Resolution of Prochlorococcus and Synechococcus Ecotypes by Using 16S-23S Ribosomal DNA Internal Transcribed Spacer Sequences (original) (raw)
Abstract
Cultured isolates of the marine cyanobacteria Prochlorococcus and Synechococcus vary widely in their pigment compositions and growth responses to light and nutrients, yet show greater than 96% identity in their 16S ribosomal DNA (rDNA) sequences. In order to better define the genetic variation that accompanies their physiological diversity, sequences for the 16S-23S rDNA internal transcribed spacer (ITS) region were determined in 32 Prochlorococcus isolates and 25 Synechococcus isolates from around the globe. Each strain examined yielded one ITS sequence that contained two tRNA genes. Dramatic variations in the length and G+C content of the spacer were observed among the strains, particularly among Prochlorococcus strains. Secondary-structure models of the ITS were predicted in order to facilitate alignment of the sequences for phylogenetic analyses. The previously observed division of Prochlorococcus into two ecotypes (called high and low-B/A after their differences in chlorophyll content) were supported, as was the subdivision of the high-B/A ecotype into four genetically distinct clades. ITS-based phylogenies partitioned marine cluster A Synechococcus into six clades, three of which can be associated with a particular phenotype (motility, chromatic adaptation, and lack of phycourobilin). The pattern of sequence divergence within and between clades is suggestive of a mode of evolution driven by adaptive sweeps and implies that each clade represents an ecologically distinct population. Furthermore, many of the clades consist of strains isolated from disparate regions of the world's oceans, implying that they are geographically widely distributed. These results provide further evidence that natural populations of Prochlorococcus and Synechococcus consist of multiple coexisting ecotypes, genetically closely related but physiologically distinct, which may vary in relative abundance with changing environmental conditions.
In open-ocean ecosystems, carbon fixation is dominated by the marine cyanobacteria Prochlorococcus and Synechococcus. Together they have been shown to contribute between 32 and 80% of the primary production in oligotrophic oceans (14, 21, 24, 60). Prochlorococcus is closely related to the marine cluster A Synechococcus, based on analyses using gene sequences from 16S rRNA (16S rDNA) and rpoC1, a subunit of DNA-dependent RNA polymerase (37, 59). However, the two genera have very different light-harvesting systems. Prochlorococcus contains divinyl chlorophyll a (chl a2) and both monovinyl and divinyl chlorophyll b (chl b) as its major photosynthetic pigments, rather than chlorophyll a and phycobiliproteins that are typical of cyanobacteria (7, 8, 13).
Cultured isolates of Prochlorococcus have been divided into two genetically and physiologically distinct groups, referred to as ecotypes because their differing physiologies have implications for their ecological distributions (28, 31, 44). High-B/A isolates have larger ratios of chl b/a2 and are able to grow at extremely low irradiances (less than 10 μmol of quanta[Q] m−2 s−1), where low-B/A isolates are incapable of growth. Low-B/A isolates have lower chl b/a2 ratios and are able to grow maximally at higher light intensities, where high-B/A isolates are inhibited (28). The ecotypes also differ in their sensitivity to copper toxicity, with low-B/A isolates able to grow at free cupric ion concentrations five times higher than those high-B/A isolates can tolerate (26). Furthermore, all low-B/A isolates tested to date are incapable of using nitrate or nitrite as a nitrogen source, while some high-B/A isolates can grow on nitrite (30).
The high- and low-B/A ecotypes were originally named for their difference in optimal growth irradiance (low- and high-light adapted, respectively [31]). However, considering that the ecotypes are differently “adapted” to a multitude of environmental factors, the pigment ratio, a property of the cell rather than of the environment in which it thrives, is a better descriptor for the ecotypes (28, 44). Furthermore, the chl b/a2 ratio, unlike the growth response to a range of irradiances, is a phenotype that can be rapidly and easily measured in a novel isolate.
The 16S rDNA sequences of the Prochlorococcus ecotypes correlate with their physiology. Strains of the low-B/A ecotype are phylogenetically very closely related (99% identity in 16S rDNA sequence) and form a clade well supported by bootstrap values (31, 44, 58). Strains of the high-B/A ecotype have a lower degree of identity in their 16S rDNA sequence (97 to 98%) and are not monophyletic but instead form at least three independent branches (44). High-B/A Prochlorococcus strains also have a higher degree of sequence identity to marine cluster A Synechococcus strains than the low-B/A ecotype. In fact, branching orders between some high-B/A Prochlorococcus strains and marine cluster A Synechococcus strains are not well resolved using 16S rDNA sequences (31, 44, 58).
The strains assigned to marine cluster A Synechococcus are genetically and physiologically diverse (64). All contain phycoerythrins as their major light-harvesting pigments, and some possess the chromophore phycourobilin (PUB), which can attach to phycoerythrin in combination with phycoerythrobilin (PEB) (33). The relative amounts of PUB and PEB vary among strains, and some strains are able to chromatically adapt by changing the ratio of their chromophores in response to different wavelengths of light (35). In addition, some isolates are capable of a novel form of swimming motility (6, 65, 66). Synechococcus strains also vary in the G+C content of their genomes and in the ability to utilize organic nutrient sources such as urea (10, 65).
Genetic diversity in marine cluster A Synechococcus strains has been examined in a few strains using 16S rDNA sequences (58) and more extensively using rpoC1 sequences (54, 55). Using rpoC1, a collection of strains from the California Current could be divided into two lineages consistent with their high or low PUB amounts. However, each of these lineages was distinct from the typical laboratory model high- and low-PUB strains (WH 8103 and WH 7803, respectively), suggesting that pigment content alone may not resolve the multiple ecotypes of marine cluster A Synechococcus. Motility, however, does appear to be correlated with phylogeny, as all motile isolates characterized to date are closely related (55). Another recently described clade consists of strains whose members are capable of altering their pigment content in response to light quality in an acclimation process known as chromatic adaptation (35).
In most eubacteria, the genes for rRNA are organized in operons, with the genes encoding the 16S, 23S, and 5S rRNAs separated by internal transcribed spacer (ITS) regions (15). The ITS contains antitermination box B-box A motifs which prevent premature termination of transcription (5) and also have a role in holding the secondary structure of the nascent rRNA for processing to mature rRNAs (1). The spacer between the 16S and 23S rRNA genes can encode 0, 1, or 2 tRNA genes. Because the ITS exhibits a great deal of length and sequence variation, it has been used in many bacterial groups to delineate closely related strains (3, 9, 19). Whole-genome sequences suggest that low-B/A Prochlorococcus strains possess a single rRNA operon, while high-B/A Prochlorococcus and marine Synechococcus strains possess two identical rRNA operons (http://www.jgi.doe.gov/JGI_microbial/html/index.html).
Here we report on use of the ITS as a phylogenetic tool to identify clades which may represent ecologically distinct populations of Prochlorococcus and Synechococcus. Although isolates in culture collections may not represent the full extent of diversity due to biases introduced by isolation protocols, they provide crucial physiological information to attach to the phylogenetic clusters. By examining a wide range of physiologically diverse isolates of Prochlorococcus and marine cluster A Synechococcus, we lay the groundwork for informed studies of genetic diversity and distributions in field populations of these oceanic cyanobacteria and provide a framework for interpreting their phenotypic evolution and delineating their taxonomy.
MATERIALS AND METHODS
Isolation of strains.
Thirty-two isolates of Prochlorococcus from diverse oceanic regions were employed in this study (Table 1). Isolation conditions, physiology, and genetic data have been reported previously for many of the strains (see Table 1). The majority were isolated by filtering seawater through two stacked 0.6-μm-pore-size filters and enriching with nutrients (7). Five (MIT 9302, MIT 9303, MIT 9311, MIT 9312, and MIT 9313) were isolated by sorting on a flow cytometer (31). Seven of the strains (SS120, SS35, SS51, SS2, MED4, SB, and GP2) have been rendered clonal by serial dilution, and one (MED4Ax) has been rendered free of heterotrophic bacteria by plating (45).
TABLE 1.
Origins and characteristics of the Prochlorococcus strains used in this studya
| Group and strain | Isolation | Chl b/a2 ratio | Strain originb | Reference | GenBank accession no. | |
|---|---|---|---|---|---|---|
| Location | Depth (m) | |||||
| Low-B/A Prochlorococcus clade I | ||||||
| MED4 | Mediterranean Sea | 5 | 0.20 | MIT | 29 | AF397673 |
| MED4Axc | Mediterranean Sea | 5 | ND | MIT | 45 | AF397674 |
| MIT 9515 | Equatorial Pacific | 15 | 0.24 | MIT | This work | AF397675 |
| MB11E08 | Monterey Bay | 0 | — | 51 | AY033307 | |
| MB11F02 | Monterey Bay | 0 | — | 51 | AY033310 | |
| Low-B/A Prochlorococcus clade II | ||||||
| AS9601 | Arabian Sea | 50 | 0.34 | R. Olson and A. Shalapyonok | 47 | AF397677 |
| GP2 | West Pacific | 150 | 0.32 | A. Shimada | 48 | AF397678 |
| MIT 9107 | South Pacific | 25 | 0.34 | MIT | 58 | AF397679 |
| MIT 9116 | South Pacific | 25 | 0.31 | MIT | This work | AF397680 |
| MIT 9123 | South Pacific | 25 | 0.34 | MIT | This work | AF397681 |
| MIT 9201 | South Pacific | 0 | 0.27 | MIT | 28 | AF397682 |
| MIT 9202 | South Pacific | 79 | 0.31 | MIT | 28 | AF397683 |
| MIT 9215 | Equatorial Pacific | 0 | 0.30 | MIT | 28 | AF397684 |
| MIT 9301 | Sargasso Sea | 90 | 0.42 | MIT | This work | AF397685 |
| MIT 9302 | Sargasso Sea | 100 | 0.51 | MIT | 31 | AF397686 |
| MIT 9311 | Gulf Stream | 135 | 0.47 | MIT | This work | AF397687 |
| MIT 9312 | Gulf Stream | 135 | 0.34 | MIT | 31 | AF397688 |
| MIT 9314 | Gulf Stream | 180 | 0.28 | MIT | This work | AF397689 |
| MIT 9321 | Equatorial Pacific | 50 | 0.27 | MIT | This work | AF397690 |
| MIT 9322 | Equatorial Pacific | 0 | 0.28 | MIT | This work | AF397691 |
| MIT 9401 | Sargasso Sea | 0 | 0.34 | MIT | This work | AF397692 |
| SB | West Pacific | 40 | 0.32 | A. Shimada | 49 | AF397693 |
| RS810 | Red Sea | 80 | ND | D. Lindell and A. Post | 23 | AF397676 |
| High-B/A Prochlorococcus clade I | ||||||
| NATL1A | North Atlantic | 30 | 0.77 | F. Partensky | 40 | AF397694 |
| NATL2A | North Atlantic | 10 | 0.97 | F. Partensky | 46 | AF397695 |
| PAC1 | Tropical Pacific | 100 | ND | L. Campbell | 39 | AF397696 |
| High-B/A Prochlorococcus clade II | ||||||
| LG | Sargasso Sea | 120 | ND | MIT, B. Palenik | 7 | AF397697 |
| SS2d | Sargasso Sea | 120 | ND | MIT | This work | AF397698 |
| SS35d | Sargasso Sea | 120 | ND | MIT | This work | AF397699 |
| SS51d | Sargasso Sea | 120 | ND | MIT | This work | AF397700 |
| SS120d | Sargasso Sea | 120 | 1.41 | MIT | 7 | AF397701 |
| High-B/A Prochlorococcus clade III | ||||||
| MIT 9211 | Equatorial Pacific | 83 | 1.37 | MIT | 28 | AF397702 |
| High-B/A Prochlorococcus clade IV | ||||||
| MIT 9303 | Sargasso Sea | 100 | 0.97 | MIT | 31 | AF397703 |
| MIT 9313 | Gulf Stream | 135 | 0.91 | MIT | 31 | AF397704 |
Twenty-two of 25 Synechococcus strains used in this study (Table 2)are clonal and have been described previously (65). Strain WH 9908 was isolated from Woods Hole in April 1999 (by M. Sullivan), when the water temperature was less than 10°C, and rendered clonal by picking a single colony from an agar plate. Strains C8015, RS9705, and RS9708 were isolated from the Gulf of Aqaba, Red Sea (23). Of the 25 strains, 22 are phycoerythrin-containing marine cluster A strains. Two strains from marine cluster B (WH 8101 and WH 5701) and one freshwater strain assigned to the Cyanobium cluster (PCC 6307) were also included (Table 2).
TABLE 2.
Origins and characteristics of the Synechococcus strains used in this studya
| Group and strain | Isolation location | _A_495/_A_545 | Other datab | Originc | Reference | GenBank accession no. |
|---|---|---|---|---|---|---|
| Marine A Synechococcus clade I | ||||||
| WH 8015 | Woods Hole | 0.44 | WH | 65 | AF397717 | |
| WH 8016 | Woods Hole | 0.4 | WH | 65 | AF397718 | |
| WH 8020 | Sargasso Sea | 0.78 | CA | WH | 65 | AF397719 |
| WH 9908 | Woods Hole | NDg | WH | This work | AF397720 | |
| Marine A Synechococcus clade II | ||||||
| WH 6501 | Tropical Atlantic | 0.43 | WH | 65 | AF397706 | |
| WH 8002 | Gulf of Mexico | 0.48 | WH | 65 | AF397707 | |
| WH 8005 | Gulf of Mexico | 0.44 | WH | 65 | AF397708 | |
| WH 8012 | Sargasso Sea | 0.4 | WH | 65 | AF397709 | |
| WH 8109 | Sargasso Sea | 0.89 | WH | 65 | AF397710 | |
| Marine A Synechococcus clade III | ||||||
| C8015 | Red Sea | >1 | Motile | D. Lindell and A. Post | 23 | AF397711 |
| WH 8102 | Western Caribbean | 2.06 | Motile | WH | 65 | AF397712 |
| WH 8103 | Sargasso Sea | 2.4 | Motile, NCA | WH | 65 | AF397713 |
| WH 8112 | Sargasso Sea | Variable | Motile, CA | WH | 65 | AF397714 |
| WH 8113 | Sargasso Sea | Variable | Motile, CA | WH | 65 | AF397715 |
| WH 8406 | Gulf of Mexico | 0.84 | Motile | WH | 65 | AF397716 |
| Marine A Synechococcus clade IV | ||||||
| EBAC392 | Monterey Bay | —d | — | 4 | AF268237 | |
| MB11A04 | Monterey Bay | — | — | 51 | AY033297 | |
| MB11E09 | Monterey Bay | — | — | 51 | AY033308 | |
| Marine A Synechococcus clade V | ||||||
| RS9705 | Red Sea | <1 | D. Lindell and A. Post | 23 | AF397725 | |
| RS9708 | Red Sea | <1 | D. Lindell and A. Post | 23 | AF397726 | |
| WH 7803 | Sargasso Sea | 0.39 | WH | 65 | AF397727 | |
| Marine A Synechococcus clade VI | ||||||
| WH 7805 | Sargasso Sea | No PUB | WH | 65 | AF397721 | |
| WH 8008 | Gulf of Mexico | No PUB | WH | 65 | AF397722 | |
| WH 8017 | Woods Hole | No PUBe | WH | 65 | AF397723 | |
| WH 8018 | Woods Hole | No PUB | WH | 65 | AF397724 | |
| Marine B Synechococcus | ||||||
| WH 8101 | Woods Hole | NAf | WH | 65 | AF397728 | |
| WH 5701 | Long Island Sound | NAf | WH | 65 | AF397729 | |
| Cyanobium Synechococcus | ||||||
| PCC 6307 | Wisconsin lake | NAf | PCC | 42 | AF397730 |
Culture conditions.
For physiology experiments, 20-ml batch cultures of Prochlorococcus strains were grown in acid-washed 50-ml glass test tubes at 24°C on a 14 h:10 h light-dark cycle under 18 μmol of Q white light m−2 s−1. This light level is roughly equivalent to 1% of surface irradiance, which corresponds to a depth of ≈100 m (assuming typical oligotrophic water values for surface irradiance of _I_0 = 2,000 μmol of Q m−2 s−1 and an extinction coefficient of k = 0.045 m−1 [18]). Medium was made from 0.2-μm-filtered, autoclaved Sargasso Sea water enriched with Pro2 nutrients (final concentrations: 10 μM NaH2PO4, 50 μM NH4Cl, 100 μM urea, 1.17 μM EDTA, 8 nM Zn, 5 nM Co, 90 nM Mn, 3 nM Mo, 10 nM Se, 10 nM Ni, and 1.17 μM Fe) (28).
For DNA extraction, Prochlorococcus strains were grown in 60-ml acid-washed polycarbonate bottles using the medium and culture conditions described above. Synechococcus strains were grown for DNA extraction in 100-ml acid-washed flasks in SN medium under constant light (65).
Pigment measurements.
The light-dependent physiology of 15 isolates of Prochlorococcus was examined by measuring their chl b/a2 ratios (28). All experiments were performed in triplicate. Cells were acclimated to the experimental conditions (see above) for at least 10 generations before measurements were taken. A known volume (18 to 22 ml) of exponential-phase culture was filtered onto a 25-mm Whatman GF/F under low vacuum, and filters were stored in liquid nitrogen until extraction. Pigments were extracted with 90% acetone according to established protocols (13, 27) and quantified on a spectrophotometer (Becton Dickinson DU640). Unlike high-pressure liquid chromatography, spectrophotometric methods cannot resolve divinyl chlorophyll b2 from “normal” monovinyl chlorophyll b; thus, total (b1 + b2) values are reported. Pigment concentrations were calculated according to the trichromatic equations of Jeffrey and Humphreys (17).
DNA isolation, PCR, and sequencing.
DNA was extracted from 50 ml of late-exponential-phase cultures by using a modified protocol involving cetyltrimethylammonium bromide, phenol, and chloroform (2). The ITS/23S fragment was amplified using primers 16S-1247f (CGTACTACAATGCTACGG) and 23S-1608r (CYACCTGTGTCGGTTT). Primer 16S-1247f was designed using available 16S rDNA sequences from Prochlorococcus and Synechococcus strains (31, 44, 58) and is a perfect match only to cyanobacterial 16S rDNA sequences, as judged by using the Probe Match function of the latest release of the Ribosomal Database Project (25).
Reactions were done in 25-μl volumes with final concentrations of reactants as follows: 0.25 mM deoxynucleoside triphosphates, 0.1 mM each primer, 0.1 to 1 μg of template DNA, and 0.1 to 0.5 U of the high-fidelity polymerase Pfu (Stratagene, La Jolla, Calif.). Cycling parameters were 94°C for 4 min, followed by 30 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 6 min, and a final extension at 72°C for 10 min using either a Robocycler (Stratagene) or a PTC100 (MJ Research). Amplified fragments were visualized on agarose gels. Only one band was observed from each culture. Control reactions lacking template DNA were always performed in parallel and gave no products.
For sequencing, PCRs were performed in quintuplicate and pooled, and primers were removed using Strataprep columns (Stratagene). Products were sequenced on an ABI377 or ABI310 (PE Biosystems) automated sequencer using Big Dye terminator sequencing kits according to the manufacturer's instructions. The ITS was sequenced bidirectionally using primer 16S-1247f and primers internal to the PCR fragment: ITS-Alaf (TWTAGCTCAGTTGGTAGAG), ITS-Alar (CTCTACCAACTGAGCTAWA), and 23S-241r (TTCGCTCGCCRCTACT).
Secondary-structure determination.
The complete rRNA operon sequences of Prochlorococcus strains MED4 and MIT 9313 and Synechococcus strains WH 8102 were obtained from the Department of Energy's Joint Genome Institute (http://www.jgi.doe.gov/JGI_microbial/html/index.html), where the genome sequences of these three strains are near completion. For each of the three strains, ITS sequences obtained from the genome project were identical to those determined independently from PCR products as described above. To simplify the construction of secondary-structure models of the ITS, the 16S rRNA, 23S rRNA, and 5S rRNA sequences were deleted from the predicted transcripts. The remaining sequences were folded using mfold (70). Structures were refined and displayed in LoopDLoop (http://iubio.bio.indiana.edu/soft/molbio/loopdloop/).
Phylogenetic analysis.
Sequences were edited and aligned manually based on the predicted secondary structures using the Genetic Data Environment (50) or BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Phylogenetic analyses and calculation of fractional identities and G+C contents used PAUP* version 4b8 (53). Phylogenetic analyses employed either 233 or 434 positions of the 16S-23S rDNA spacer and did not include sequences from the tRNAs.
Distance trees were inferred using minimum evolution as the objective criterion and paralinear (logdet) or HKY85 distances. Distance and maximum-parsimony bootstrap analyses (1,000 resamplings) were performed with heuristic searches utilizing random addition and tree-bisection reconnection branch-swapping methods. Maximum-likelihood analyses used the HKY85 model of nucleotide substitution with rate heterogeneity and empirical nucleotide frequencies. The gamma shape parameter and the transition-transversion ratio were initially estimated from a distance topology and refined by iterative likelihood searches. Likelihood bootstrap analyses (100 resamplings) were performed with heuristic searches and tree-bisection reconnection branch-swapping methods starting from a neighbor-joining tree. Phylogenetic trees were visualized with Treeview (34).
RESULTS AND DISCUSSION
Physiology of Prochlorococcus strains.
In order to expand the physiological framework for our phylogenetic analyses from that established by Moore and Chisholm (28) based on 10 strains of Prochlorococcus, we analyzed the chl b/a2 ratio of 15 additional Prochlorococcus strains (Table 1) grown at 18 μmol of Q m−2 s−1 illumination. This growth irradiance was chosen based on previous physiological data (28) because it was a level at which all strains were capable of growth and where ecotypic differences in the chl b/a2 ratio were pronounced.
At this light level, chl b/a2 ratios for 13 of the strains fell between 0.2 and 0.5 (Table 1), placing them physiologically in the low-B/A ecotype. The remaining two strains, NATL1A and NATL2A, had much higher chl b/a2 ratios (0.77 and 0.97, respectively), classifying them as high-B/A strains. In addition, NATL1A and NATL2A exhibited growth and pigment responses over a range of irradiances that were similar to those of the other high-B/A strains (E. Cohen, G. Rocap, and S. W. Chisholm, unpublished data).
With the addition of these pigment data for these 15 strains, it is clear that the majority of the Prochlorococcus isolates in culture collections are of the low-B/A ecotype (19 compared to only 6 high-B/A strains among the 25 physiologically characterized strains in this study). This is in spite of concentrated efforts to isolate additional high-B/A strains into culture by sampling from the deep euphotic zone, where they are presumed to predominate. This suggests that there may be an isolation bias for the low-B/A strains. For example, filtration steps used to eliminate Synechococcus cells, which are larger, could also preferentially remove larger Prochlorococcus cells. Indeed, based on flow cytometric measurements, the high-B/A clade IV strains MIT 9303 and MIT 9313 are the largest Prochlorococcus isolates we have in our collection. It is noteworthy in this regard that both of these isolates were obtained by flow cytometric sorting, not filtration. In addition, low-B/A strains have a higher μmax at their optimum growth light intensity than do high-B/A strains (28), which could allow them to take over mixed enrichment cultures if care is not taken to avoid these relatively high light levels.
Sequences of ITS.
Sequences of the 16S-23S rDNA ITS region were determined for 32 strains of Prochlorococcus and 25 clonal strains of Synechococcus (Tables 1 and 2). Although the majority of the Prochlorococcus isolates have not been rendered clonal, PCR amplifications yielded single-band products, and few sequence ambiguities were observed. Spacer sequences of all of the strains examined contained genes encoding two tRNAs, for isoleucine and alanine, as has been observed in freshwater Synechococcus sp. strain PCC 6803 (56).
Sequencing of the 57 strains resulted in 45 unique ITS sequences. Prochlorococcus strains SS120, SS2, SS35, and SS51 (Table 1) are clonal derivatives of the primary culture LG, and all five strains had identical sequences. The axenic strain MED4Ax also had an identical sequence to its parent strain MED4. Strains MIT 9107, MIT 9116, and MIT 9123, coisolates from the same water sample from the South Pacific, were identical to each other, as were coisolates MIT 9312 and MIT 9311 from the Gulf Stream. Strains MIT 9321, MIT 9322, and MIT 9401 also possessed identical ITS sequences, although MIT 9401 was isolated from the Sargasso Sea while the other two are from the Equatorial Pacific. Synechococcus strains WH 7805, WH 8008, and WH 8018, isolated from the Sargasso Sea, the Gulf of Mexico, and Woods Hole, respectively (Table 2), also possess identical ITS sequences.
Since the same set of primers was used repeatedly on different templates, it could be argued that the identical sequences are the result of contamination in the PCR. However, many of the identical sequences were prepared several months apart, and other cultures were amplified in the interim that yielded different sequences. Furthermore, in no case did a Prochlorococcus culture yield a sequence phylogenetically affiliated with sequences from Synechococcus cultures or vice versa, nor did any Prochlorococcus culture yield a sequence which was incongruent with its pigment phenotype, as might be expected to have occurred if the identical sequences were the result of chance contamination.
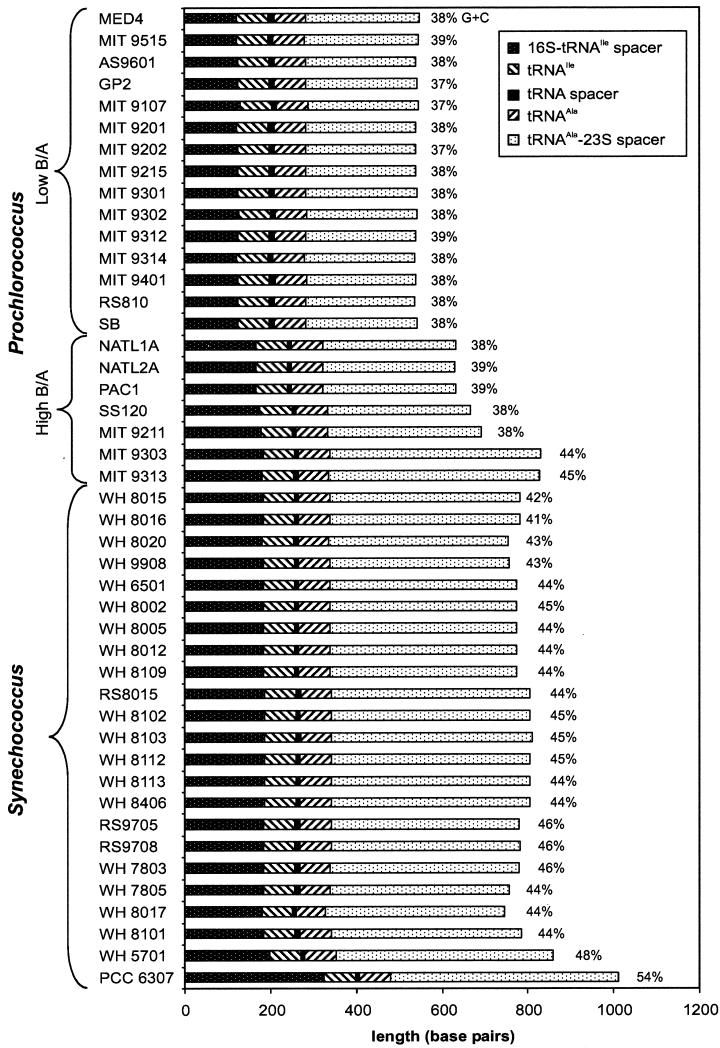
Marked differences in the length of the ITS were observed among the 45 unique sequences (Fig. 1). Lengths of the ITS ranged from 537 bp in Prochlorococcus strain MIT 9314 to 1,012 bp in Synechococcus sp. strain PCC 6307. Within the Prochlorococcus strains, the length differences were strongly correlated with ecotype. All of the low-B/A Prochlorococcus strains had ITS regions ranging in length from 537 to 548 bp. The high-B/A Prochlorococcus strains had much longer ITS sequences, and there was a larger range of lengths among their sequences (Fig. 1). NATL1A, NATL2A, PAC1, SS120, and MIT 9211 ITS sequences ranged in length from 632 to 693 bp, while MIT 9303 and MIT 9313 had spacers of 831 and 829 bp, respectively. The ITS in marine cluster A Synechococcus strains ranged from 747 bp (WH 8017) to 810 bp (WH 8103). The majority of the length difference was in the 3′ end of the spacer (tRNAAla-23S spacer), which ranged from 255 to 531 bp among the strains examined.
FIG. 1.
Length of the ITS region between the 16S and 23S rDNAs in Prochlorococcus and Synechococcus isolates. Low-B/A Prochlorococcus isolates all have short spacers, while high-B/A isolates have longer spacer lengths and are not as similar to one another. The freshwater strain PCC 6307 has a much longer spacer than the marine Synechococcus and Prochlorococcus isolates. The G+C content of the 16S-23S rDNA ITS is indicated to the right of each bar. Low-B/A Prochlorococcus and four high-B/A Prochlorococcus isolates have a lower G+C content than two high-B/A Prochlorococcus and Synechococcus isolates. For sets of isolates with identical ITS sequences (see text), only one isolate is represented here.
Substantial differences also existed in the G+C content of the ITS (Fig. 1). Whereas the low-B/A Prochlorococcus ITS sequences had the lowest G+C content (37 to 39%), the high-B/A Prochlorococcus spanned a range of values (37 to 45%). The G+C content of sequences from NATL1A, NATL2A, PAC1, SS120, and MIT 9211 was quite similar to that of the low-B/A isolates, ranging from 37 to 39%. However, MIT 9303 and MIT 9313 had higher G+C contents (44 and 45%, respectively). Values for MIT 9303 and MIT 9313 were in the range of the majority of the marine Synechococcus strains (41 to 46% G+C). As with its much longer length, the G+C content of the ITS sequence from Cyanobium cluster Synechococcus sp. strain PCC 6307 was markedly different (54% G+C) from all of the Prochlorococcus and marine Synechococcus strains. The majority of the strains had lower G+C contents in the 16S-tRNAIle spacer and the tRNAAla-23S spacer than in the more highly conserved tRNAs. In fact, for MED4 the spacer sequence without the tRNA has a G+C content that is the same (30%) as that of the whole genome (http://www.jgi.doe.gov/JGI_microbial/html/index.html).
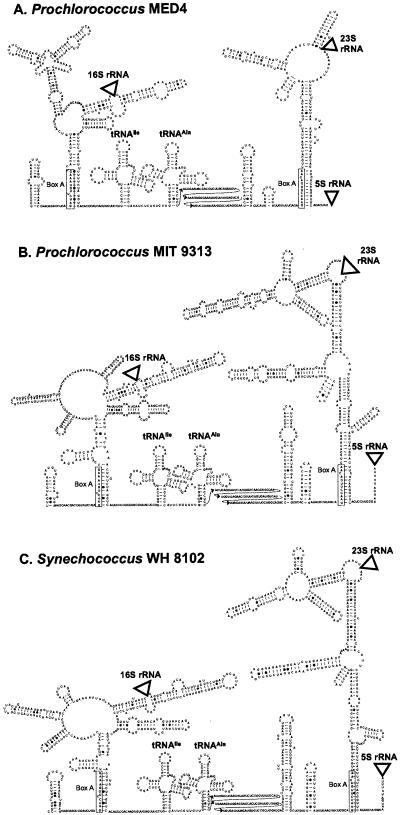
ITS secondary structure.
Complete rRNA operon sequences (derived from the whole genome sequence) from Prochlorococcus strains MED4 and MIT 9313 and Synechococcus strain WH 8102 were used to predict the secondary structure of the ITS in these three strains (Fig. 2). These proposed structures show conserved features observed in other bacteria (16, 32) and are consistent with general rRNA processing patterns in prokaryotes (1). For example, the spacers on both sides of the 16S rRNA (the 16S upstream spacer and 16S-tRNAIle spacer) are capable of base pairing with each other (Fig. 2), which may help bring the 3′ and 5′ ends of the 16S rRNA sequence together so it can be cleaved to a mature 16S rRNA (1).
FIG. 2.
Predicted secondary structures of the ITS regions of the rRNA operon in (A) Prochlorococcus strain MED4, (B) Prochlorococcus strain MIT 9313, and (C) Synechococcus strain WH 8102. Locations of the 16S rRNA, 23S rRNA, and 5S rRNA are represented by triangles. All strains have antiterminator box B loops and box A sequences conserved in most bacterial ITS sequences, as well as genes for tRNAs isoleucine and alanine typical of cyanobacterial ITS sequences. Sequence corresponding to the box A motif is enclosed in a rectangle. The 5′ region of the tRNAAla-23S rRNA spacer (between the tRNAAla and the box B loop) for which no structure was inferred is shown in three rows of text to save space.
Similarly, the tRNAAla-23S spacer and the spacer downstream of the 23S rRNA are also capable of base pairing (Fig. 2), which may promote processing of the 23S rRNA. Antitermination motifs (5) can also be identified in these three strains, including the box B, a stem-loop of unconserved sequence which precedes the conserved sequence motif box A. In the region upstream from the 16S rRNA, the box A-like sequence GAUC(C/U)UGGAAAG can base pair with the 16S-tRNAIle spacer to form a double-stranded processing site. Similarly, in the tRNAIle-23S spacer, there is a box B spacer loop preceding a box A sequence, GAACCUUGACAA. This box A and the region immediately downstream from it are involved in base pairing with the region downstream of the 23S rRNA to form a double-stranded processing site.
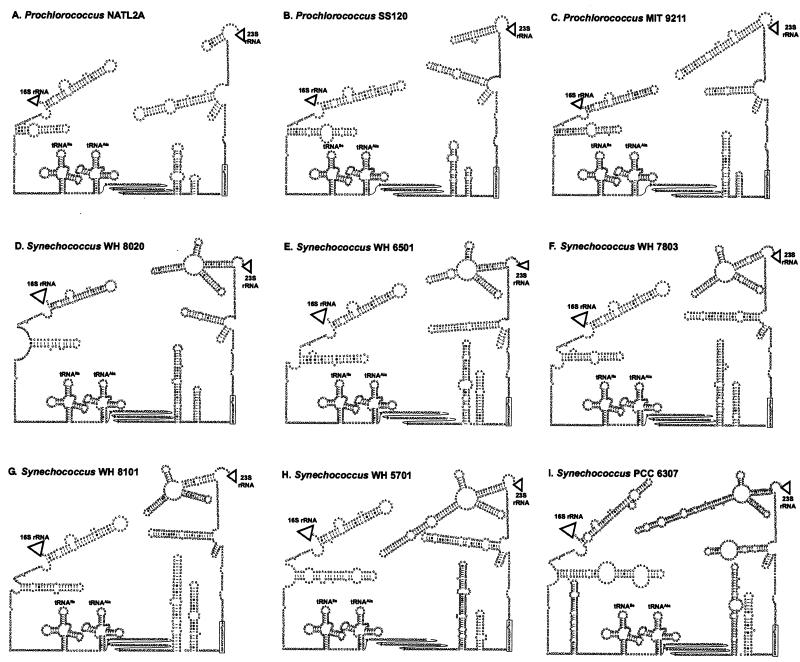
The secondary structures of these three strains and the identification of the double-stranded processing sites allowed the prediction of structures for strains in which sequence upstream of the 16S rDNA and downstream of 23S rDNA was not determined (Fig. 3). The nine strains depicted in Fig. 3 were chosen based on preliminary phylogenetic analyses as being representative of the ITS sequence types in Prochlorococcus and Synechococcus. As with the three strains described above, a box B loop preceding a box A sequence [GAACCUUGA(C/A)AA] could be identified in all these strains.
FIG. 3.
Predicted secondary structures of the 16S-23S rRNA ITS in three high-B/A Prochlorococcus strains and six Synechococcus strains identified in this study. (A) Prochlorococcus strain NATL2A, (B) Prochlorococcus strain SS120, (C) Prochlorococcus strain MIT 9211, (D) marine cluster A Synechococcus strain WH 6501, (E) marine cluster A Synechococcus strain WH 8020, (F) marine cluster A Synechococcus strain WH 7803, (G) marine cluster B Synechococcus strain WH 8101, (H) marine cluster B Synechococcus strain WH 5701, and (I) Cyanobium cluster Synechococcus sp. strain PCC 6307. Locations of the 16S rRNA, 23S rRNA, and 5S rRNA are represented by triangles. All strains have an antiterminator box B loop and box A sequence conserved in most bacterial ITS sequences as well as genes for tRNAs isoleucine and alanine typical of cyanobacterial ITS sequences. Sequence corresponding to the box A motif is enclosed in a rectangle. The 5′ region of the tRNAAla-23S rRNA spacer (between the tRNAAla and the box B loop) for which no structure was inferred is shown in three rows of text to save space.
Structural details evident in the predicted structures help explain the differences in ITS length between the ecotypes and may be informative characters in their own right. For example, the region immediately upstream of the 23S rRNA forms a cloverleaf in Synechococcus sp. strain PCC 6307, which is smaller in the marine Synechococcus strains (Fig. 3). This structure is also present in Prochlorococcus strain MIT 9313 (Fig. 2B), but is reduced to a single stem-loop in Prochlorococcus strain MIT 9211, and this stem-loop has become successively smaller in Prochlorococcus strains SS120 and NATL2A and is not present in MED4. The progression of shorter ITS sequences is consistent with data from the whole-genome sequences of MED4 and MIT 9313, suggesting that there has been an overall genome minimization in MED4 (http://www.jgi.doe.gov/JGI_microbial/html/index.html).
Phylogenetic relationships among Prochlorococcus strains.
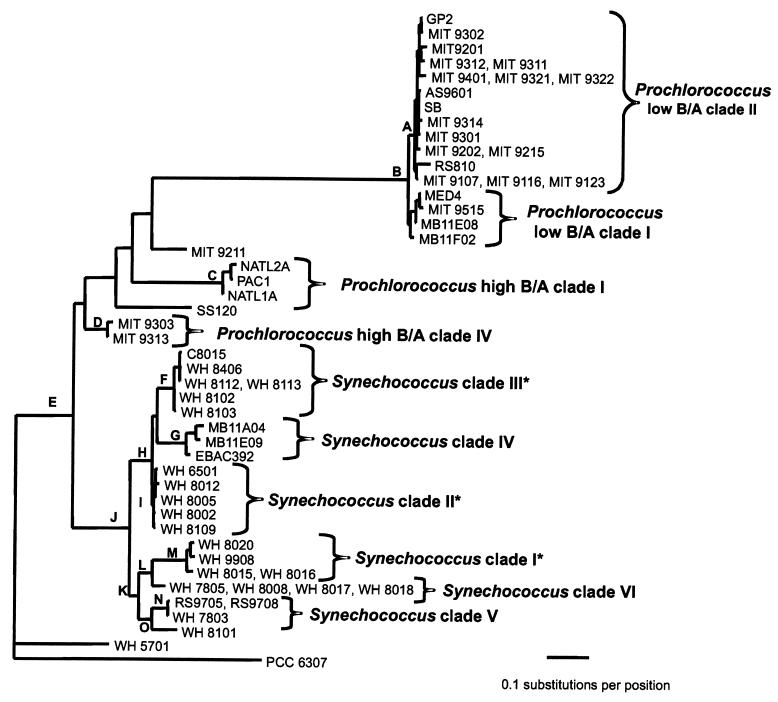
Phylogenetic analyses used alignments based on the inferred secondary structures of the ITS. In phylogenetic analyses confined to 233 bp of the 16S-23S spacer, the low-B/A Prochlorococcus strains form a clade (node B) well supported by bootstrap values (Fig. 4A). All of the low-B/A isolates show greater than 93% identity. Within the low-B/A clade there is evidence for at least two subdivisions. One of these, labeled low-B/A II in Fig. 4A, is equivalent to the Prochlorococcus low-B/A clade II defined previously by 16S rDNA sequences (44, 67). This clade has high bootstrap support in these analyses, and its members show greater than 95% identity in the ITS sequence.
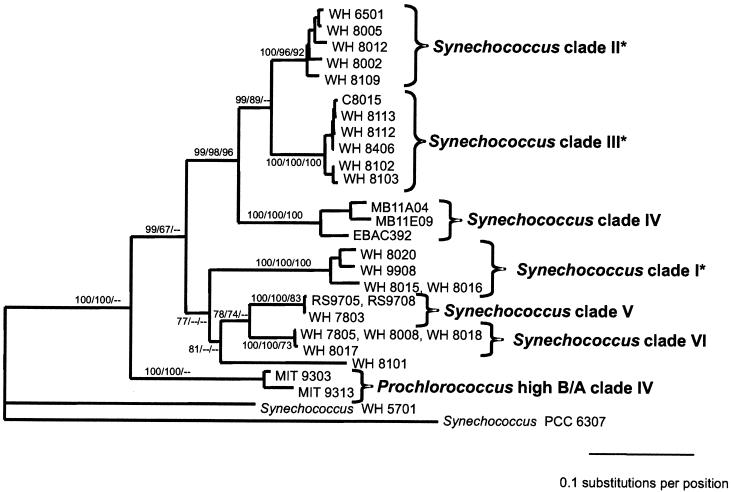
FIG. 4.
(A) Evolutionary relationships of Prochlorococcus and Synechococcus isolates and environmental sequences from Monterey Bay inferred using 233 positions of the 16S-23S rDNA spacer. The tree shown was inferred by maximum likelihood (−ln likelihood = 2,364.57) using the HKY85 model of nucleotide substitution with rate heterogeneity (gamma shape parameter = 0.383), empirical nucleotide frequencies, and a transition-transversion ratio of 1.156. The tree shown was not significantly worse (as determined by the Kishino-Hasegawa test) than the best tree found by likelihood (−ln likelihood = 2,362.09) in which the Prochlorococcus low-B/A clade I was not monophyletic. The Prochlorococcus low-B/A clade I was monophyletic in the best trees inferred using HKY85 or paralinear (logdet) distances and minimum evolution as the objective function. The tree is rooted with Cyanobium cluster Synechococcus sp. strain PCC 6307. Bootstrap values from distance/maximum-parsimony/maximum-likelihood analyses are as follows, with values less than 50 indicated by dashes: A, 79/72/81; B, 100/100/100; C, 100/100/99; D, 100/99/51; E, 64/51/—; F, 100/88/78; G, 100/98/93; H, 97/95/80; I, 100/69/—; J, 90/93/71; K, 55/—/—; L, 60/52/57; M, 100/99/99; N, 99/99/82; and O, 91/87/83. (B) Evolutionary relationships of marine Synechococcus isolates inferred using 434 positions of the 16S-23S rDNA spacer. Two strains of high-B/A Prochlorococcus and three environmental sequences from Monterey Bay were also included. Asterisks denote clades which are congruent with those identified using rpoC1. The phylogenetic framework was determined using paralinear distances (logdet) and minimum evolution as the objective function. Trees inferred using HKY85 distances resulted in identical clades with essentially similar branching orders. Bootstrap values from distance/maximum-parsimony/maximum-likelihood analyses are listed to the left of each node, with values less than 50 indicated by dashes. The tree is rooted with Cyanobium cluster Synechococcus sp. strain PCC 6307.
The remaining low-B/A isolates form a second clade in distance-based analyses but are not monophyletic in the best likelihood tree. On the other hand, likelihood trees in which this group is constrained to be monophyletic are not significantly worse than the best tree, as determined by the Kishino-Hasegawa test. But the monophyly of this clade has low bootstrap support by all methods. Thus, we conclude that there are at least two, and perhaps more, low-B/A clades of Prochlorococcus. This is consistent with previous work using 16S rDNA analysis (44, 67), in which two low-B/A clades were resolved.
The high-B/A Prochlorococcus strains are not monophyletic, but are dispersed in four distinct lineages (Fig. 4A). One clade, designated high-B/A I, contains isolates NATL1A, NATL2A, and PAC1, which consistently branch together and show a high sequence identity (97%). Isolates SS120 and MIT 9211 have a lower degree of sequence identity to each other (80%) and to the other five high-B/A isolates (less than 83%), and they have each been assigned to a different clade (Fig. 4A). Finally, isolates MIT 9303 and MIT 9313 (99% identity) make up Prochlorococcus high-B/A clade IV. This is consistent with the four lineages resolved by 16S rDNA in this ecotype (44).
Given the phylogenetic divergence of the Prochlorococcus high-B/A isolates into four clades, the question arises whether the designation high- and low-B/A ecotypes is still adequate for Prochlorococcus. Recently, Rippka et al. (41) proposed a subspecies, pastoris, to describe the low-B/A Prochlorococcus strain PCC 9511, which has a 16S rDNA sequence identical to that of the well-characterized low-B/A isolate MED4. In fact, the high- and low-B/A ecotypes each probably represent a separate species, with each of the clades identified here being a subspecies within the two species. However, until the taxonomy is fully delineated, the designations high- and low-B/A ecotype remain useful. They accurately describe culture physiologies that are correlated with phylogenetic relationships and are convenient terms, to be used with the caveat that the high-B/A ecotype consists of a wider range of physiologies and genetic diversity than the low-B/A ecotype.
The branching order of the low-B/A Prochlorococcus clade with respect to the high-B/A clades and Synechococcus suggests that the low-B/A clade is more recently arisen, consistent with 16S rDNA analyses (58). If the ability to synthesize divinyl chlorophylls a and b has been acquired only once (i.e., if the root of the tree is outside the Prochlorococcus clade, which is reasonable to assume), then the general possession of divinyl chlorophyll a and b is a primitive state shared by all Prochlorococcus strains and the low-B/A clade is a derived state. This is consistent with a possible evolutionary scenario in which a phycobilisome-containing ancestor acquired the ability to synthesize divinyl chlorophylls a and b, allowing it to colonize the deep euphotic zone, where competition for nutrients is reduced, as few other photosynthetic organisms are known to thrive there. Then later modifications, including the ability to survive in higher light and higher copper concentrations characteristic of surface waters, gave rise to the low-B/A clade, which is more competitive throughout the water column.
Phylogenetic relationships among Synechococcus strains.
Six clades can be identified within the marine cluster A Synechococcus strains based on high bootstrap support values and a large degree of within-clade sequence identity (Fig. 4A), which are also well supported in analyses using an expanded subset of 434 positions of the 16S-23S rDNA spacer (Fig. 4B). Three of these clades are congruent with those identified using rpoC1. The first, designated clade I (Fig. 4), contains four strains with 98% identity, one of which (WH 8020) is capable of chromatic adaptation. The second clade (II) consists of low-PUB strains, with 96 to 99% sequence identity. A third well-supported clade (III) consists of motile high-PUB strains that have very high sequence identity (99%).
The remaining three clades cannot yet be compared to those defined by rpoC1 because there are not enough strains in common. Synechococcus clade IV is made up of three environmental sequences that were cloned from Monterey Bay (4, 51). A fifth clade consists of the low-PUB strains WH 7803, RS9705, and RS9708. Finally, clade VI consists of four strains which all lack PUB. Three of these, WH 7805, WH 8008, and WH 8018, have identical sequences, while the fourth, WH 8017, is 99% identical. Strain WH 8017 was originally reported to have small amounts of PUB (65), but the reexamination of its pigment content revealed that it lacks PUB and has an absorption spectrum identical to that of WH 8018 (data not shown).
At present it is not possible to discern phenotypes associated with all of the clades of Synechococcus. The phycoerythrin composition varies within the clades, suggesting that, with the exception of Synechococcus clade IV, whose members all lack PUB, the ratio of PUB to PEB cannot be used as a defining feature within marine cluster A of Synechococcus. Without PUB, strains in clade VI are not as efficient in absorbing the blue light characteristic of open ocean waters and may predominate in coastal waters (69).
Two other clades are also associated with characteristic phenotypes. All of the strains in clade III have the ability to swim, consistent with analyses using rpoC1 in which all motile isolates form a single clade (55). One of the strains in clade I is capable of chromatic adaptation, and this clade may be equivalent to the clade of chromatically adapting strains defined by using rpoC1 sequences (35). Unfortunately, it is not yet possible to completely compare the clades identified using the ITS with those delineated by rpoC1 because the majority of the strains used in the two studies are different. Sequencing of both loci in additional strains should enable us in the near future to propose genus and species boundaries within marine cluster A of Synechococcus.
Clusters of strains that show greater than 99% identity in their 16S rDNA sequences, such as the low-B/A Prochlorococcus strains, are a common feature in the microbial world (62). This may be due to the asexual nature of bacterial reproduction, by which a novel beneficial mutation can sweep through a population, purging it of diversity at most loci while not affecting populations which occupy a different ecological niche and thus are not in direct competition. These adaptive sweeps would decrease diversity within a population and increase diversity between populations. Clusters in which the average sequence divergence between strains of different clusters is more than twice as great as the average sequence divergence between strains of the same cluster may represent ecologically distinct populations (38), and it has been suggested that such sequence similarity clusters should be at the heart of a natural species concept for bacteria (61). Indeed, in these six clades of Synechococcus, the average divergence between clades is always more than twice as great as that within each clade (data not shown), suggesting that the clades do represent ecologically distinct populations, even if we do not at present know how they are all differentiated from one another phenotypically.
Interestingly, the marine cluster B Synechococcus strain WH 8101 often branches within the marine cluster A clade V or VI. Although support for these branching orders varies, in no tree topology was WH 8101 more closely related to the marine cluster B strain WH 5701 than to the marine cluster A Synechococcus strains. This suggests that the strains now classified as marine cluster B Synechococcus may also consist of a number of genetically distinct clades that will require further work to fully characterize.
Global distribution of clades.
A striking feature of the clades of both Prochlorococcus and marine cluster A Synechococcus is their lack of correlation with geographic location of isolation (Tables 1 and 2). This is consistent with results obtained for low-B/A Prochlorococcus strains using 16S rDNA sequences (31, 44, 58). At first glance this conclusion is at odds with the detection of low-B/A I but not low-B/A II type sequences in surface waters in the North Atlantic by using probes to whole cells or to amplified 16S rDNA (67, 68). However, the presence of both types of low-B/A sequences in a single water sample has been detected in environmental libraries constructed by using the intergenic region between the photosynthetic electron transport chain genes petB and petD and ITS sequences (43, 57). Taken together, these data suggest that all of the clades may be distributed globally but that their relative abundances may change depending on local conditions, as well as seasonally and with depth within a given geographic location.
The examination of sequences directly from the environment, thus avoiding culturing biases, has been widespread in recent years and has given new insights into the diversity of environmental bacteria (12, 63), including marine cyanobacteria (11, 36, 57). In this study one clade of marine cluster A Synechococcus (clade IV) is made up entirely of environmental sequences. Thus, even in this well-studied group, culture collections may not yet represent the full extent of genetic and physiological diversity present in natural populations, and additional direct examination of oceanic populations is required.
The ITS is an excellent candidate for direct sequence diversity studies of field populations of marine cyanobacteria because it is variable enough to differentiate the ecotypes unambiguously using restriction fragment length polymorphism (RFLP) or terminal-RFLP analyses. In fact, all six Prochlorococcus clades yield distinct RFLP patterns when their amplified ITS sequences are cut with _Hae_II (43). Furthermore, the sequence data presented here allow the design of oligonucleotides specific for each clade that can be used as primers in quantitative PCR (20, 22, 52) to determine the abundances of each clade in the environment.
Concluding remarks.
Using the sequences of the 16S-23S rDNA ITS region, we have successfully delineated two important groups of marine cyanobacteria into strain clades which likely represent ecological units. For some of the clades, for example, the low-B/A Prochlorococcus strains, some inferences can be made about the nature of the niche that these organisms occupy (high-light surface waters). In other cases, there is not yet an obvious phenotype to explain the different niches these lineages may occupy. The contribution of this work is in identifying the sequence similarity clusters that potentially correspond to ecologically distinct units. This will allow more informed selection of strains in laboratory experiments, as representative strains from each lineage can now be employed in further physiological studies to discover the features of each clade which may have led to niche differentiation. Furthermore, the genetic differences that identify the clades can be used as specific markers to examine their distribution and relative abundances under different environmental conditions, ultimately providing a better understanding of the forces affecting the evolution and population dynamics of this globally successful group of cyanobacteria.
Acknowledgments
This work was supported by an NSF graduate fellowship to G.R., by NASA grant NAG5-3727 and NSF grant OCE9820035 to S.W.C., and by NSF grant OCE9315895 to D.L.D. and J.B.W.
We thank the researchers listed in Tables 1 and 2 for cultures and DNA from Prochlorococcus and Synechococcus strains not in the MIT or Woods Hole culture collections. We also thank Marcelino Suzuki and Ed DeLong for sharing sequence data prior to publication, Nathan Ahlgren for sequencing assistance, and Mitch Sogin and Lisa Moore for helpful discussions. Preliminary sequence data for Prochlorococcus strains MED4 and MIT 9313 and Synechococcus strain WH 8102 were obtained from the DOE Joint Genome Institute (JGI) at http://www.jgi.doe.gov/JGI_microbial/html/index.html.
REFERENCES
- 1.Apirion, D., and A. Miczak. 1993. RNA processing in prokaryotic cells. BioEssays 15**:**113-120. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Barry, T., G. Colleran, M. Glennon, L. R. Dunican, and F. Gannon. 1991. The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1**:**51-56. [DOI] [PubMed] [Google Scholar]
- 4.Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2**:**516-529. [DOI] [PubMed] [Google Scholar]
- 5.Berg, K. L., C. Squires, and C. L. Squires. 1989. Ribosomal RNA operon antitermination. Function of leader and spacer region box B-box A sequences and their conservation in diverse microorganisms. J. Mol. Biol. 209**:**345-358. [DOI] [PubMed] [Google Scholar]
- 6.Brahamsha, B. 1996. An abundant cell surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium Synechococcus. Proc. Natl. Acad. Sci. USA 93**:**6504-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chisholm, S. W., S. L. Frankel, R. Goericke, R. J. Olson, B. Palenik, J. B. Waterbury, L. West-Johnsrud, and E. R. Zettler. 1992. Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Arch. Microbiol. 157**:**297-300. [Google Scholar]
- 8.Chisholm, S. W., R. J. Olson, E. R. Zettler, R. Goericke, J. Waterbury, and N. Welschmeyer. 1988. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 334**:**340-343. [Google Scholar]
- 9.Christensen, H., K. Jorgensen, and J. E. Olsen. 1999. Differentiation of Campylobacter coli and C. jejuni by length and DNA sequence of the 16S-23S rRNA internal spacer region. Microbiology 145**:**99-105. [DOI] [PubMed] [Google Scholar]
- 10.Collier, J., B. Brahamsha, and B. Palenik. 1999. The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as a nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology 145**:**447-459. [DOI] [PubMed] [Google Scholar]
- 11.Ferris, M. J., and B. Palenik. 1998. Niche adaptation in ocean cyanobacteria. Nature 396**:**226-228. [Google Scholar]
- 12.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345**:**60-63. [DOI] [PubMed] [Google Scholar]
- 13.Goericke, R., and D. J. Repeta. 1992. The pigments of Prochlorococcus marinus: the presence of divinyl-chlorophyll a and b in a marine prokaryote. Limnol. Oceanogr. 37**:**425-433. [Google Scholar]
- 14.Goericke, R., and N. A. Welschmeyer. 1993. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the Sargasso Sea. Deep Sea Res. 40**:**2283-2294. [Google Scholar]
- 15.Gürtler, V., and V. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142**:**3-16. [DOI] [PubMed] [Google Scholar]
- 16.Iteman, I., R. Rippka, N. Tandeau de Marsac, and M. Herdman. 2000. Comparison of conserved structural and regulatory domains within divergent 16S rRNA-23S rRNA spacer sequences of cyanobacteria. Microbiology 146**:**1275-1286. [DOI] [PubMed] [Google Scholar]
- 17.Jeffrey, S. W., and G. F. Humphrey. 1975. New spectrophotometric equations for determining chlorophyll a, b, _c_1 and _c_2 in higher plants, algae, and natural phytoplankton. Biochem. Physiol. Pflanz. 168**:**191-194. [Google Scholar]
- 18.Kirk, J. T. O. 1994. Light and photosynthesis in aquatic ecosystems. Cambridge University Press, Cambridge, England.
- 19.Leblond-Bourget, N., H. Philippe, I. Mangin, and B. Decaris. 1996. 16S rRNA and 16S to 23S internal transcribed spacer sequence analyses reveal inter- and intraspecific Bifidobacterium phylogeny. Int. J. Syst. Bacteriol. 46**:**102-111. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S.-Y., J. Bollinger, D. Bezdicek, and A. Ogram. 1996. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl. Environ. Microbiol. 62**:**3787-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, W. K. W. 1995. Composition of ultraphytoplankton in the central North Atlantic. Mar. Ecol. Prog. Ser. 122**:**1-8. [Google Scholar]
- 22.Lim, E. L., A. V. Tomita, W. G. Thilly, and M. F. Polz. 2001. Combination of competitive quantitative PCR and constant-denaturant capillary electrophoresis for high-resolution detection and enumeration of microbial cells. Appl. Environ. Microbiol. 67**:**3897-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindell, D. 2000. Assessing the nitrogen status of marine prokaryotic phytoplankton populations using molecular methods. Ph.D. thesis. Hebrew University, Jerusalem, Israel.
- 24.Liu, H., H. A. Nolla, and L. Campbell. 1997. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat. Microb. Ecol. 12**:**39-47. [Google Scholar]
- 25.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29**:**173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann, E. L., N. Ahlgren, J. W. Moffett, and S. W. Chisholm. 2002. Copper toxicity and cyanobacterial ecology in the Sargasso Sea. Limnol. Oceanogr., in press.
- 27.Moore, L. R. 1997. Physiological ecology of Prochlorococcus: a comparison of isolates from diverse oceanic regimes. Ph.D. thesis. Massachusetts Institute of Technology, Cambridge, Mass.
- 28.Moore, L. R., and S. W. Chisholm. 1999. Photophysiology of the marine cyanobacterium Prochlorococcus: ecotypic differences among cultured isolates. Limnol. Oceanogr. 44**:**628-638. [Google Scholar]
- 29.Moore, L. R., R. E. Goericke, and S. W. Chisholm. 1995. Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar. Ecol. Prog. Ser. 116**:**259-275. [Google Scholar]
- 30.Moore, L. R., A. F. Post, G. Rocap, and S. W. Chisholm. 2002. Utilization of different nitrogen sources by the marine cyanobacteria, Prochlorococcus and Synechococcus. Limnol. Oceanogr., in press.
- 31.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393**:**464-467. [DOI] [PubMed] [Google Scholar]
- 32.Nour, M. 1998. 16S-23S and 23S-5S intergenic spacer regions of lactobacilli: nucleotide sequence, secondary structure and comparative analysis. Res. Microbiol. 149**:**433-448. [DOI] [PubMed] [Google Scholar]
- 33.Ong, L. J., and A. N. Glazer. 1991. Phycoerythrins of marine unicellular cyanobacteria I bilin types and locations and energy transfer pathways in Synechococcus spp. phycoerythrins. J. Biol. Chem. 266**:**9515-9527. [PubMed] [Google Scholar]
- 34.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biol. Sci. 12**:**357-358. [DOI] [PubMed] [Google Scholar]
- 35.Palenik, B. 2001. Chromatic adaptation in marine Synechococcus strains. Appl. Environ. Microbiol. 67**:**991-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palenik, B. 1994. Cyanobacterial community structure as seen from RNA polymerase gene sequence analysis. Appl. Environ. Microbiol. 60**:**3212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palenik, B., and R. Haselkorn. 1992. Multiple evolutionary origins of prochlorophytes, the chlorophyll _b_-containing prokaryotes. Nature 355**:**265-267. [DOI] [PubMed] [Google Scholar]
- 38.Palys, T., L. K. Nakamura, and F. M. Cohan. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47**:**1145-1156. [DOI] [PubMed] [Google Scholar]
- 39.Parpais, J., D. Marie, F. Partensky, P. Morin, and D. Vaulot. 1996. Effect of phosphorus starvation on the cell cycle of the photosynthetic prokaryote Prochlorococcus spp. Mar. Ecol. Prog. Ser. 132**:**265-274. [Google Scholar]
- 40.Partensky, F., N. Hoepffner, W. K. W. Li, O. Ulloa, and D. Vaulot. 1993. Photoacclimation of Prochlorococcus sp. (Prochlorophyta) strains isolated from the North Atlantic and Mediterranean Sea. Plant Physiol. 101**:**285-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rippka, R., T. Coursin, W. Hess, C. Lichtle, D. Scanlan, K. Palinska, I. Iteman, F. Partensky, J. Houmard, and M. Herdman. 2000. Prochlorococcus marinus Chisholm et al. 1992 subsp. pastoris subsp. nov. strain PCC 9511, the first axenic chlorophyll _a_2/_b_2-containing cyanobacterium (Oxyphotobacteria). Int. J. Syst. Evol. Microbiol. 50**:**1833-1847. [DOI] [PubMed] [Google Scholar]
- 42.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111**:**1-61. [Google Scholar]
- 43.Rocap, G. 2000. Genetic diversity and ecotypic differentiation in the marine cyanobacteria Prochlorococcus and Synechococcus. Ph.D. thesis. Massachusetts Institute of Technology and Woods Hole Oceanographic Institute, Cambridge, Mass.
- 44.Rocap, G., L. R. Moore, and S. W. Chishom. 1999. Molecular phylogeny of Prochlorococcus ecotypes, p. 107-116. In L. Charpy and A. W. D. Larkum (ed.), Marine cyanobacteria. Bulletin de l'Institut Océanographique, Monaco, Special Issue 19.
- 45.Saito, M. 2000. The role of cobalt in the ecology of Prochlorococcus. Ph.D. thesis. Massachusetts Institute of Technology and Woods Hole Oceanographic Institute, Cambridge, Mass.
- 46.Scanlan, D., W. Hess, F. Partensky, J. Newman, and D. Vaulot. 1996. High degree of genetic variation in Prochlorococcus (Prochlorophyta) revealed by RFLP analysis. Eur. J. Phycol. 31**:**1-9. [Google Scholar]
- 47.Shalapyonok, A., R. J. Olson, and L. S. Shalapyonok. 1998. Ultradian growth in Prochlorococcus spp. Appl. Environ. Microbiol. 64**:**1066-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimada, A., S. Kanai, and T. Maruyama. 1995. Partial sequence of ribulose-1,5-bisphosphate carboxylase/oxygenase and the physiology of Prochloron and Prochlorococcus (Prochlorales). J. Mol. Evol. 40**:**671-677. [DOI] [PubMed] [Google Scholar]
- 49.Shimada, A., M. Nishijima, and T. Maruyama. 1995. Seasonal appearance of Prochlorococcus in Suruga Bay, Japan, in 1992-1993. J. Oceanogr. 51**:**289-300. [Google Scholar]
- 50.Smith, S. W., R. Overbeek, C. R. Woese, and W. Gilbert. 1994. The genetic data environment: an expandable GUI for multiple sequence analysis. Comput. Appl. Biol. Sci. 10**:**671-675. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki, M. T., O. Béjà, L. T. Taylor, and E. F. DeLong. 2001. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ. Microbiol. 3**:**323-331. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66**:**4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swofford, D. L. 1999. PAUP*: phylogenetic analysis using parsimony (* and other methods). Sinauer Associates, Sunderland, Mass.
- 54.Toledo, G., and B. Palenik. 1997. Synechococcus diversity in the California Current as seen by RNA polymerase (rpoC1) gene sequences of isolated strains. Appl. Environ. Microbiol. 63**:**4298-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toledo, G., B. Palenik, and B. Brahamsha. 1999. Swimming marine Synechococcus strains with widely different photosynthetic pigment ratios form a monophyletic group. Appl. Environ. Microbiol. 65**:**5247-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomioka, N., and M. Sugiura. 1984. Nucleotide sequence of the 16S-23S spacer region in the rrnA operon from a blue-green alga. Anacystis nidulans. Mol. Gen. Genet. 193**:**427-430. [DOI] [PubMed] [Google Scholar]
- 57.Urbach, E., and S. W. Chisholm. 1998. Genetic diversity in Prochlorococcus populations flow cytometrically sorted from the Sargasso Sea and the Gulf Stream. Limnol. Oceanogr. 43**:**1615-1630. [Google Scholar]
- 58.Urbach, E., D. Distel, D. J. Scanlan, J. B. Waterbury, and S. W. Chisholm. 1998. Rapid diversification of marine picophytoplankton with dissimilar light harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria). J. Mol. Evol. 46**:**188-201. [DOI] [PubMed] [Google Scholar]
- 59.Urbach, E., D. Robertson, and S. W. Chisholm. 1992. Multiple evolutionary origins of prochlorophytes within the cyanobacterial radiation. Nature 355**:**267-270. [DOI] [PubMed] [Google Scholar]
- 60.Veldhuis, M. J. W., G. W. Kraay, J. D. L. Van Bleijswijk, and M. A. Baars. 1997. Seasonal and spatial variation in phytoplankton biomass, productivity and growth in the northwestern Indian Ocean: the southwest and northeast monsoon, 1992-1993. Deep Sea Res. 44**:**425-449. [Google Scholar]
- 61.Ward, D. M. 1998. A natural species concept for prokaryotes. Curr. Opin. Microbiol. 1**:**271-277. [DOI] [PubMed] [Google Scholar]
- 62.Ward, D. M., M. J. Ferris, S. C. Nold, and M. M. Bateson. 1998. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62**:**1353-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345**:**63-65. [DOI] [PubMed] [Google Scholar]
- 64.Waterbury, J. B., and R. Rippka. 1989. Subsection I. Order Chroococcales Wettstein 1924, Emend. Rippka et al., 1979, p. 1728-1745. In J. T. Staley, M. P. Bryant, N. Pfennig, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 3. Williams and Wilkins, Baltimore, Md. [Google Scholar]
- 65.Waterbury, J. B., S. W. Watson, F. W. Valois, and D. G. Franks. 1986. Biological and ecological characterization of the marine unicellular cyanobacteria Synechococcus. Can. Bull. Fish. Aquat. Sci. 214**:**71-120. [Google Scholar]
- 66.Waterbury, J. B., J. M. Willey, D. G. Franks, F. W. Valois, and S. W. Watson. 1985. A cyanobacterium capable of swimming motility. Science 30**:**74-76. [DOI] [PubMed] [Google Scholar]
- 67.West, N. J., and D. J. Scanlan. 1999. Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl. Environ. Microbiol. 65**:**2585-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.West, N. J., W. A. Schonhuber, N. J. Fuller, R. I. Amann, R. Rippka, A. F. Post, and D. J. Scanlan. 2001. Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology. 147**:**1731-1744. [DOI] [PubMed] [Google Scholar]
- 69.Wood, A. M., D. A. Phinney, and C. S. Yentsch. 1998. Water column transparency and the distribution of spectrally distinct forms of phycoerythrin containing organisms. Mar. Ecol. Prog. Ser. 162**:**25-31. [Google Scholar]
- 70.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Amsterdam, The Netherlands.