450. doi: 10.1083/jcb.132.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bevilacqua M P, Pober J S, Mendrick D L, Cotran R S, Gimbrone M A. Proc Natl Acad Sci USA. 1987;84:9238–9242. doi: 10.1073/pnas.84.24.9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dittel B N, Wayner E A, McCarthy J B, LeBien T W. Blood. 1993;81:2272–2282. [PubMed] [Google Scholar]
- 21.Weinrauch Y, Foreman A, Shu C, Zarember K, Levy O, Elsbach P, Weiss J. J Clin Invest. 1995;95:1916–1924. doi: 10.1172/JCI117873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick B A, Colgan S P, Delp-Archer C, Miller S I, Madara J L. J Cell Biol. 1993;123:895–907. doi: 10.1083/jcb.123.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colgan S P, Blancquaert M A, Thrall M A, Bruyninckx M A. Vet Immunol Immunopathol. 1992;31:205–227. doi: 10.1016/0165-2427(92)90010-n. [DOI] [PubMed] [Google Scholar]
- 24.Colgan S P, Serhan C N, Parkos C A, Delp-Archer C, Madara J L. J Clin Invest. 1993;92:75–82. doi: 10.1172/JCI116601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weersink A J, van Kessel K P, van den Tol M E, van Strijp J A, Torensma R, Verhoef J, Elsbach P, Weiss J. J Immunol. 1993;150:253–263. [PubMed] [Google Scholar]
- 26.Weiss J, Elsbach P, Olsson I, Odeberg H. J Biol Chem. 1978;253:2664–2672. [PubMed] [Google Scholar]
- 27.Gottadi C J, Caplan M J. J Tissue Cult Methods. 1992;14:173–180. [Google Scholar]
- 28.Pugin J, Schurer-Maly C C, Leturcq D, Moriarity A, Ulevitch R L, Tobias P S. Proc Natl Acad Sci USA. 1993;90:2744–2748. doi: 10.1073/pnas.90.7.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cotran R S, Mayadas-Norton T. Pathol Biol. 1998;46:164–170. [PubMed] [Google Scholar]
- 30.Ouellette A J. Am J Physiol. 1999;277:G257–G261. doi: 10.1152/ajpgi.1999.277.2.G257. [DOI] [PubMed] [Google Scholar]
- 31.Harwig S S, Tan L, Qu X D, Cho Y, Eisenhauer P B, Lehrer R I. J Clin Invest. 1995;95:603–610. doi: 10.1172/JCI117704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imler J L, Hoffmann J A. Trends Cell Biol. 2001;11:304–311. doi: 10.1016/s0962-8924(01)02004-9. [DOI] [PubMed] [Google Scholar]
- 33.Cario E, Podolsky D K. Infect Immun. 2000;68:7010–7017. doi: 10.1128/iai.68.12.7010-7017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raud J, Palmertz U, Dahlen S-E, Hedqvist P. In: Cell–Cell: Interactions in the Release of Inflammatory Mediators. Wong P Y-K, Serhan C N, editors. New York: Plenum; 1991. pp. 185–192. [Google Scholar]
- 35.Levy B D, Clish C B, Schmidt B, Gronert K, Serhan C N. Nat Immun. 2001;2:612–619. doi: 10.1038/89759. [DOI] [PubMed] [Google Scholar]
- 36.Pouliot M, Clish C B, Petasis N A, Van Dyke T E, Serhan C N. Biochemistry. 2000;39:4761–4768. doi: 10.1021/bi992551b. [DOI] [PubMed] [Google Scholar]
- 37.Dubois R N, Abramson S B, Crofford L, Gupta R A, Simon L S, Van De Putte L B, Lipsky P E. FASEB J. 1998;12:1063–1073. [PubMed] [Google Scholar]
- 38.Nisengard R J, Newman M G, Zambon J J. In: Oral Microbiology and Immunology. Nisengard R J, Newman M G, editors. Vol. 2. Philadelphia: Saunders; 1994. pp. 360–384. [Google Scholar]
- 39.Sartor R B. In: Inflammatory Bowel Disease. Kirsner J B, Shorter R J, editors. Baltimore: Williams & Wilkins; 1995. pp. 96–124. [Google Scholar]
- 40.Monajemi H, Meenan J, Lamping R, Obradov D O, Radema S A, Trown P W, Tytgat G N, Van Deventer S J. Gastroenterology. 1996;110:733–739. doi: 10.1053/gast.1996.v110.pm8608882. [DOI] [PubMed] [Google Scholar]
- 41.Haapamaki M M, Haggblom J O, Gronroos J M, Pekkala E, Alanen K, Nevalainen T J. Hepato-Gastroenterology. 1999;46:2273–2277. [PubMed] [Google Scholar]
- 42.Stoffel M P, Csernok E, Herzberg C, Johnson T, Carroll S F, Gross W L. Clin Exp Immunol. 1996;104:54–59. doi: 10.1046/j.1365-2249.1996.d01-654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levin M, Quint P A, Goldstein B, Barton P, Bradley J S, Shemie S D, Yeh T, Kim S S, Cafaro D P, Scannon P J, et al. Lancet. 2000;356:961–967. doi: 10.1016/s0140-6736(00)02712-4. [DOI] [PubMed] [Google Scholar]
- 44.Levy O, Elsbach P. Curr Infect Dis Reports. 2001;3:407–412. doi: 10.1007/s11908-007-1007-y. [DOI] [PubMed] [Google Scholar]