The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis (original) (raw)
Abstract
The covalent attachment of ubiquitin is an important determinant for selective protein degradation by the 26S proteasome in plants and animals. The specificity of ubiquitination is often controlled by ubiquitin-protein ligases (or E3s), which facilitate the transfer of ubiquitin to appropriate targets. One ligase type, the SCF E3s are composed of four proteins, cullin1/Cdc53, Rbx1/Roc1/Hrt1, Skp1, and an F-box protein. The F-box protein, which identifies the targets, binds to the Skp1 component of the complex through a degenerate N-terminal ≈60-aa motif called the F-box. Using published F-boxes as queries, we have identified 694 potential F-box genes in Arabidopsis, making this gene superfamily one of the largest currently known in plants. Most of the encoded proteins contain interaction domains C-terminal to the F-box that presumably participate in substrate recognition. The F-box proteins can be classified via a phylogenetic approach into five major families, which can be further organized into multiple subfamilies. Sequence diversity within the subfamilies suggests that many F-box proteins have distinct functions and/or substrates. Representatives of all of the major families interact in yeast two-hybrid experiments with members of the Arabidopsis Skp family supporting their classification as F-box proteins. For some, a limited preference for Skps was observed, suggesting that a hierarchical organization of SCF complexes exists defined by distinct Skp/F-box protein pairs. Collectively, the data shows that Arabidopsis has exploited the SCF complex and the ubiquitin/26S proteasome pathway as a major route for cellular regulation and that a diverse array of SCF targets is likely present in plants.
Protein degradation is an important posttranscriptional regulatory process that allows cells to respond rapidly to intracellular signals and changing environmental conditions by adjusting the levels of key proteins. One major proteolytic route in eukaryotes involves the ubiquitin (Ub)/26S proteasome pathway (1, 2). Here, proteins destined for degradation become modified by the covalent attachment of multiple Ubs. The ubiquitinated substrates are then recognized by the 26S proteasome and degraded while the Ub moieties are recycled. In both yeast and animals, the Ub/26S proteasome pathway is responsible for removing most abnormal polypeptides and many short-lived cell regulators, which in turn control numerous processes, including the cell cycle, signal transduction, transcription, stress responses, and defense (1). Recent studies infer similar roles for the pathway in plants. For example, mutations in specific components of the Arabidopsis Ub/26S proteasome pathway block embryogenesis, hormonal responses, floral homeosis, photomorphogenesis, circadian rhythms, senescence, and pathogen invasion (2). Although many of the components of this pathway have been described, the mechanisms for target recognition remain poorly defined.
Ub conjugation is achieved through an ATP-dependent reaction cascade involving the sequential action of three enzymes, E1, E2s, and E3s (1). As the final enzyme in the cascade, the E3s or Ub-protein ligases are responsible for recognizing the substrate and facilitating Ub transfer, which results in the formation of an isopeptide bond between a lysl ɛ-amino group of the target and the C-terminal glycine carboxyl group of the Ub. Often a chain of Ubs is attached, typically using lysine-48 within the Ub moieties as the acceptor site for polymerization.
Currently, five types of E3s have been identified that differ according to their subunit organization and/or mechanism of Ub transfer (1, 3, 4). One important E3 type is the SCF complex, which in yeast (Saccharomyces cerevisiae) is composed of four primary subunits—cullin1/Cdc53, Rbx1/Roc1/Hrt1, Skp1, and an F-box protein (3, 4). The cullin1, Rbx1, and Skp1 subunits appear to form the core ligase activity, with Rbx1 recruiting the E2 bearing an activated Ub. F-box proteins perform the crucial role of delivering appropriate targets to the complex. These proteins all contain a degenerate ≈60-aa N-terminal motif called the F-box, which is used to interact with the rest of the SCF complex by binding to the Skp subunit. The C-terminal portions of F-box proteins typically contain a variable protein-interaction domain that binds the target and thus confers specificity to the SCF complex. In yeast and animals, a number of F-box proteins are present, easily classified by the nature of this interaction domain (3, 4). In yeast, for example, the F-box protein Grr1p contains C-terminal leucine-rich repeats (LRRs) that recruit the phosphorylated forms of the cyclins Cln1p and Cln2p to the SCFGrr1 complex, whereas the Cdc4 F-box protein contains WD-40 repeats that recruit the phosphorylated form of the cyclin-dependent kinase inhibitor Sic1 to the SCFcdc4 complex. The existence of multiple F-box proteins, each with unique specificity or specificities would allow SCFs to ubiquitinate a diverse array of substrates.
Recent studies in Arabidopsis thaliana indicate that F-box proteins and SCF complexes play critical roles in various aspects of plant growth and development (2). ASK1, one of the 19 Skp proteins in Arabidopsis (5), is involved in male gametogenesis and floral organ identity (6, 7). Arabidopsis F-box (FBX) genes have been implicated in auxin (TIR1) and jasmonate signaling (COI1) (8, 9), floral homeosis (UFO) (10), flowering time and circadian clocks (ZTL/FKF/LKP2) (11–13), leaf senescence and lateral shoot branching (ORE1/MAX2) (14, 15), and photomorphogenesis (EID1) (16). Cursory searches of the Arabidopsis genome detected many other FBX loci, suggesting that they comprise a large gene family (17, 18). For example, Andrade et al. (19) identified 48 genes encoding proteins with Kelch repeats following a degenerate F-box motif. To further determine the complexity of the F-box protein family in plants, we conducted an exhaustive search for _FBX_-related loci in the Arabidopsis genome. This search identified 694 potential FBX genes, making this superfamily one of the largest found so far in plants. The size and diversity of this collection suggest that SCF E3s impact most aspects of plant cell biology presumably by recognizing and ubiquitinating a wide array of protein targets.
Materials and Methods
Prediction of F-Box Proteins.
All F-box proteins published as of 01/17/01 were used as queries in multiple blast searches (20) against the Arabidopsis genome (annotated 01/17/2001). Sequences predicted to encode F-boxes by the smart and pfam databases (http://smart.embl-heidelberg.de) were then used in blast searches against the complete, nonredundant, re-annotated Arabidopsis genome (released 02/17/01). One set of blast searches used only the F-box motifs and had an E value cutoff of 10, whereas the other set used the full-length proteins with a cutoff of 1e-10. Of the 1,849 nonredundant sequences retrieved, 650 were annotated by smart/pfam as containing an F-box motif. Fifteen of the 650 loci were re-annotated as two F-box genes and another locus as three. Hand analysis led to the identification of 27 additional loci. Four of those loci are missing in the current annotation and have been named using the Arabidopsis Genome Initiative (AGI) numbers of the genes that bracket the locus. The additional 44 genes and the revised annotations are described in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org. Four genes (At1g51290, At5g02700, At3g28410, and At3g49030) appear to contain two F-boxes; in these cases, the domain with a better match to the consensus F-box motif was used.
Alignments, Phylogenetic Analysis, and EST Identification.
Intron/exon organizations were based on the Arabidopsis genome annotation, our re-annotation data, and analysis of ESTs. F-box sequences were retrieved and aligned using CLUSTALX PC V.1.81 (21). An unrooted phylogenetic tree of the alignment was created by MEGA V.2.1 (22), using the _p_-distance method with gaps treated by pairwise deletion and a 1,000 bootstrap replicate. Alignments and trees of selected groups were generated by CLUSTALX MAC V.1.6b (21). Percent identities and similarities were calculated using MACBOXSHADE V.2.11 (Institute of Animal Health, Pirbright, U.K.). Additional domains were predicted using smart and pfam databases, blast searches, and sequence alignments. Chromosomal maps were generated using the Genome Pixelizer Tcl/Tk script [www.atgc.org/GenomePixelizer (released 02/15/2002)]. The predicted mRNA sequences were used to conduct blast searches against the GenBank Arabidopsis EST dataset of 07/18/2001. EST sequences with an 80–100% match were retrieved, checked for redundancy, and analyzed to verify the match.
Yeast Strains, Media, and Yeast Two-Hybrid (Y2H) Technique.
Two haploid yeast strains, YPB2 and LB414α (like YPB2 but Lys+ and MATα), were from (8). FBX cDNAs were expressed as C-terminal fusions to Gal4-BD in plasmid pBI770 (8) and transformed into YPB2. ASK cDNAs were expressed as C-terminal fusions to the Gal4-AD in pBI771 (23) and transformed into LB414α. The following constructs, pBI770, cruc-pBI770, pBI771, cruc-pBI771 (23), UFO-pBI770, ASK1-pBI771, and ASK2-pBI771 (10), were from William Crosby (PBI, Saskatoon, SK, Canada). Yeast were grown at 28°C on YPAD or synthetic complete (SC) media (23) and were transformed according to ref. 24. Yeast strains were mated on YPAD for 24 h and then grown on SC medium lacking leucine and tryptophan. Strains were tested for β-galactosidase activity by using an overlay assay (25) and for resistance to 10 mM 3-amino-1′,2′,4′ triazole (3-AT) added to selective SC medium minus histidine (23). For each mating, 10 μl of culture with an OD600 1 × 10−2 were spotted and grown at 28°C for 4 days.
Construction of GAL4-BD and GAL4-AD Fusions.
Coding regions of FBX and ASK genes were PCR amplified from Arabidopsis thaliana ecotype Col-1. Templates included a 2–3-kb size-selected Arabidopsis lambda ZAP cDNA library, total RNA converted to cDNA by RT-PCR, genomic DNA (for intron-less genes), or when available full-length ESTs provided by the Arabidopsis Biological Resource Center, Columbus, OH. Primers were designed to add 30–33 nucleotides to allow the in-frame insertion of the products into the desired AD and BD plasmids by homologous recombination. The PCR products were transformed into the yeast strains along with their respective plasmids (linearized with _Sal_I and _Not_I) as described at ratios of 3:1 (insert to plasmid) (26). Plasmids were identified by PCR and verified as correct by DNA sequence analysis.
Results
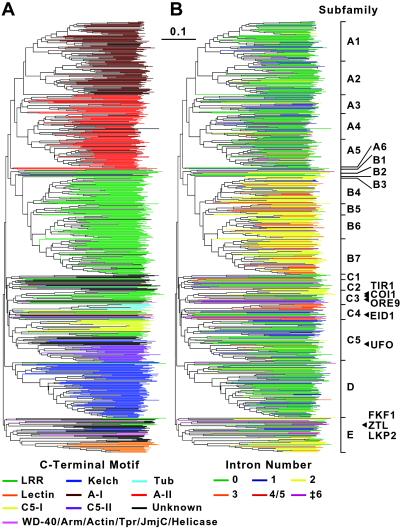
From reiterative blast searches of the Arabidopsis genome, using as queries a variety of known F-box proteins from animals, yeast and plants, we identified a large collection of putative Arabidopsis FBX loci. Subsequent analysis condensed this collection to 694 unique FBX genes predicted to encode proteins with an F-box motif. When compared with closely related genes, a few of the 694 loci have characteristics of pseudogenes—i.e., they potentially contain premature translation stop codons and/or errors in splicing. Because we are not yet able to confirm these possibilities, these potentially aberrant loci were included in the analysis. Using the F-box motif sequences alone for the alignments, an unrooted phylogenetic tree of the entire collection was created (Fig. 1). The F-box of each protein was defined as the region corresponding to residues 107–168 in human Skp2 (an F-box protein), which includes most of the important contacts for Skp1 binding (27, 28). The first ≈40 residues represent the core of the Skp-binding site and are followed by an ≈20-residue variable domain with additional contacts that may help confer a Skp binding preference (27, 28).
Fig 1.
Phylogenetic tree of the F-box protein superfamily from Arabidopsis. The 60-aa F-box motifs from the 694 potential F-box proteins were aligned by clustalx. The alignment then was used to generate an unrooted phylogenetic tree with MEGA 2.1, using the _p_-distance method and a bootstrap value of 1,000. Individual members of the tree are color-coded by (A) the nature of the domain(s) C-terminal to the F-box or (B) the number of introns within the respective genes. Unknown represents F-box proteins that are truncated or have no obvious C-terminal interaction domain. The 20 groups identified from the phylogenetic analysis are marked on the right. Arrowheads denote the position of Arabidopsis F-box proteins identified previously by genetic analyses. The bar represents the branch length equivalent to 0.1 amino acid changes per residue. Expanded versions of the trees bearing the AGI numbers for each locus can be found in Figs. 7 and 8, which are published as supporting information on the PNAS web site.
Hand analysis of the phylogenetic tree revealed five distinct clades of F-box proteins, which we have tentatively assigned to five families designated A-E (Fig. 1). The A-C families were further divided into 18 subfamilies, giving 20 distinct groups of proteins. Additionally, At1g27490 forms its own clade near the A2 subfamily and appears to be a pseudogene. Both the families and subfamilies vary substantially in size—e.g., the A1 subfamily contains 64 members whereas the A6 subfamily contains only 3 (Table 1). A sequence alignment of the F-box cores from representative members of the 20 groups revealed conserved islands separated by regions with weak homology (Fig. 2). Importantly, many of the conserved residues correspond with those known by x-ray crystallographic analysis to be important for Skp association (27, 28), strongly suggesting that this collection can also bind Skps. When members of each of the 20 F-box groups were aligned individually, additional conservation within both the core and variable F-box domains was evident, which may be important for preferential binding of the different Skps (Fig. 1 and data not shown).
Table 1.
Organization and expression of the Arabidopsis F-box protein subfamilies
| Subfamily | A1 | A2 | A3 | A4 | A5 | A6 | B1 | B2 | B3 | B4 | B5 | B6 | B7 | C1 | C2 | C3 | C4 | C5 | D | E |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. genes | 64 | 53 | 30 | 41 | 46 | 3 | 6 | 5 | 2 | 43 | 18 | 38 | 57 | 8 | 18 | 22 | 26 | 66 | 91 | 56 |
| No. expressed | 6 | 9 | 10 | 3 | 5 | 2 | 2 | 2 | 0 | 6 | 6 | 9 | 11 | 7 | 13 | 16 | 19 | 12 | 31 | 22 |
| No. ESTs | 19 | 61 | 25 | 3 | 9 | 3 | 4 | 8 | 0 | 8 | 10 | 19 | 25 | 31 | 51 | 111 | 138 | 361 | 34 | 72 |
Fig 2.
Sequence alignment of representative Arabidopsis F-box motifs. The 42-aa core F-box sequences from UFO, the human F-box protein Skp2, and from representatives of each of the 20 F-box protein groups from Arabidopsis were aligned by clustalx and displayed with macboxshade, using a threshold of 55%. Conserved and similar amino acids are shown in black and gray boxes, respectively. Dots denote gaps. Designations on the left identify the group and AGI number for each protein. Arrowheads mark the amino acids positions important for the Skp/F-box interactions between human Skp1 and Skp2 (27, 28). The alignment of all 694 F-box motifs can be found in Table 3, which is published as supporting information on the PNAS web site.
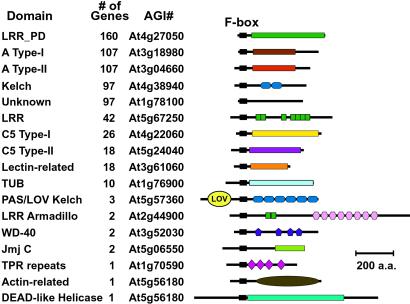
Most Arabidopsis F-box proteins are predicted to contain a long region C-terminal to the F-box. Analysis of these C-terminal regions by smart and pfam revealed a diverse array of potential protein-interaction domains that presumably participate in substrate recognition, including leucine-rich (LRR), Kelch, WD-40, Armadillo (Arm), and tetratricopeptide (TPR) repeats, and Tub, actin, DEAD-like helicase, and jumonji (Jmj)-C domains (Fig. 3). The most abundant F-box types are those containing LRR or Kelch repeats. LRRs are 20- to 29-aa motifs with positionally conserved leucines or other aliphatic residues. Multiple LRRs assemble together to form an arched docking structure (29). Forty-two of the F-box proteins, including TIR1, COI, and ORE9/MAX2 (8, 9, 14, 15), matched one of the LRR consensus sequences in the smart database. An additional 160 proteins were found to contain a plant-specific derivative of the cysteine-containing LRR (designated here LRR_PD). We identified 100 Kelch repeat-containing F-box proteins, including 48 that were previously described (19). Individual Kelch repeats form four-stranded β-sheets that assemble together to create a β-propeller tertiary structure (11, 19). Additionally, three of the Kelch/F-box proteins, LKP2, ZTL, and FKF, previously shown to be required for flowering and circadian rhythms in Arabidopsis, have an N-terminal PAS/LOV domain and may function as flavin-containing photoreceptors (11–13).
Fig 3.
Diagrams of representative Arabidopsis F-box proteins with information on the structure and position of the C-terminal interaction domains. Shown on the left are the types of C-terminal domains, the number of F-box proteins predicted to have those domains, and the AGI number of the representative diagramed on the right.
Ten Arabidopsis F-box proteins were predicted to contain a C-terminal Tub domain. This 120-aa domain was first identified in mouse TUB1, which is involved in controlling obesity (30). With the exception of At1g61940, which appears to be truncated, the Arabidopsis Tub domains are 44–54% similar to their mouse TUB1 counterpart, including the presence of several residues invariant among TUB1 homologs from other organisms (30). Two proteins (At5g21040 and At3g52030) contain a string of WD-40 repeats and two genes (At2g44900 and At3g60350) contain a string of Arm repeats (29). The Jmj C domain, predicted to be a metal-binding site (31), was found in two F-box proteins, At1g78280 and At5g06550. Finally, single FBX genes encoded one of the following: an actin-related domain (At5g56180), a tandem array of the 34-aa TPR (At1g70590) (29), or a potential DEAD-Box helicase domain (At5g56180) (32).
A large number of the Arabidopsis F-box proteins (374) had C-terminal regions with no obvious similarity to motifs in the smart and pfam databases. We used blast searches and large-scale alignments among members of this group to uncover C-terminal structures. These alignments identified five potentially unique domains (Fig. 1A). The A Type-I and -II domains, which are mainly found in the A1–2 subfamilies and the A3–6 subfamilies, respectively, are related 200-aa hydrophobic regions with positionally conserved tryptophans and can be distinguished from each other by additional regions of conservation. The C5 Type-I and -II motifs are enriched in a number of positionally conserved bulky hydrophobic and charged residues and appear to be plant-specific (data not shown). We identified 18 F-box proteins mainly in the E family that have C-terminal homology to the squash lectin PP2 (33), suggesting that these F-box proteins may detect glycosylated substrates. For the remaining 97 in the “unknown” group, no consensus C-terminal domains were detected, suggesting they are either improperly annotated, are pseudogenes, or use novel domains to interact with their respective targets.
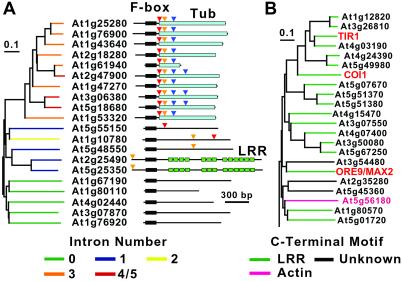
When the phylogenetic tree of F-box sequences was color-coded to reflect the nature of the C-terminal domain, a striking clustering of protein-interaction domains was evident (Fig. 1A). All but 12 of the 100 Kelch-containing F-box proteins are in the D family and all but two of the 160 LRR_PD-containing F-box proteins are in the B family. With only one exception, the new domains present in the A1/2 and A3–6 subfamilies are restricted to those subfamilies. Likewise, all of the F-box-Tub and most of the F-box-lectin-related domain proteins are localized to small clades within the C4 and E subfamilies, respectively. This correlation further supports the phylogenetic relationships in the F-box tree and suggests a co-evolution of the F-box motif with the target-interaction domain. However, regions of the tree can be found where proteins with similar F-boxes have widely different C-terminal domains (Fig. 1A). Examples of this phenomenon can be seen in the C3 and C4 subfamilies where clusters of F-box proteins enriched in LRRs and Tubs or LRRs and actin-related F-box proteins are on adjacent branches (Fig. 4 A and B).
Fig 4.
Expanded sections of the Arabidopsis F-box protein phylogenetic tree. The sections were extracted from the complete tree created using the 60-aa F-box motif for alignment (see Fig. 1). (A) Section of the tree showing part of the C3 subfamily containing members with a C-terminal Tub domain. The phylogeny on the left is color-coded based on the number of introns in the respective genes. The diagrams on the right show the predicted organization of the corresponding proteins showing the Tub and LRR domains in cyan and green, respectively. The arrowheads in the protein diagrams mark the intron positions. Yellow, blue, and red arrowheads denote whether the intron interrupts the gene after the first, second or third nucleotide of the codon, respectively. (B) Section of the tree showing part of the C4 subfamily containing the TIR1, COI1, and ORE9/MAX2 proteins. The tree is color-coded based on the nature of the region C-terminal to the F-box. The bars represent the branch length equivalent to 0.1 amino acid changes per residue.
Further support for the phylogenetic tree was provided by analysis of the intron/exon organization. Surprisingly, 45% of the FBX genes were predicted to be intron-free, a much higher percentage than predicted for Arabidopsis genes overall (20.7%) (18). When the phylogenetic tree is color-coded based on intron number a similar clustering within the families and subfamilies is evident (Fig. 1B). For example, almost all of the genes in Family B have two introns with many having identical insertion sites for one or both. Within the C4 subfamily, many of the Tub FBX genes have similarly positioned introns (Fig. 4A). Given that the F-box families and subfamilies can be organized in similar ways by using either the F-box motif, the nature of the variable C-terminal domain, or the positions of introns/exons, the tree presented in Fig. 1 likely represents a good approximation of the relationships within this superfamily.
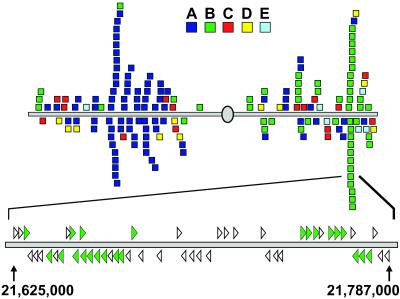
The sheer size of the FBX superfamily suggests that it evolved in Arabidopsis through a large number of duplication events. To address this, we examined the locations of the FBX loci on the Arabidopsis chromosomes. Analysis of all five chromosomes can be found in Fig. 9, which is published as supporting information on the PNAS web site, with a view of Chromosome III shown in Fig. 5. When the locations are colored according to the F-box type, a substantial clustering of related FBX genes is evident. For example, chromosome III contains two regions concentrated in members of the A and B families. In one 150-kbp region alone, 22 of the 45 total predicted open-reading frames are members of the B7 subfamily. Overall, 35.9% of the 694 FBX genes are arranged in tandem repeats of two to seven genes. Additionally, approximately 29% of the 694 genes are within five genes of a closely related gene in the phylogenetic tree and approximately 17% are the adjacent gene. This finding suggests that tandem duplications of chromosomal regions played a major role in creating the large array of FBX loci in Arabidopsis.
Fig 5.
Location of various FBX genes in chromosome III of Arabidopsis. Each gene is color-coded according to its family as shown in Fig. 1. Below is an expanded view of a ≈150-kbp segment of chromosome III showing the position and orientation of several clusters of B7 FBX genes (green arrowheads). The direction of the arrowheads indicate the 5′ to 3′ orientation of each gene. Open arrowheads identify non-FBX genes. The nucleotide location of the chromosomal segment is shown below. Similar descriptions of the other Arabidopsis chromosomes can be found in Fig. 9.
Despite the significant relationships among the F-box proteins within each subfamily, direct sequence alignments between close relatives suggest that most of the Arabidopsis F-box proteins do not have obvious functional paralogs. As a consequence, we predict that many will have unique substrates and thus perform different functions in Arabidopsis. This can best be illustrated by analysis of an expanded region of the C3 subfamily containing TIR1, COI1, and ORE9/MAX2 (Fig. 4B). Despite conservation of the F-box motif, presence of similar C-terminal LRRs, and similar intron/exon positions, genetic analysis indicates that the corresponding proteins are functionally distinct (8, 9, 14, 15).
Evidence of the expression of the Arabidopsis FBX genes was provided by searches against the Arabidopsis EST database. ESTs were detected for 27.5% of the FBX loci, including representatives from each of the 20 groups except the two loci of the B3 subfamily (Table 1). Subfamilies C and E had the highest levels of apparent expression with 47.9% and 39.3% of their genes having ESTs. Overall, the number of FBX genes with ESTs was much lower than that estimated for whole genome (60.3%) (18), suggesting that many are expressed at low levels or restricted to specific cells or developmental stages.
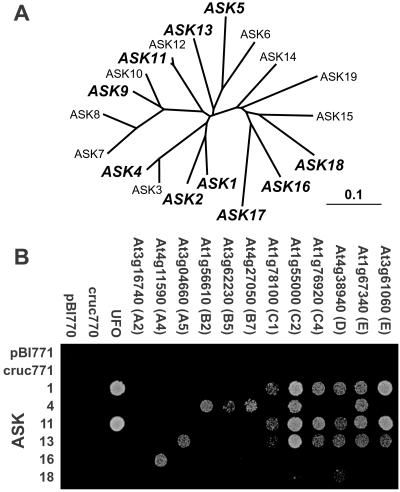
Final confirmation that the Arabidopsis FBX genes actually encode F-box proteins will require biochemical analyses. As a first step, Y2H analysis was used to test whether a sampling of F-box proteins from each of the five families could interact with Arabidopsis orthologs of Skp1. Arabidopsis contains 19 Skp1-related proteins, designated ASK1–19 (Fig. 6A; ref. 5). Ten representative ASKs (ASK1, 2, 4, 5, 9, 11, 13, and 16–18) were tested pairwise with 23 representative F-box proteins by Y2H for β-galactosidase activity and for survival on 3-amino-1′, 2′, 4′ triazole (3-AT) (see Table 4). In all cases, full-length F-box and ASK proteins were expressed as C-terminal fusions with the Gal4 DNA-binding (BD) and activation (AD) domains, respectively. As shown in Fig. 6B and Table 4, Y2H interactions were detected between 8 of the 10 ASKs tested and representatives of all five F-box protein families, A–E. These interactions were similar to that of the BD-UFO/AD-ASK1 combination, which was used as the control Skp/F-box protein pair (10). No activity was detected on expression of any BD-F-box or AD-ASK fusion protein alone, demonstrating that the interactions between the pairs are significant. Importantly, some of the F-box proteins showed an ASK preference. For example, representative members of the A4 (At4g11590) and A2 (At3g16740) subfamilies preferred ASK16 and ASK11, respectively, whereas four of the five representatives of the B family interacted only with ASK4. Also, even though both members of the E family interacted with ASKs 1, 2, 11, and 13, they displayed distinct preferences; At3g61060 interacted with ASK5 and At1g67340 interacted with ASK4. These results suggest that there is an ASK specificity among the F-box proteins.
Fig 6.
Interaction of representative Arabidopsis F-box proteins with various Arabidopsis Skps (ASKs) by Y2H analysis. (A) Phylogenetic analysis of the 19 ASK proteins. Those in bold were used as bait in the Y2H analysis. (B) Y2H analyses of representative F-box protein/ASK pairs by growth selection at 28°C for 4 days on 10 mM 3-AT. See Table 4, which is published as supporting information on the PNAS web site, for complete results.
Discussion
Previous genetic analyses indicated that F-box proteins and SCF complexes are critical regulators in plants (2). Here we provide further support for their importance with the discovery of almost 700 possible FBX genes in the Arabidopsis genome. This superfamily represents ≈2.7% of the predicted genes in Arabidopsis, making it one of the largest so far described in plants. The size of this superfamily also represents a substantial expansion relative to other eukaryotes. Yeast is predicted to have 14 FBX genes, Drosophila melanogaster to have 24, humans to have at least 38, and Caenorhabditis elegans to have 337 (refs. 3 and 18, and data not shown). A compensatory expansion is also evident for the Skps, with 1 SKP gene in yeast, 6 in Drosophila, 8 in humans, 21 in C. elegans, and 19 in Arabidopsis (5, 34, 35), suggesting that nematodes and plants have evolved an extensive combinatorial system of SCF complexes beyond that used by these other eukaryotes.
Phylogenetic analysis supports the division of the Arabidopsis F-box protein superfamily into 5 distinct families and 20 subfamilies. Given the substantial diversity among the protein sequences, this analysis was based on the F-box motif sequence alone. However, further analysis of the tree with respect to three other criteria, potential target-interaction domains, intron/exon organization, and chromosomal clustering of the corresponding genes, supports the groupings. It appears then that this tree provides a fairly accurate representation of the F-box protein superfamily as a whole.
Y2H analysis showed that representative members of the five major families of F-box proteins could bind to one or more members of the Arabidopsis ASK family, providing support that many of these proteins can function in SCF complexes. Importantly, some, and possibly all of the F-box proteins appear to preferentially associate with specific ASKs. Similar binding studies of various Skp/F-box protein pairs from C. elegans revealed a comparable preference (34, 35). These results support the notion that through combinatorial arrangements of the various subunits, specific types of SCF complexes can be formed. Considering that Arabidopsis is predicted to encode 2 Rbx subunits, over 5 cullins, 19 ASKs, and 694 F-box proteins, a wide array of specialized SCF combinations is possible. These complexes could offer not only greater substrate selectivity, but also an additional way to regulate the SCF complex by controlling levels of subunits other than F-box proteins.
Five of the 23 F-box constructs tested showed no significant interactions with the 10 ASKs. This failure could indicate that these proteins are not bona fide F-Box proteins or that they function in other related Ub ligation complexes. As an example of the latter, a set of target recognition factors within the mammalian Von-Hippel Lindau ubiquitination complex use an motif functionally similar to the F-Box to promote their association with the rest of the ligase machinery through a Skp1-like protein (36). However, more likely possibilities are that these proteins (i) interact with members of the Arabidopsis ASK family not tested here, (ii) are not active in the Y2H system, and/or (iii) require either the target (37) or additional factors to associate with the rest of the SCF complex. Recently, it has become apparent that some SCF complexes require additional proteins [e.g., Cks1 (38)], which may help stabilize Skp/F-Box protein binding.
The chromosomal arrangements of the FBX genes implicate tandem duplications in the expansion of the gene family. Additionally, given the placement of proteins with similar F-box motifs but dissimilar C-terminal interaction domains within the same subfamily, it appears that domain shuffling has occurred to further expand F-box protein diversity (39). Direct sequence comparisons suggest that most of the F-box proteins do not have functional paralogs. This diversity is also supported by genetic analysis of the C3 subgroup genes TIR1, COI1, and ORE9/MAX2. The phenotypes of mutants in these genes suggest that the wild-type proteins have unique functions even though they have similar F-box motifs and C-terminal domains (8, 9, 14, 15).
Extrapolated further, one can imagine that many of the 694 F-box proteins have distinct targets and thus as a whole could impact many facets of Arabidopsis biology. Certainly, the diverse array of C-terminal interaction domains present suggests that F-box proteins use a broad palette of mechanisms for target recognition. Some exploit protein-interaction domains common in F-box proteins from yeast and animals (e.g., LRR, WD-40), whereas others appear to employ domains unique to plant F-box proteins, such as the actin-like, lectin-like, and Tub domains, and the four domains found in the A and C families. Such diversity would then imply that the recognition sites within the targets are also heterogeneous, which in turn may explain the apparent absence of a common degradation signal among Ub/26S proteasome targets. For many of the SCF/target interactions characterized thus far, phosphorylation of the target appears to be an essential determinant in F-box protein recognition (3, 4). Consequently, it is possible that many of the protein kinases in plants actually function to modulate target/F-box protein interactions, thus intimately connecting these two protein superfamilies in proteolytic control. Based on the genomic complexity revealed thus far, the Ub/26S proteasome pathway is easily one of the most elaborate regulatory systems in Arabidopsis, further supporting the importance of protein turnover in plant cell control.
Supplementary Material
Supporting Information
Acknowledgments
We thank Drs. Eddy Risseeuw and William Crosby for the Y2H strains and control plasmids. This work was supported by U.S. Department of Agriculture–National Research Initiative Competitive Grants Program Grant 00-35301-9040 and National Science Foundation Arabidopsis 2010 Program Grant MCB-0115870 (both to R.D.V.).
Abbreviations
- AGI, Arabidopsis Genome Initiative
- LRR, leucine-rich repeat
- Ub, ubiquitin
- Y2H, yeast two-hybrid
References
- 1.Hershko A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67**,** 425-479. [DOI] [PubMed] [Google Scholar]
- 2.Callis J. & Vierstra, R. D. (2000) Curr. Opin. Plant Biol. 3**,** 381-386. [DOI] [PubMed] [Google Scholar]
- 3.Kipreos E. T. & Pagano, M. (2000) Genome Biol. 5**,** 1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deshaies R. J. (1999) Annu. Rev. Cell Dev. Biol. 15**,** 435-467. [DOI] [PubMed] [Google Scholar]
- 5.Farras R., Ferrando, A., Jasik, J., Kleinow, T., Okresz, L., Tiburcio, A., Salchert, K., del Pozo, C., Schell, J. & Koncz, C. (2001) EMBO J. 20**,** 2742-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang M., Hu, Y., Lodhi, M., McCombie, W. R. & Ma, H. (1999) Proc. Natl. Acad. Sci. USA 96**,** 11416-11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao D., Yang, M., Solava, J. & Ma, H. (1999) Dev. Genet. 25**,** 209-223. [DOI] [PubMed] [Google Scholar]
- 8.Gray W. M., del Pozo, J. C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W. L., Yang, M., Ma, H. & Estelle, M. (1999) Genes Dev. 13**,** 1678-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie D. X., Feys, B. F., James, S., Nieto-Rostro, M. & Turner, J. G. (1998) Science 280**,** 1091-1094. [DOI] [PubMed] [Google Scholar]
- 10.Samach A., Klenz, J. E., Kohalmi, S. E., Risseeuw, E., Haughn, G. W. & Crosby, W. L. (1999) Plant J. 20**,** 433-445. [DOI] [PubMed] [Google Scholar]
- 11.Somers D. E., Schultz, T. F., Milnamow, M. & Kay, S. A. (2000) Cell 101**,** 319-329. [DOI] [PubMed] [Google Scholar]
- 12.Nelson D. C., Lasswell, J., Rogg, L. E., Cohen, M. A. & Bartel, B. (2000) Cell 101**,** 331-340. [DOI] [PubMed] [Google Scholar]
- 13.Schultz T. F., Kiyosue, T., Yanovsky, M., Wada, M. & Kay, S. A. (2001) Plant Cell 13**,** 2659-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stirnberg P., van De Sande, K. & Leyser, H. M. (2002) Development (Cambridge, U.K.) 129**,** 1131-1141. [DOI] [PubMed] [Google Scholar]
- 15.Woo H. R., Chung, K. M., Park, J. H., Oh, S. A., Ahn, T., Hong, S. H., Jang, S. K. & Nam, H. G. (2001) Plant Cell 13**,** 1779-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dieterle M., Zhou, Y. C., Schafer, E., Funk, M. & Kretsch, T. (2001) Genes Dev. 15**,** 939-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao W. & Jang, J. (2000) Trends Plant Sci. 5**,** 454-457. [DOI] [PubMed] [Google Scholar]
- 18.Arabidopsis Genome Initiative (2000) Nature (London) 408**,** 796-815. [DOI] [PubMed] [Google Scholar]
- 19.Andrade M. A., Gonzalez-Guzman, M., Serrano, R. & Rodriguez, P. L. (2001) Plant Mol. Biol. 46**,** 603-614. [DOI] [PubMed] [Google Scholar]
- 20.Altschul S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25**,** 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 24**,** 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17**,** 1244-1245. [DOI] [PubMed] [Google Scholar]
- 23.Kohalmi S. E., Nowak, J. & Crosby, W. L. (1997) in Differentially Expressed Genes in Plants: A Bench Manual, eds. Hansen, E. & Harper, G. (Taylor & Francis, London), pp. 63–82.
- 24.Gietz R. D. & Woods, R. A. (1994) in Molecular Genetics of Yeast: Practical Approaches, ed. Johnston, J. A. (Oxford Univ. Press, New York), pp. 121–134.
- 25.Duttweiler H. M. (1996) Trends Genet. 12**,** 340-341. [DOI] [PubMed] [Google Scholar]
- 26.Oldenburg K. R., Vo, K. T., Michaelis, S. & Paddon, C. (1997) Nucleic Acids Res. 25**,** 451-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulman B. A., Carrano, A. C., Jeffrey, P. D., Bowen, Z., Kinnucan, E. R., Finnin, M. S., Elledge, S. J., Harper, J. W., Pagano, M. & Pavletich, N. P. (2000) Nature (London) 408**,** 381-386. [DOI] [PubMed] [Google Scholar]
- 28.Zheng N., Schulman, B., Song, L., Miller, J. J., Jeffrey, P. D., Wang, P., Chu, C., Koepp, D. M., Elledge, S. J., Pagano, M., et al. (2002) Nature (London) 416**,** 703-709. [DOI] [PubMed] [Google Scholar]
- 29.Andrade M. A., Perez-Iratxeta, C. & Ponting, C. P. (2001) J. Struct. Biol. 134**,** 117-131. [DOI] [PubMed] [Google Scholar]
- 30.Kleyn P. W., Fan, W., Kovats, S. G., Lee, J. J., Pulido, J. C., Wu, Y., Berkemeier, L. R., Misumi, D. J., Holmgren, L., Charlat, O., et al. (1996) Cell 85**,** 281-290. [DOI] [PubMed] [Google Scholar]
- 31.Clissold P. M. & Ponting, C. P. (2001) Trends Biochem. Sci. 26**,** 7-9. [DOI] [PubMed] [Google Scholar]
- 32.Kim, J., Kim, J.-H., Kim, D.-H., Kang, H.-Y., Bae, S.-H., Pan, Z.-C. & Seo, Y.-S. (2002) J. Biol. Chem., in press.
- 33.Golecki B., Schulz, A. & Thompson, G. A. (1999) Plant Cell 11**,** 127-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nayak S., Santiago, F. E., Jin, H., Lin, D., Schedl, T. & Kipreos, E. T. (2002) Curr. Biol. 12**,** 277-287. [DOI] [PubMed] [Google Scholar]
- 35.Yamanaka A., Yada, M., Imaki, H., Koga, M., Ohshima, Y. & Nakayama, K. I. (2002) Curr. Biol. 12**,** 267-275. [DOI] [PubMed] [Google Scholar]
- 36.Kamura T., Sato, S., Haque, D., Liu, L., Kaelin, W. G., Jr., Conaway, R. C. & Conaway, J. W. (1998) Genes Dev. 12**,** 3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galan J. M. & Peter, M. (1999) Proc. Natl. Acad. Sci. USA 96**,** 9124-9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganoth D., Bornstein, G., Ko, T. K., Larsen, B., Tyers, M., Pagano, M. & Hershko, A. (2001) Nat. Cell Biol. 3**,** 321-324. [DOI] [PubMed] [Google Scholar]
- 39.Morgenstern B. & Atchley, W. R. (1999) Mol. Biol. Evol. 16**,** 1654-1663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information