Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth (original) (raw)
Abstract
Although hundreds of evolutionarily conserved microRNAs have been discovered, the functions of most remain unknown. Here, we describe the embryonic spatiotemporal expression profile, transcriptional regulation, and loss-of-function phenotype of Drosophila miR-1 (DmiR-1). DmiR-1 RNA is highly expressed throughout the mesoderm of early embryos and subsequently in somatic, visceral, and pharyngeal muscles, and the dorsal vessel. The expression of DmiR-1 is controlled by the Twist and Mef2 transcription factors. DmiR-1KO mutants, generated using ends-in gene targeting, die as small, immobilized second instar larvae with severely deformed musculature. This lethality is rescued when a DmiR-1 transgene is expressed specifically in the mesoderm and muscle. Strikingly, feeding triggers _DmiR-1KO_-associated paralysis and death; starved first instar DmiR-1KO larvae are essentially normal. Thus, DmiR-1 is not required for the formation or physiological function of the larval musculature, but is required for the dramatic post-mitotic growth of larval muscle.
Keywords: MicroRNA, miR-1, muscle, Drosophila, larval development
MicroRNAs (miRNAs) are small, ∼21-nucleotide (nt), noncoding RNAs that post-transcriptionally regulate gene expression. They do so by base-pairing with sequences in the 3' untranslated regions (UTRs) of target messenger RNAs (mRNAs); complete base-pairing results in target mRNA degradation whereas incomplete base-pairing results in target mRNA translational repression. Biochemical approaches have provided insight into the numbers of miRNAs encoded within individual genomes and the principles that govern miRNA biogenesis and miRNA:target mRNA interactions. However, our understanding of the biological roles of miRNAs is still quite limited and for only a few miRNAs have functions been assigned by genetic analysis (Ambros 2004).
One approach to explore the extent of miRNA function has been genetic studies removing the function of Dicer, an RNase III enzyme required for miRNA biogenesis. Zebrafish embryos devoid of both maternal and zygotic Dicer (and hence lacking mature miRNAs) proceed quite far through embryogenesis but arrest with specific defects in morphogenesis, somitogenesis, heart development, and brain formation (Giraldez et al. 2005). This and other studies have led to the suggestion that miRNAs are not involved in building the basic animal body plan but rather regulate the “fine tuning” or diversification of cell types within organs and tissues. A second approach to explore miRNA function has been the analysis of the phenotypic consequences of ectopic expression or overexpression of specific miRNAs. Such approaches have indicated that certain vertebrate miRNAs play crucial roles in insulin secretion (Poy et al. 2004) or the regulation of B- and T-cell lineage development (Chen et al. 2004). One major caveat of ectopic expression and overexpression analysis is that the phenotypic consequences may not be directly relevant to the function of the miRNA at normal physiological levels and in its normal context. Therefore, an unequivocal understanding of the function of individual miRNAs requires the isolation of loss-of-function mutations. To date, no knockouts of vertebrate miRNA genes have been reported. However, mutations in a handful of worm and fly miRNAs that have been isolated by forward genetic screens reveal that miRNAs play diverse roles in development (Ambros 2004). Analysis of miRNA loss-of-function mutations indicate that in worms, lin-4 and let-7 regulate developmental timing (Lee et al. 1993; Reinhart et al. 2000) and that lsy-6 governs neuronal cell identity (Johnston and Hobert 2003), and in flies, bantam (Brennecke et al. 2003) and miR-14 (Xu et al. 2003) regulate apoptosis, cell growth, and fat metabolism.
MiR-1 is an evolutionarily conserved miRNA whose tissue-specific expression pattern also appears to be phylogenetically conserved. The worm and fly genomes each possess a single miR-1 gene (Cel-miR-1 and DmiR-1, respectively) while the zebrafish, mouse, and human genomes each contain two miR-1 loci (Dre-miR-1-1 and Dre-miR-1-2, Mmu-miR-1-1 and Mmu-miR-1-2, and Hsa-miR-1-1 and Hsa-miR-1-2, respectively). Northern blot analysis indicated that miR-1 is expressed specifically in mouse and human heart and skeletal muscle (Lee and Ambros 2001; Sempere et al. 2004). This result has been further confirmed using three different in vivo techniques to examine miRNA expression; a murine Mmu-miR-1-1 and Mmu-miR-1-2 “sensor” transgene was repressed specifically in the adult heart (Mansfield et al. 2004); promoter fusion constructs to Mmu-miR-1-1 and Mmu-miR-1-2 were both expressed in cardiac and skeletal muscle precursor cells (Zhao et al. 2005); in situ hybridization using locked nucleic acid (LNA) probes detected Dre-miR-1-1 and Dre-miR-1-2 expression in muscle (Wienholds et al. 2005). Two recent studies suggest that miR-1 might possess a variety of in vivo functions. Microarray analysis of HeLa cells transfected with Hsa-miR-1 indicated that miR-1 could function to maintain muscle cell identity by repressing the expression of nonmuscle genes (Lim et al. 2005). Overexpression analysis of Mmu-miR-1 indicated that it may regulate the proliferation of cardiomyoctes by controlling the expression of the Hand2 transcription factor (Zhao et al. 2005).
To directly assess the in vivo function of miR-1, we examined the expression pattern, transcriptional regulation, and loss-of-function phenotype of Drosophila miR-1 (DmiR-1). We find that, as in zebrafish, mice, and humans, DmiR-1 is specifically expressed in muscle cells. We show that DmiR-1 expression is regulated by the promesodermal transcription factor Twist and the promyogenic transcription factor Mef2. We also find that the muscles form normally in DmiR-1KO mutant embryos and function normally in first instar DmiR-1KO mutant larvae. However, when larval growth is initiated by feeding, DmiR-1KO mutant larvae become paralyzed, arrest their growth and ultimately die as small, second instar larvae with massively disrupted somatic musculature.
Results
DmiR-1 primary transcript and mature DmiR-1 miRNA are expressed specifically in the mesoderm and its muscle cell derivatives
In situ hybridization was performed using probes to detect the full-length primary transcript (pri-DmiR-1) from which the DmiR-1 21mer is processed (Fig. 1A-F). Pri-DmiR-1 was initially detected ventrally in the presumptive mesoderm of stage 5 embryos (Fig. 1A) in a pattern similar to that of the snail and twist gene products (Leptin 1991). Direct comparison of the pri-DmiR-1 expression pattern along the anterior-posterior axis with the snail and twist mRNA pattern in the cellular blastoderm indicated that pri-DmiR-1 shares the sharp posterior border of snail RNA but its anterior boundary is posterior to that of both twist and snail RNA (Fig. 1A; data not shown). Pri-DmiR-1 was expressed in the ventral-most cells of the cellular blastoderm, in a swath that is 18-20 cells wide (data not shown). During gastrulation and after invagination of the ventral cells of the cellular blastoderm, pri-DmiR-1 was expressed exclusively in mesodermal cells (Fig. 1B). After subdivision of the mesoderm, pri-DmiR-1 expression was observed in the cephalic mesoderm and the primordia of the visceral and somatic musculature (Fig. 1C). After germ-band retraction, pri-DmiR-1 was detected in the pharyngeal and visceral musculature (Fig. 1E), the myocardial cells of the dorsal vessel (Fig. 1F) and in the segmentally repeated clusters of mesodermal cells that give rise to the somatic muscles (Fig. 1D). Notably, pri-DmiR-1 was restricted to muscle progenitors and was not expressed in other mesodermal derivatives such as the fat body, gonadal mesoderm, and midline glia.
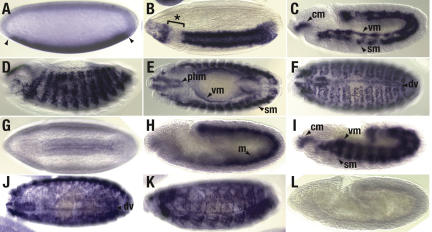
Figure 1.
DmiR-1 is detected initially in the presumptive mesoderm and later in the pharyngeal, somatic, and visceral musculature as well as the dorsal vessel. Wild-type (_A_-K) or DmiR-1KO mutant (L) embryos were hybridized with a 1.1-kb riboprobe to detect the full-length pri-DmiR-1 transcript (_A_-F) or an LNA oligo to detect the processed DmiR-1 21mer (_G_-L). In all figures, embryos are oriented with anterior to the left and dorsal up. (A) Lateral view, stage 5 embryo with DmiR-1 expression in the presumptive mesoderm. Anterior and posterior limits of expression are indicated (arrowheads). (B) Lateral view, gastrulating stage 6 embryo with DmiR-1 expression in most of the invaginating mesoderm except in a portion of the head mesoderm (aster isked bracket). (C) Lateral view, stage 11 embryo with DmiR-1 expression restricted to the cephalic (cm), somatic (sm), and visceral (vm) mesoderm. (D) Lateral view, stage 13 embryo with punctate, segmental repeated expression of DmiR-1 in the forming somatic musculature. (E) Dorsal view of the same embryo presented in D showing expression in the forming pharyngeal (phm), somatic (sm), and visceral (vm) musculature. (F) Dorsal view, stage 15 embryo with DmiR-1 expression in clusters of fused somatic mesodermal cells as well as in the two rows of heart precursor cells comprising the forming dorsal vessel (dv). (G) Ventral view, gastrulating stage 6 embryo with processed DmiR-1 expression faintly detected in the invaginating presumptive mesodermal layer. (H) Lateral view, stage 10 embryo with DmiR-1 expression throughout the mesoderm (m). (I) Lateral view, stage 11 embryo with processed DmiR-1 expression restricted to the cephalic (cm), somatic (sm), and visceral (vm) mesoderm. (J) Dorsolateral view, stage 15 embryo with processed DmiR-1 expression in the somatic musculature and dorsal vessel (dv). (K) Lateral view, stage 16 embryo with cytoplasmic staining of processed DmiR-1 in syncytial fibers of differentiating somatic muscles. (L) Lateral view, stage 10 DmiR-1KO mutant embryo in which processed DmiR-1 expression is absent, confirming the specificity of the DmiR-1 LNA oligo.
The expression pattern of the mature 21 nt form of DmiR-1 was directly assayed using a 21-nt digoxigenin-labeled LNA oligonucleotide probe complementary to DmiR-1 (Fig. 1G-K). The similarity between the two expression patterns (Fig. 1, cf. A-F and G-K) confirmed that DmiR-1 RNA is expressed specifically in the mesoderm and its muscle cell derivatives. However, there were two key differences between the expression patterns. First, while pri-DmiR-1 was detected in a punctate, nuclear-staining pattern, DmiR-1 21mer was cytoplasmic (Fig. 1, cf. F and K). This is consistent with the rapid processing of miRNA transcripts into mature miRNAs in conjunction with transport to the cytoplasm (Lee et al. 2002). Second, while robust expression in the presumptive mesoderm was detected with the pri-DmiR-1 probes, the DmiR-1 21mer was first faintly detected later at gastrulation (Fig. 1G). This could reflect stage-specific differences in probe accessibility. Alternatively, pri-DmiR-1 may be transcribed beginning at the cellular blastoderm stage but not processed until gastrulation.
Twist is necessary and sufficient for DmiR-1 expression
Formation of the dorsoventral axis is controlled by a nuclear gradient of the Dorsal protein (for review, see Stathopoulos and Levine 2002a). In the ventral-most cells of the cellular blastoderm, high levels of Dorsal activates two genes, twist and snail, which are required for the formation of the mesoderm (Leptin 1991). Twist, a basic helix-loop-helix (bHLH) transcription factor, in turn positively regulates the expression of mesodermal-specific transcripts (Furlong et al. 2001), whereas Snail transcriptionally represses genes ordinarily expressed in the more lateral neuroectoderm. To determine whether the Dorsal signaling pathway is involved in the regulation of DmiR-1, we examined DmiR-1 expression in twist and snail mutant embryos (Fig. 2A-D). While DmiR-1 was clearly detected in the mesoderm of stage 10 wild-type embryos (Fig. 2A) and snail18 mutant embryos (Fig. 2C), DmiR-1 expression was not detected in twist1 or twist3 snail18 mutant embryos (Fig. 2B,D). Thus, Twist but not Snail is required for DmiR-1 expression.
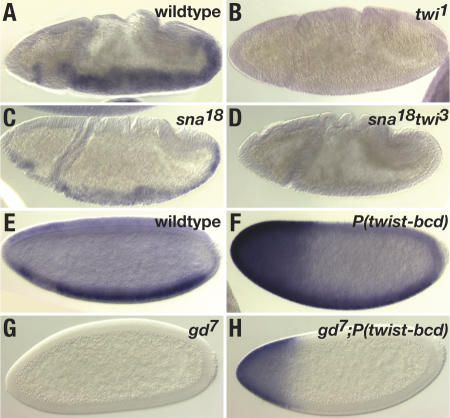
Figure 2.
Twist is necessary and sufficient for DmiR-1 expression. Embryos were hybridized with a digoxigenin-labeled 1.1-kb riboprobe to detect pri-DmiR-1. All panels are lateral views, anterior to the left and dorsal up. (A) Stage 10 wild-type embryo with DmiR-1 expression in the invaginated mesodermal layer. (B) Stage 10 twi1 mutant embryo with absence of DmiR-1 expression. (C) Stage 10 sna18 mutant embryo with DmiR-1 expression. (D) Stage 10 sna18 twi3 mutant embryo with absence of DmiR-1. (E) Stage 5 wild-type embryo with DmiR-1 expression in the presumptive mesoderm. (F) Stage 5 embryo that contains a twist-bcd transgene and consequently expresses ectopic Twist at the anterior pole of the embryo (Stathopoulos and Levine 2002b). DmiR-1 expression is detected in the presumptive mesoderm as well as at the anterior pole. (G) Stage 5 gd7 embryo that completely lacks Dorsal nuclear protein. DmiR-1 is not detected. (H) Stage 5 gd7 embryo containing a twist-bcd transgene. DmiR-1 expression is only detected at the anterior pole.
To determine whether ectopic Twist can activate DmiR-1 expression, we took advantage of a construct in which the Twist ORF was fused to the 3′ untranslated region (UTR) of the bicoid transcript and that consequently causes Twist protein to be ectopically expressed at the anterior pole of the cellularizing embryo (Stathopoulos and Levine 2002b). In an otherwise wild-type stage 5 embryo carrying this P(twist-bcd) transgene, DmiR-1 was detected not only in the presumptive mesoderm but also ectopically at the anterior pole (Fig. 2F). This finding indicated that even in dorsal portions of the embryo where the Dorsal nuclear protein is present at only low levels, Twist is capable of promoting DmiR-1 expression. To investigate whether Twist alone was sufficient to drive DmiR-1 expression in the absence of other promesodermal factors, we examined the DmiR-1 expression pattern in gastrulation defective (gd) embryos carrying the P(twist-bcd) transgene (Fig. 2G,H). In gd mutant embryos, activation of the Toll receptor is blocked and consequently neither Dorsal nor any other promesodermal factors are expressed. DmiR-1 expression was not detected in gd mutant embryos (Fig. 2G). However, in gd mutant embryos carrying the P(twist-bcd) transgene, DmiR-1 was detected at the anterior pole (Fig. 2H). This data indicated that Twist is the only promesodermal factor required for DmiR-1 expression.
The DmiR-1 genomic locus contains functional Twist-binding sites
The DmiR-1 genomic locus is located on the left arm of chromosome two at cytological location 38C. The closest predicted or known proximal gene (CG15476) is located ∼14 kb away and the closest predicted or known distal gene (CG2493) is located ∼24 kb away. To map full-length pri-DmiR-1, we performed 5′RACE on total larval RNA and amplified a fragment extending 440 bp upstream of the first nt of the DmiR-1 21mer. This placed the putative DmiR-1 transcription start site downstream (17 and 65 nt, respectively) from two TATA boxes. Both of these TATA boxes are conserved across five sequenced Drosophila genomes and the TATA box located at -17 nt exactly matches the TATA box consensus sequence (STATAWAAR) of Drosophila core promoters (Ohler et al. 2002). Two evolutionarily conserved polyadenylation sites (AATAAA) were identified 108 and 552 bp downstream of the DmiR-1 21mer, indicating predicted pri-DmiR-1 transcripts of either 548 or 992 nt in length. However, full-length pri-DmiR-1 was not detected by Northern blot, presumably because it is processed very quickly (data not shown).
To investigate whether Twist could directly activate DmiR-1, the DmiR-1 genomic locus was examined for Twist-binding sites. While bHLH transcription factors are known to bind to the consensus hexanucleotide sequence E-box, CANNTG (Castanon and Baylies 2002), we focused only on the varieties of E-boxes found in known mesodermal targets of Twist, mef2 (CATGTG; Cripps et al. 1998), snail (CACGTG; Ip et al. 1992) and tinman (CATGTG and CATATG; Yin et al. 1997). Thirteen such E-boxes were identified within a 4 kb region surrounding DmiR-1 (Fig. 3A). Eight of these E-boxes are evolutionarily conserved across five Drosophila genomes. Six of the eight evolutionarily conserved E-boxes contain a TG core (CATGTG) while two contain a CG or a TA core. Finally, the DmiR-1 locus of the more distant mosquito genome contains a cluster of three TG core E-boxes within 100 bp of each other and ∼2.7 kb upstream of the mosquito miR-1 21mer.
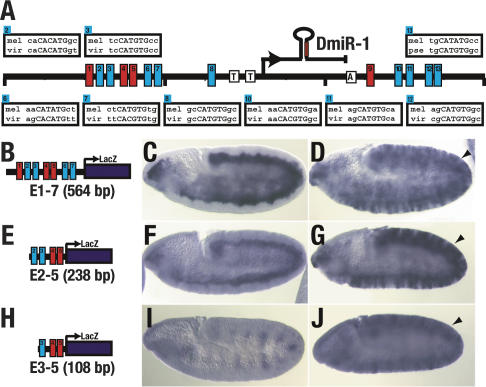
Figure 3.
E-Box fragments from the DmiR-1 genomic locus are sufficient to recapitulate the mesodermal expression pattern of DmiR-1. (A) Schematic representation of a 4-kb region of the DmiR-1 genomic locus. The transcription start site of pri-DmiR-1 as determined by 5′ RACE is indicated with an arrow. Two evolutionarily conserved TATA boxes (T) and polyadenylation site (A) are also indicated. Thirteen Twist-binding sites (CANNTG), termed E-boxes, were identified (boxes numbered 1-13) and the evolutionary conservation of nine E-boxes, from Drosophila melanogaster (mel) to either Drosophila virilis (vir) or Drosophila pseudoobscura (pse), are shown. (B,E,H) Schematic representations of enhancer-LacZ constructs containing either seven E-boxes (E1-7 shown in B), four E-boxes (E2-5 shown in E), or three E-boxes (E3-5 shown in H). (C,D,F,G,I,J) Embryos were hybridized with a LacZ RNA probe and are oriented with anterior to the left and dorsal up. (C) Stage 10 embryo containing the E1-7 transgene with LacZ expression in the mesoderm. (F) Stage 10 embryo containing the E2-5 transgene with LacZ expression in the mesoderm. (I) Stage 10 embryo containing the E3-5 transgene with LacZ expression in the ectoderm but not the mesoderm. (D,G,J) daughterless-Gal4 driven expression of UAS-Twist (Baylies and Bate 1996) leads to ectopic expression of the E1-7 transgene (D, arrowhead), the E2-5 transgene (G, arrowhead), and the E3-5 transgene (J, arrowhead).
Seven E-boxes are clustered within a 381-bp region upstream of the putative DmiR-1 transcription start site (Fig. 3A). To determine whether this cluster is a functional Twist enhancer, a series of constructs were built in which fragments containing all seven (E1-7, Fig. 3B), four (E2-5, Fig. 3E) or three (E3-5, Fig. 3H) of the E-boxes were fused to a lacZ reporter gene and assayed for the ability to direct lacZ expression in the mesoderm. The E1-7 fragment drove lacZ expression throughout embryogenesis in a pattern almost identical to the DmiR-1 expression pattern (Fig. 3C). In contrast, the E2-5 fragment drove lacZ expression throughout the mesoderm early in embryogenesis (Fig. 3F) but, like twist expression itself, faded such that by stage 13, lacZ was only detected in a few of the cells of the somatic mesoderm (Thisse et al. 1988; data not shown). The E3-5 fragment did not direct expression of lacZ in the mesoderm though some ectodermal lacZ expression was detected (Fig. 3I).
To determine whether the E1-7, E2-5, and E3-5 enhancer fragments contained sites that can respond to Twist protein, we examined lacZ driven expression from all three constructs in embryos in which twist was ectopically expressed. In all three cases, ectopic ectodermal expression of lacZ was detected indicating that all three fragments contained functional Twist responsive sites (Fig. 3D,G,J). Thus, enhancer fragments containing known Twist-binding sites are responsive to Twist and recapitulate the endogenous mesodermal DmiR-1 expression pattern, supporting the conclusion that Twist is the only promesodermal factor required for DmiR-1 expression.
Mef2 is required for DmiR-1 expression
While the data presented above indicated that Twist regulates DmiR-1 expression in early embryos, additional factors must also be involved in controlling DmiR-1 expression at later stages. DmiR-1 is expressed in all the myogenic cells of late embryos (see Fig. 1), whereas Twist expression is maintained in only a small subset of cells of the somatic and visceral musculature (Thisse et al. 1988; Bate et al. 1991). Mef2 is a good candidate to regulate late DmiR-1 expression: Mef2 protein is expressed in an almost identical pattern to DmiR-1 in late-stage embryos (Fig. 1; Bour et al. 1995) and is known to regulate genes involved in the terminal differentiation of muscles (Kelly et al. 2002). Therefore, we examined DmiR-1 expression in mef2 mutant embryos (Fig. 4A,B). While DmiR-1 is initially expressed in mef2 mutant embryos, DmiR-1 expression is not maintained indicating that mef2 is required for DmiR-1 expression. We furthermore identified a site that perfectly matched the decanucleotide consensus binding site for Mef2 proteins (YTAWWWWTAR) (Andres et al. 1995) and found that this site was evolutionarily conserved across the five sequenced Drosophila genomes (Fig. 4C). To determine whether this might be a functional Mef2-binding site, we compared the expression of two enhancer/lacZ fusion constructs carrying either the 564-bp E1-7 fragment that contained the Mef2 site or the 338-bp E1-5 fragment that did not (Fig. 4C). The E1-7 fragment directed late-stage expression throughout the developing somatic musculature in a pattern reminiscent of mef2 expression (Fig. 4D). In contrast, the E1-5 fragment directed lacZ expression in only a few cells of the somatic musculature in a pattern reminiscent of twist (Fig. 4E). This data indicated that Mef2 regulates late-stage DmiR-1 expression through an evolutionarily conserved upstream Mef2-binding site.
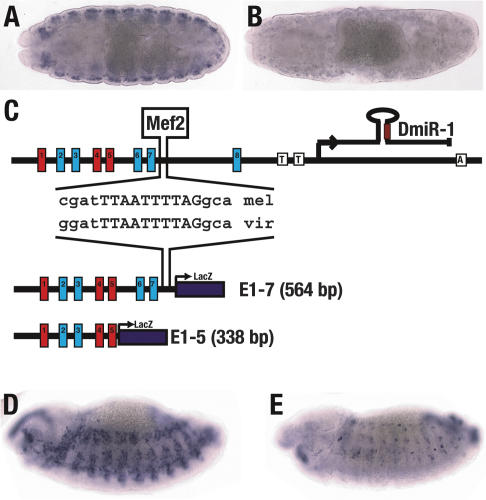
Figure 4.
DMef2 is required for DmiR-1 expression. (A,B) Stage 15 wild-type (A) and Dmef222-21 mutant (B) embryos were hybridized with a digoxigenin-labeled 1.1-kb riboprobe to detect pri-DmiR-1. Expression is detected in the wild-type embryo (A) but not in the Dmef222-21 mutant embryo (B). (C) A DMef2-binding site (YTAWWWWTAR) was identified next to E-box 7 in the DmiR-1 genomic locus and is evolutionarily conserved between D. melanogaster (mel) and D. virilis (vir). This DMef2-binding site is contained in the E1-7-LacZ enhancer construct but not the E1-5-LacZ enhancer construct. (D,E) Stage 13 embryos containing either the E1-7 transgene (D) or the E1-5 transgene (E) were stained with a LacZ RNA probe. (D) Embryo containing the E1-7 transgene with LacZ expression in all somatic mesodermal cells. (E) Embryo containing the E1-5 transgene with LacZ expression in just a few somatic mesodermal cells. The remaining _lacZ_-positive cells are presumably cells with persistent Twist expression (Bate et al. 1991).
DmiR-1 is essential and is required in muscle cells
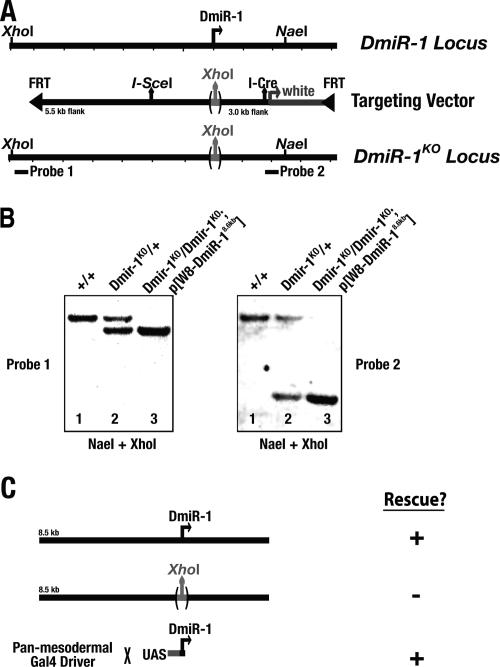
We used ends-in gene targeting (Rong and Golic 2000) to generate a null mutation in DmiR-1. Accordingly, a targeting vector containing ∼8.6 kb of the DmiR-1 locus was engineered in which (1) 57 bp, including the entire region encoding the DmiR-1 21mer, were deleted and replaced with a XhoI site; (2) an I-SceI site was inserted ∼400 bp upstream of the DmiR-1 putative transcription start site; and (3) this fragment was flanked by FRT sites (Fig. 5A). The targeting vector was transformed into flies and the transgenic flies were used to generate a DmiR-1KO mutation by the two-step ends-in method (see Materials and Methods; Rong and Golic 2000). Southern blot analysis confirmed that the mutant copy was successfully identified (Fig. 5B).
Figure 5.
DmiR-1 is required in the mesoderm for viability. (A) Schematic representations of the wild-type DmiR-1 genomic locus; the ends-in targeting fragment in which 57 bp of the DmiR-1 genomic locus, including the sequence for the entire DmiR-1 21mer, was replaced with a XhoI site; and the mutated DmiR-1 genomic locus after targeting. Locations of the probes and restriction enzyme sites used in the Southern blots in B are also shown. (B) Southern blot analysis indicates successful targeting of the DmiR-1 genomic locus and the replacement of DmiR-1 with a XhoI Site. Genomic DNA from wild-type (+/+), heterozygous DmiR-1KO (DmiR-1KO/+), or homozygous DmiR-1KO (DmiR-1KO/DmiR-1KO; _P[W8-DmiR-18.6kb_]) flies was hybridized with either a distal (Probe 1, left panel) or proximal (Probe 2, right panel) probe. Genomic DNA in both panels was double-digested with restriction enzymes NaeI and XhoI. Both probes detected a 13.5-kb XhoI → NaeI fragment in wild type (lane 1, left and right panels). Insertion of the XhoI site of the targeting vector into the DmiR-1 locus was predicted to cleave the 13.5-kb XhoI → NaeI into a 10-kb XhoI → XhoI fragment and a 3.5-kb XhoI → NaeI fragment. In addition to the wild-type 13.5-kb fragment, Probe 1 detected the 10-kb fragment in heterozygous DmiR-1KO genomic DNA (lane 2, left panel) and likewise Probe 2 detected the 3.5-kb fragment in heterozygous DmiR-1KO genomic DNA (lane 2, right panel). Since both Probes 1 and 2 lie outside of the DNA encoded by the _P(W8-DmiR-18.6kb)_-rescuing transgene (cf. probe locations in A and rescuing transgene in C), Probe 1 detected only the 10-kb fragment in homozygous DmiR-1KO genomic DNA (lane 3, left panel), and likewise Probe 2 detected only the 3.5-kb fragment in homozygous DmiR-1KO genomic DNA (lane 3, right panel). (C) Flies homozygous for the mutated DmiR-1KO locus were inviable. This lethality was rescued with an 8.6-kb fragment containing the wild-type DmiR-1 genomic locus (P[W8-DmiR-18.6kb]) but was not rescued with the same 8.6-kb fragment in which DmiR-1 was replaced with a XhoI site (P[W8-DmiR-18.6kb → XhoI]). Finally, _DmiR-1KO_-associated lethality was also rescued when DmiR-1 was expressed specifically in the mesoderm and its muscle cell derivatives using the panmesodermal how24B Gal4 line to drive expression of a UAS-DmiR-1 transgene.
Homozygous mutant DmiR-1KO adults never eclosed, indicating that DmiR-1 is required for viability. Four lines of evidence indicated that the lethality associated with the DmiR-1KO allele is due specifically to the loss of DmiR-1. First, the processed DmiR-1 21mer was not detected in homozygous DmiR-1KO mutant embryos by in situ hybridization (Fig. 1L). Second, homozygous mutant DmiR-1KO animals could be rescued to adulthood with an 8.6-kb transgene encoding the entire DmiR-1 locus (Fig. 5C). Third, homozygous mutant DmiR-1KO animals could not be rescued with an identical 8.6-kb transgene in which 57 bp, including the sequence of the entire DmiR-1 21mer, had been replaced with a XhoI site (Fig. 5C). This finding indicated that the 57 bp encoding the DmiR-1 21mer contained the rescuing activity. Finally, homozygous mutant DmiR-1KO animals could be rescued to adulthood when a UAS-DmiR-1 transgene containing 354 bp surrounding the DmiR-1 21mer was specifically expressed in the mesoderm and its muscle cell derivatives (Fig. 5C). These observations strongly support the conclusion that DmiR-1 activity is required specifically in the mesoderm and/or its muscle cell derivatives for viability, a finding consistent with the observed DmiR-1 expression pattern (Fig. 1).
Analysis of the DmiR-1 lethal phase revealed that the majority of DmiR-1KO mutants die as small second instar larvae 2-7 d after hatching (Fig. 6A,E). This phenotype was highly penetrant; few DmiR-1KO mutant larvae died as first instars and none progressed beyond second instar. DmiR-1KO second instar larvae displayed three distinctive characteristics. First, they never grew larger than their size as young second instars (Fig. 6A). Second, they became increasingly lethargic and compromised in their movement (for example, unable to return to their ventral sides when placed on their dorsal sides). Prior to death, most older second instar DmiR-1KO mutant larvae stopped moving entirely and their only signs of life were their responsiveness to touch as well as the contractions of their dorsal vessel. Finally, the body architecture of second instar DmiR-1KO mutant larvae markedly deteriorated. While the bodies of wild-type second instar larvae were cylindrical in shape, the bodies of DmiR-1KO mutant larvae appeared to collapse and flatten.
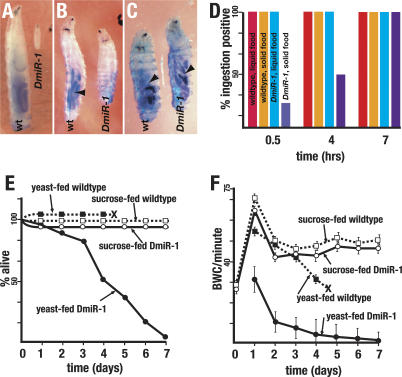
Figure 6.
Feeding triggers _DmiR-1KO_-associated lethality. (A) DmiR-1KO mutants arrest and die as second instar larvae. Compare a 4-d-old wild-type (wt) larva and a 6-d-old DmiR-1KO mutant larva (DmiR-1). (B) DmiR-1KO mutant larvae displayed delayed solid food uptake. Note that the blue food in the gut of the 4-h-old wild-type (wt) larva (arrowhead) and its absence in the gut of an identically aged DmiR-1KO mutant larva. (C) DmiR-1KO mutant larvae displayed normal liquid food uptake. Note that the blue food in the gut of the 1-h-old wild-type (wt) and DmiR-1KO mutant (DmiR-1) larvae (arrowheads). (D) Solid and liquid food uptake rates of newly hatched wild-type larvae (n = 50 per media) and DmiR-1KO mutant larvae (n = 50 per media). DmiR-1KO mutant larvae displayed delayed solid food uptake rates but normal liquid food uptake rates. (E) Lethality rates of yeast-fed wild-type larvae (▪, n = 50), sucrose-fed wild-type larvae (□, n = 50), yeast-fed DmiR-1KO mutant larvae (•, n = 50), and sucrose-fed DmiR-1KO mutant larvae (○, n = 50). Time of puparium formation indicated by ×. (F) Body wall contraction rates (BWC/minute) over time of yeast-fed wild-type larvae (▪, n = 10), sucrose-fed wild-type larvae (□, n = 10), yeast-fed DmiR-1KO mutant larvae (•, n = 10), and sucrose-fed DmiR-1KO mutant larvae (○, n = 10). Time of puparium formation indicated by ×. Vertical lines indicate standard deviations.
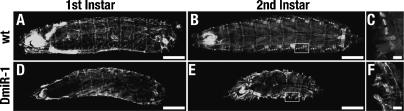
To characterize at the cellular level the body wall muscle of live wild-type and mutant larvae, we examined the expression pattern of a UAS-GFP transgene under the control of the muscle-cell-specific how24B Gal4 driver in wild-type and mutant larvae. The organization and morphology of the musculature of DmiR-1KO first instar larvae appeared normal (Fig. 7A,D). In contrast, the musculature of DmiR-1KO second instar larvae was dramatically disordered (Fig. 7E,F). In comparison to the well-ordered network of body wall muscles of a wild-type second instar larva (Fig. 7B,C), the muscles of a DmiR-1KO mutant appeared as an indistinct mass of GFP fluorescence (Fig. 7E,F). Despite the apparent complete collapse of the body wall musculature, the DmiR-1KO second instar larva in Figure 7E was still alive and, though locomotion defective, was responsive to touch.
Figure 7.
Body wall musculature of second instar DmiR-1KO mutant larvae is severely disrupted. Dorsal views of the expression pattern of UAS-GFP driven by the muscle-cell-specific how24B Gal4 driver in first instar (A,D) and second instar (B,E), wild-type (A,B) or DmiR-1KO mutant (D,E) larvae. (A) Muscle pattern of a wild-type first instar larva. (B) Well-organized network of the somatic musculature of a second instar wild-type larva. Interweaving dorsal muscles are clearly seen in the center of the animal as well as the bright, punctate spots of the lateral transverse muscle down each side of the animal. (C) Close-up (rotated 90° clockwise), indicated by the white box in B, of the lateral transverse muscles of a single hemisegment. (D) Muscle pattern of DmiR-1KO mutant larva. Interweaving dorsal as well as the punctate lateral transverse muscle are clearly essentially normal. (E) Severely disrupted musculature of a second instar DmiR-1KO mutant larvae. The GFP pattern appears as amorphous, irregular condensations. (F) Closeup (rotated 90° clockwise), indicated by the white box in E, of the disorganized muscles of a single hemisegment. Bars: A,D, 100 μm; B,E, 200 μm; C,F, 30 μm.
Newly hatched DmiR-1KO larvae have essentially normal musculature
We next determined that the _DmiR-1KO_-terminal phenotype, death associated with massive disruption of the body wall musculature, was not the result of defective differentiation or development of the embryonic mesoderm or muscle system. No penetrant embryonic defect was identified using numerous mesodermal and muscle cell markers (data not shown) and ∼80% of DmiR-1KO mutant embryos hatched into motile first instar larvae. Although the in situ hybridization data (above) and Northern blot data (data not shown) suggested that the DmiR-1 transcript was zygotically transcribed and not maternally inherited, we nevertheless tested whether the survival of DmiR-1 embryos was caused by maternal rescue of an embryonic defect. Accordingly, we examined embryos in which both the zygotic and any maternal contribution of DmiR-1 were removed. We found that maternal/zygotic DmiR-1KO embryos (produced from females carrying DmiR-1KO germline clones mated to heterozygous DmiR-1KO males) displayed the identical phenotype to zygotic _DmiR-1KO_-null embryos. This result confirmed that DmiR-1 is not required for embryonic development, and that the second instar larval lethal phenotype reflected the earliest acting function of zygotic DmiR-1.
Although the muscle systems of _DmiR-1KO_-null embryos formed normally, we wondered whether a general physiological defect in the larval musculature underlay the second instar locomotion phenotype. If so, newly hatched larvae should display phenotypes associated with defective muscles. To address this, we quantitatively assayed the function of the somatic musculature, the dorsal vessel, and the visceral musculature in newly hatched wild-type and mutant first instar larvae. Strikingly, the muscle function of newly hatched DmiR-1KO mutant larvae, as described below, was essentially normal.
The embryonic somatic musculature gives rise to the larval body wall muscle that is required for larval locomotion. Forward movement is initiated by a contraction of the posterior body wall muscles that propagates anteriorly, narrowing and lengthening the larval body. Body wall contractions (BWC) were quantified by counting the number of end-to-end contractions per minute executed by individual newly hatched larvae. BWC were unaffected in newly hatched DmiR-1KO mutant larvae (Fig. 6F, day 0).
The fly dorsal vessel is the functional equivalent of the vertebrate heart and propels circulation of the hemolymph by periodic lateral constrictions. Dorsal vessel function was assayed by counting the number of dorsal vessel contractions (DVC) per second. Newly hatched wild-type and DmiR-1KO DVC rates and behavior were virtually indistinguishable; the wild-type DVC rate was 1.87 beats per second (n = 10 larvae, standard deviation [SD] is ±0.1) while the DmiR-1KO DVC rate was 1.86 beats per second (n = 10, SD is ±0.33).
Two components of the alimentary tract are the pharyngeal and visceral musculature. The pharyngeal muscles allow the larvae to take up food while the peristaltic contractions of the visceral muscles push the food along the tract. Food uptake and excretion assays were employed to assay the function of the pharyngeal and visceral musculature, respectively. Yeast paste dyed with Brilliant Blue is readily detectable through the larval cuticle and is apparently nontoxic (Fig. 6A). In a typical assay, 100% of the newly hatched wild-type larvae scored positive for ingestion within 30 min (Fig. 6D). In contrast, only 15% of the DmiR-1KO mutant larvae scored positive for ingestion within the same time (Fig. 6B). However, all DmiR-1KO mutant larvae scored positive for ingestion by 7 h after being exposed to food (Fig. 6D). To address whether the delay in food uptake might reflect a weakness in the pharyngeal muscle to suck up solid food, we determined the ingestion rates of wild-type and mutant larvae cultured on Whatman paper soaked with liquid growth media containing yeast extract, sucrose, and blue dye. On this media, DmiR-1KO mutant larvae ingested food at the same rate as wild type; all wild-type and mutant larvae scored positive for ingestion after 30 min (n = 50) (Fig. 6D). This indicated that DmiR-1KO mutants larvae may have weakened pharyngeal muscles that are compromised in their ability to suck up solid food but are able to ingest liquid food at wild-type rates. While wild-type larvae raised on liquid growth media grew and eventually pupariated, DmiR-1KO mutants arrested as second instars. This data indicated that the second instar growth arrest is not a result of a food intake problem since DmiR-1KO mutant larvae can uptake liquid food normally yet exhibited the same second instar growth arrest as mutant larvae fed a diet of solid food.
Furthermore, the mild delay in solid food intake did not reflect a general defect in the passage of food through the digestive tract, since DmiR-1KO larval excretion rates were normal. Wild-type and DmiR-1KO mutant larvae that had scored positive for blue food ingestion after 4 h were transferred to a new plate and scored for the complete loss of the dye. In this assay, DmiR-1KO mutants exhibited the same rate of excretion as wild type (data not shown). This result indicated that the visceral musculature of the mutant larvae functioned normally.
Feeding-dependent muscle disruption, paralysis, and death of DmiR-1KO larvae
The data presented above indicated that newly hatched DmiR-1KO mutant larvae were essentially normal in their muscle function. This finding suggested that the DmiR-1KO mutant phenotype was triggered only after first instar larvae were fed a nutritional food source sufficient to support progression to the second instar. To directly address this hypothesis, we compared the behavior of wild-type and mutant larvae fed a diet of sucrose or liquid growth media. Sucrose-fed wild-type larvae arrest development as first instars but can survive for as long as 2 wk, actively foraging for food (Britton and Edgar 1998; Zinke et al. 2002). Strikingly, DmiR-1KO mutant larvae that were fed only sucrose exhibited the same foraging behavior and longevity as sucrose-fed wild-type larvae and continued to crawl incessantly and at normal rates long after age-matched, yeast-fed DmiR-1KO mutant larvae had become immobilized and died (Fig. 6E,F). While the BWC rates of yeast-fed DmiR-1KO mutant larvae gradually decreased, the BWC rates of similarly aged sucrose-fed DmiR-1KO mutant larvae were maintained at nearly wild-type levels (Fig. 6F). This finding was significant for two reasons. First, it indicated that not only is muscle development normal in first instar DmiR-1KO mutant larvae but the physiology of their muscular system is also essentially wild type. Second, it indicated that food and, by implication growth, is a trigger for the second instar lethality of DmiR-1KO mutant larvae.
Discussion
We present four lines of evidence that DmiR-1 is essential for myofiber function during larval development. First, consistent with the muscle- and heart-specific expression of miR-1 in zebrafish, mouse, and humans (Mansfield et al. 2004; Sempere et al. 2004; Wienholds et al. 2005; Zhao et al. 2005), zygotically expressed DmiR-1 is expressed in most, if not all, the myogenic cells of the larval muscle system. Second, the genetic removal of zygotic DmiR-1 results in a highly penetrant and temporally specific locomotion defect; first instar DmiR-1KO larvae become increasingly lethargic and they eventually die as small immobilized, second instar larvae. Third, the body wall muscles of second instar DmiR-1KO mutant larvae are massively disrupted, presumably causing the locomotion defect. Finally, the larval DmiR-1KO mutant phenotypes, gradual paralysis, and death, can be rescued when DmiR-1 transcript is expressed specifically in muscle cells.
Strikingly, the DmiR-1KO larval phenotype is only manifest after feeding. Newly hatched DmiR-1KO larvae that have not yet begun to feed are nearly wild type in their body wall contractions, dorsal vessel contractions, and excretion. Furthermore, first instar DmiR-1 mutant larvae that are cultured with only sucrose for an energy source but no nutritional supplement to support growth, and consequently do not proceed through larval development to the second instar, are essentially wild type in their movement and longevity. This growth-dependent larval muscle phenotype of DmiR-1KO animals is in contrast to previously described mutations that more generally disrupt larval muscle function. For example, mutations in the α-actinin and ryanodine receptor genes, both of which are expressed throughout the larval musculature, cause growth-independent phenotypes; muscle cell function is strongly compromised even in newly hatched larvae (Fyrberg et al. 1998; Sullivan et al. 2000). From this, we conclude that DmiR-1 function and hence the regulation of gene expression by the DmiR-1 miRNA, is critical for maintaining muscle integrity during the dramatic, post-mitotic growth in muscle mass of wild-type larvae.
Wild-type newly hatched Drosophila larvae encounter a developmental decision: Larvae that are fed a sucroseonly diet arrest development as first instars and can remain so, vigorously searching for food, for up to 2 wk (Britton and Edgar 1998; Zinke et al. 2002). By contrast, larvae that are fed a nutritional food source grow dramatically, increasing their body mass ∼200-fold in 4 d. For most terminally differentiated larval-specific tissues, including the gut, epidermis, fat body, trachea and salivary glands, larval growth is accomplished by expansion of cell size rather than by cell proliferation. For these cells, feeding triggers a specialized cell cycle, the endocycle, in which cells undergo rounds of DNA replication without division (Edgar and Orr-Weaver 2001). In contrast, the precursors of adult structures, including imaginal disc cells and neuroblast cells, grow via conventional diploid cell cycles.
Little is known about the cell biology of larval muscle growth. While neither cell division nor nuclear division takes place, each myofiber expands at least 100-fold in size, presumably also involving endocyclic DNA replication (Haas 1950; Johansen et al. 1989; Bate et al. 1991). Our data do not distinguish whether the DmiR-1KO mutation disrupts entry into the endocycle or some other step in muscle cell growth. Genes that act throughout the larva to control entry into the endocycle have been identified as mutations that cause growth arrest after feeding (Galloni and Edgar 1999; Migeon et al. 1999; Pierce et al. 2004). But unlike DmiR-1KO, endocycle mutants display wild-type larval behavior and locomotion. Hence the DmiR-1KO phenotype cannot be explained as simply a consequence of muscle cells failing to enter the endocycle.
Our understanding of the range of functions that miRNAs perform is still emerging. Some miRNAs act as developmental switches, for example, by controlling stage-specific cell fate choice or by modulating cell division versus cell death (Ambros 2004). Our analysis of the DmiR-1KO phenotype indicates that DmiR-1 does not play such a role in controlling mesodermal cell fates or cell proliferation during embryogenesis, though it is possible that DmiR-1 might function redundantly in such roles with other proteins and/or other mesodermally expressed miRNAs.
DmiR-1 could play a role in triggering growth of muscle in response to feeding, for example by repressing one or more inhibitors of cell growth. However, the myofiber disruption in DmiR-1KO mutants also suggests a role for DmiR-1 in promoting muscle integrity. It has been proposed that some miRNAs could act to reinforce and maintain cell identity during phases of rapid cell division (Bartel and Chen 2004). DmiR-1 could play an analogous role, in this case maintaining muscle tissue identity during a phase of rapid post-mitotic increase in cell mass. A role for DmiR-1 in reinforcing muscle cell identity during the extreme demands of rapid growth could involve the global repression of translation of mRNAs with muscle-toxic products. In a genome-wide microarray analysis of mRNA levels in cells transfected with specific miRNAs, Lim et al. (2005) found that vertebrate miR-1 can post-transcriptionally reduce levels of at least a hundred target mRNAs approximately two- to threefold, possibly as an indirect consequence of the translational repression of these targets by miR-1. The mRNAs that were down-regulated by miR-1 were enriched for mRNAs not normally expressed in muscle. Thus, miR-1 could function generally to maintain muscle cell identity by ensuring that mRNAs from promiscuously transcribed nonmuscle genes remain inactive. Recent findings suggest that this generalization can be extended to flies as well (A. Stark, J. Brennecke, N. Bushati, R.B. Russell, and S.M. Cohen, in prep.).
From our analysis of the top 45 predicted DmiR-1 targets (A. Stark, J. Brennecke, N. Bushati, R.B. Russell, and S.M. Cohen, in prep.), we find that of the 21 genes whose expression has been characterized, none are expressed in the mesoderm (expression patterns were queried at http://www.flybase.org). For example, a number of neurally expressed genes {prospero, pdm-2, suppressor of hairless [_Su(H)_] and lola} contain putative _DmiR-1_-binding sites. prospero and pdm-2 are two transcription factors that regulate the expression of neuroblast cell fate (for review, see Karcavich 2005). A second class of predicted DmiR-1 targets include genes that promote or maintain the polarization of differentiating (par-6) or differentiated (gliotactin and neurexin) neuronal and ectodermal cells. Gliotactin and Neurexin promote the formation of septate junctions, a structure which is never found in mesodermal cells (Tepass and Hartenstein 1994). Perhaps expression of these molecules singly or in combination could derail the maintenance of muscle cell identity and hence their misexpression in muscle would be catastrophic to muscle cell physiology. Promiscuous transcription may be a characteristic of exceedingly active nuclei such as those in muscles undergoing rapid growth, requiring the additional safeguard provided by the translational repression of toxic targets, in this case by DmiR-1.
The protein products of DmiR-1 targets whose expression in DmiR-1KO larvae leads to muscle disruption, paralysis, and death would be expected to be elevated in DmiR-1KO mutants. We stained DmiR-1KO mutant embryos with antibodies to many of the putative targets (Prospero, Su(H), Gliotactin, SoxNeuro, Lola) but no inappropriate expression in the mesoderm or its derivatives was detected (data not shown). However, first and second instar larvae, where the DmiR-1KO phenotype is manifest, are relatively intractable to the standard methods of in vivo analysis of gene expression. The small size and thick cuticle of these larvae render them essentially impervious to dissection or fixation and consequently to antibody staining. Thus, for technical reasons we have been unable to verify specific predicted targets by directly assaying target protein levels in larvae.
Recent studies involving overexpression of mouse miR-1 indicated that Mmu-miR-1 could function in mice to regulate the proliferation of cardiomyocytes in the developing heart (Zhao et al. 2005). We did not observe severe or highly penetrant defects in the formation of the dorsal vessel in DmiR-1KO embryos and the dorsal vessels of mutant larvae appeared to function normally. However, our analysis may not have detected subtle defects in the formation or function of the dorsal vessel, for example in the allocation of pericardial and myocardial cells or in the strength of the dorsal vessel muscle. The Notch pathway has been shown to regulate the differentiation of cardiac progenitors into either myocardial or pericardial cells in flies (Han and Bodmer 2003). Su(H), a component of the Notch pathway and also a putative target of DmiR-1 (A. Stark, J. Brennecke, N. Bushati, R.B. Russell, and S.M. Cohen, in prep.), promotes the differentiation of pericardial cells in Drosophila. Thus, DmiR-1, which is expressed in myocardial cells and not in pericardial cells, could reinforce proper differentiation of cardiac progenitors by repressing Su(H) activity specifically in myocardial cells.
We present evidence that places DmiR-1 within the established mesodermal and myogenic transcriptional networks. Twist and Dorsal are known to coregulate a number of mesodermal genes (Stathopoulos and Levine 2002a) and we observe what could be two weak Dorsal-binding sites in the DmiR-1 enhancer/lacZ fusion transgenes tested here. However, the finding that ectopic Twist in a gd mutant background is sufficient to direct DmiR-1 expression indicates that Twist alone, without Dorsal, is sufficient for DmiR-1 expression. Although Twist activates DmiR-1 transcription throughout the embryonic mesoderm we nevertheless find that embryos entirely depleted of DmiR-1 appear normal. The expression of DmiR-1 in larval muscle, where it carries out its critical function, is likely to be maintained by factors downstream of Twist, particularly Mef2. So, what might be the role for Twist-mediated embryonic expression of DmiR-1? The early expression of DmiR-1 throughout the mesoderm could be gratuitous for the embryo per se but nevertheless could reflect a role for Twist-activated DmiR-1 at a later developmental stage. For example, Twist expression is maintained during larval development in clusters of undifferentiated cells that will give rise to adult myoblasts and ultimately to adult muscles (Bate et al. 1991). Interestingly, the Notch signaling pathway down-regulates Twist expression and thereby promotes the formation of embryonic somatic muscle founder cells at the expense of adult myoblast progenitor cells (Anant et al. 1998). Perhaps Twist-activated expression of DmiR-1 counteracts Notch signaling and hence helps maintain adult myoblast progenitor cells during embryogenesis and larval development.
The identification and analysis of loss-of-function mutations in miRNA genes is an essential tool for identifying the precise role of each miRNA, and for defining the range of roles carried out by miRNAs in general. The value of this approach is underscored by the fact that two evolutionary conserved miRNAs have been analyzed thus far by loss-of-function mutations, Caenorhabditis elegans let-7 (Reinhart et al. 2000) and DmiR-1 (this study), and both mutations exhibit strong phenotypes. That the miR-1 sequence and tissue-specific expression profile is evolutionarily conserved from insects to vertebrates suggests that the developmental and physiological function of miR-1 could also be conserved. If so, our analysis of DmiR-1 larval mutants would predict that vertebrate miR-1 should play a critical role in regulating gene expression within muscle cells at times of growth or during other conditions of developmental and physiological challenge.
Materials and methods
Strains and genetics
Fly stocks were maintained at 22°C or 25°C on standard media. Drosophila stocks that were used included sna18cn1bw1sp1/SM6b P(eve-lacZ), cn1twi1bw1sp1/SM6b P(eve-lacZ), sna18twi3 cn1bw1sp1/SM6b P(eve-lacZ), gd7/FM3, gd7/FM3; P(Twist-bcd) (a gift from A. Stathopoulos and M. Levine, University of California at Berkeley, Berkeley, CA; Stathopoulos and Levine 2002b), mef222.21/CyO P(wg-lacZ) (a gift from H. Nguyen, Albert Einstein College of Medicine, Bronx, NY; Bour et al. 1995), P(GawB)how24B (Bloomington stock 1767), P(Gal4-da.G32)UH1 (Bloomington stock 5460), y1w*; P(ry+,70 FLP)23, P(v+,70 I-SceI)4A/TM6 (Bloomington stock 6935), w1118; P(v+ hs I-CreI.R)1A Sb1/TM6 (Bloomington stock 6937), y1w1118 P(ry+,70 FLP)/Dp(1;Y)y+; nocSco/SM6a (Bloomington stock 6416), P(UAS-GFP-CD8)LL6, and P(UAS-twist)II (a gift from M. Baylies, Memorial Sloan-Kettering Cancer Center, New York; Baylies and Bate 1996). w1118 was used as the control strain in all experiments.
DmiR-1KO mutant flies were generated using ends-in homologous recombination (Rong and Golic 2000). Briefly, an X-chromosome insertion of P(Target70-DmiR-1KO) was used as a donor. y1w*; P(ry+,70 FLP)23, P(v+,70 I-SceI)4A/TM6 virgins were crossed to P(Target70-DmiR-1KO) males and their progeny were heat-shocked for 1 h at 38°C on the third and fourth days of development. Targeting events were screened for by the loss of red/white mosaicism induced by FLP. Southern blotting and PCR was used to identify and verify the targeted mutant using standard methods. I-CreI-mediated reduction to a single allele was carried out using the following cross: targeted Target70-DmiR-1KO virgins were crossed to yw; nocSco/CyO; 70I-CreI, Sb/TM6 males. Target70-DmiR-1KO/CyO;70I-CreI, Sb females were crossed to yw; nocSco/Cyo males. After 3 d, the parents were removed and the vials heat-shocked (1 h at 38°C). Heterozygous female heat-shocked progeny were crossed to yw; noc-Sco/CyO males, and the white-eyed, Sb+ progeny were selected as reduction allele candidates. Southern blotting and PCR was used to identify and verify that both alleles were recovered, using standard methods.
To generate _DmiR-1_KO-null mutant embryos and larvae lacking both zygotic DmiR-1 and any DmiR-1 potentially supplied maternally, _DmiR-1_KO was recombined onto the FRT40A chromosome, females harboring germline clones were generated using the dominant female sterile technique (Chou et al. 1993) and crossed to DmiR-1KO/SM6b, P(eve-lacZ) or DmiR-1KO/CyO, P(Gal4-Kr.C)DC3, P(UAS-GFP.S65T)DC7 males, and homozygous mutant embryos were selected based on the absence of either LacZ or GFP expression.
Plasmid construction
The DmiR-1 locus was initially subcloned as an 8628-bp NheI → XbaI fragment from Bac clone BACR08D17 (purchased from BACPAC Resources) into the XbaI site of pBluescript SK+ generating pBS-DmiR-18.6kb. The DmiR-1 targeting vector was constructed in two steps. First, the BglII site located 802 bp upstream of DmiR-1 was converted into an I-SceI site and 57 bp of the DmiR-1 coding region, including the entire DmiR-1 21mer, was substituted with a XhoI site using the Transformer Site-Directed Mutagenesis Kit (BD Biosciences) using pBS-DmiR-18.6kb as the template. Then, the mutated DmiR-1 locus was subcloned as a NotI → KpnI fragment from pBluescript into pP(Target70) (a gift from J. Sekelsky, University of North Carolina, Chapel Hill, NC) to generate pP(Target70-DmiR-1). pP(W8-DmiR-18.6kb) was generated by subcloning the wild-type DmiR-1 locus as a NotI → KpnI fragment from pBS-DmiR-18.6kb into pP(W8). pP(W8-DmiR-18.6kb → XhoI) was generated by subcloning the mutated DmiR-1 site from pP(Target70-DmiR-1) into pP(W8-DmiR-18.6kb). UAS-DmiR-1 was constructed by PCR amplifying a 354-bp fragment containing the entire DmiR-1 hairpin and subcloning the PCR fragment into pUASP (Rorth 1998). To generate the LacZ reporter constructs, fragments containing various E-box clusters were PCR amplified from pBS-DmiR-18.6kb using a 5′ oligo containing an EcoRI site and a 3′ oligo containing a XhoI site and subcloned into the EcoRI and XhoI sites of pAOE1 (a gift from S. Small, New York University, New York). The PCR-amplified enhancers were sequenced to confirm the absence of PCR-induced errors. P-element-mediated germline transformation was performed according to standard procedures. For the various LacZ reporters, at least three independent lines were examined for expression.
Southern blotting
Genomic DNA was prepared using standard techniques. Genomic DNA was digested with XhoI and NaeI (New England Biolabs), electrophoresed and blotted onto Hybond N+ filter (Amersham). Filters were prehybridized in prehybridization buffer (0.7% SDS, 50% formamide, 5× SSC, 50 mM Na-phosphate buffer at pH 7.0, 1% Blocking reagent) for 1-4 h at 42°C prior to incubation with DIG-labeled PCR probes at 42°C overnight. After washing, the filters were analyzed by the DIG detection chemiluminescent assay according to manufacturer's recommendations (Roche).
In situ hybridization
Embryos were collected on grape juice plates and processed for staining according to standard procedures (Tautz and Pfeifle 1989). Digoxigenin-labeled antisense DmiR-1 riboprobes were generated by PCR-amplifying either a 1130- or 354-bp fragment spanning DmiR-1 from pBS-_DmiR-1_8.6kb. The T7 RNA polymerase promoter was included on the reverse primer and the PCR product was used as a template for in vitro transcription using the T7 MAXIscript kit (Ambion). DmiR-1 LNA probes were obtained from Exiqon, endlabeled using DIG Oligonucleotide 3′-End Labeling Kit (Roche), and purified on G-25 Microspin Columns (Amersham Biosciences).
Larval behavioral assays
Collection of larvae. Adults were allowed to lay embryos on apple juice agar plates for 4 h. These embryos were then collected, dechorionated, and lined-up on new apple juice agar plates. After incubation for an additional 12 h, _DmiR-1_KO embryos were collected based on the absence of GFP expression and placed on new plates. These plates were inspected every 2 h and hatched larvae were collected for muscle contraction assays.
BWC assay. Newly hatched larvae were placed on apple juice agar plates. For a single larva, the number of BWCs that occurred in three different 30-sec periods were recorded, averaged, and converted to BWC per minute.
Ingestion assay. Newly hatched larvae were placed directly on apple juice agar plates supplemented with food. Solid food was a paste of bakers yeast and water plus Brilliant Blue dye. Liquid food was 10% sucrose, 10% yeast extract, and Brilliant Blue dye dissolved in water. Sucrose solution was 10% sucrose in water. Over time, larvae were removed from food and scored for dye uptake. Those positive for ingestion had dark-blue staining in the first section of the midgut, those negative had no or very light-blue staining.
Excretion assay. Larvae were fed blue yeast paste for 4 h and those with blue dye in the hindgut were transferred to a new apple juice agar plate. Larvae scored positive for excretion if the dye had been completely eliminated from their systems.
DVC assay. Digital movies were collected of individual larvae, viewed in slow motion and dorsal vessel contractions were counted. At least 1 min of total footage was collected per larva tested.
Acknowledgments
We thank Mary Baylies, Sakari Kauppinen, Hanh Nguyen, Jeff Sekelsky, Steve Small, Angela Stathopoulos, and Michael Levine for reagents; Julius Brennecke, Alexander Stark, and Stephen Cohen for sharing their target prediction lists prior to publication; members of the Ambros lab and Albert Erives for helpful discussion; and Yashi Ahmed and Claudio Pikielny for use of their fly room and injection facility. N.S.S. was supported by a post-doctoral fellowship from the Damon Runyon Cancer Research Fund (1728-02) and an equipment grant from the Hitchcock Foundation. The Drosophila project in V.A.'s laboratory was supported by a grant from the National Institutes of Health (GM066826).
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1356105.
References
- Ambros V. 2004. The functions of animal microRNAs. Nature 431**:** 350-355. [DOI] [PubMed] [Google Scholar]
- Anant S., Roy, S., and VijayRaghavan, K. 1998. Twist and Notch negatively regulate adult muscle differentiation in Drosophila. Development 125**:** 1361-1369. [DOI] [PubMed] [Google Scholar]
- Andres V., Cervera, M., and Mahdavi, V. 1995. Determination of the consensus binding site for MEF2 expressed in muscle and brain reveals tissue-specific sequence constraints. J. Biol. Chem. 270**:** 23246-23249. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. and Chen, C.Z. 2004. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5**:** 396-400. [DOI] [PubMed] [Google Scholar]
- Bate M., Rushton, E., and Currie, D.A. 1991. Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development 113**:** 79-89. [DOI] [PubMed] [Google Scholar]
- Baylies M.K. and Bate, M. 1996. twist: A myogenic switch in Drosophila. Science 272**:** 1481-1484. [DOI] [PubMed] [Google Scholar]
- Bour B.A., O'Brien, M.A., Lockwood, W.L., Goldstein, E.S., Bodmer, R., Taghert, P.H., Abmayr, S.M., and Nguyen, H.T. 1995. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes & Dev. 9**:** 730-741. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113**:** 25-36. [DOI] [PubMed] [Google Scholar]
- Britton J.S. and Edgar, B.A. 1998. Environmental control of the cell cycle in Drosophila: Nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development 125**:** 2149-2158. [DOI] [PubMed] [Google Scholar]
- Castanon I. and Baylies, M.K. 2002. A Twist in fate: Evolutionary comparison of Twist structure and function. Gene 287**:** 11-22. [DOI] [PubMed] [Google Scholar]
- Chen C.Z., Li, L., Lodish, H.F., and Bartel, D.P. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303**:** 83-86. [DOI] [PubMed] [Google Scholar]
- Chou T.B., Noll, E., and Perrimon, N. 1993. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development 119**:** 1359-1369. [DOI] [PubMed] [Google Scholar]
- Cripps R.M., Black, B.L., Zhao, B., Lien, C.L., Schulz, R.A., and Olson, E.N. 1998. The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes & Dev. 12**:** 422-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A. and Orr-Weaver, T.L. 2001. Endoreplication cell cycles: More for less. Cell 105**:** 297-306. [DOI] [PubMed] [Google Scholar]
- Furlong E.E., Andersen, E.C., Null, B., White, K.P., and Scott, M.P. 2001. Patterns of gene expression during Drosophila mesoderm development. Science 293**:** 1629-1633. [DOI] [PubMed] [Google Scholar]
- Fyrberg C., Ketchum, A., Ball, E., and Fyrberg, E. 1998. Characterization of lethal Drosophila melanogaster α-actinin mutants. Biochem. Genet. 36**:** 299-310. [DOI] [PubMed] [Google Scholar]
- Galloni M. and Edgar, B.A. 1999. Cell-autonomous and non-autonomous growth-defective mutants of Drosophila melanogaster. Development 126**:** 2365-2375. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli, R.M., Glasner, M.E., Enright, A.J., Thomson, J.M., Baskerville, S., Hammond, S.M., Bartel, D.P., and Schier, A.F. 2005. MicroRNAs regulate brain morphogenesis in zebrafish. Science 308**:** 833-838. [DOI] [PubMed] [Google Scholar]
- Haas J.N. 1950. Cytoplasmic growth in the muscle fibers of larvae of Drosophila melanogaster. Growth 14**:** 277-294. [PubMed] [Google Scholar]
- Han Z. and Bodmer, R. 2003. Myogenic cells fates are antagonized by Notch only in asymmetric lineages of the Drosophila heart, with or without cell division. Development 130**:** 3039-3051. [DOI] [PubMed] [Google Scholar]
- Ip Y.T., Park, R.E., Kosman, D., Yazdanbakhsh, K., and Levine, M. 1992. dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes & Dev. 6**:** 1518-1530. [DOI] [PubMed] [Google Scholar]
- Johansen J., Halpern, M.E., Johansen, K.M., and Keshishian, H. 1989. Stereotypic morphology of glutamatergic synapses on identified muscle cells of Drosophila larvae. J. Neurosci. 9**:** 710-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J. and Hobert, O. 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426**:** 845-849. [DOI] [PubMed] [Google Scholar]
- Karcavich R.E. 2005. Generating neuronal diversity in the Drosophila central nervous system: A view from the ganglion mother cells. Dev. Dyn. 232**:** 609-616. [DOI] [PubMed] [Google Scholar]
- Kelly K.K., Meadows, S.M., and Cripps, R.M. 2002. Drosophila MEF2 is a direct regulator of Actin57B transcription in cardiac, skeletal, and visceral muscle lineages. Mech. Dev. 110**:** 39-50. [DOI] [PubMed] [Google Scholar]
- Lee R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294**:** 862-864. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75**:** 843-854. [DOI] [PubMed] [Google Scholar]
- Lee Y., Jeon, K., Lee, J.T., Kim, S., and Kim, V.N. 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 21**:** 4663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leptin M. 1991. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes & Dev. 5**:** 1568-1576. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., and Johnson, J.M. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433**:** 769-773. [DOI] [PubMed] [Google Scholar]
- Mansfield J.H., Harfe, B.D., Nissen, R., Obenauer, J., Srineel, J., Chaudhuri, A., Farzan-Kashani, R., Zuker, M., Pasquinelli, A.E., Ruvkun, G., et al. 2004. MicroRNA-responsive `sensor' transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 36**:** 1079-1083. [DOI] [PubMed] [Google Scholar]
- Migeon J.C., Garfinkel, M.S., and Edgar, B.A. 1999. Cloning and characterization of peter pan, a novel Drosophila gene required for larval growth. Mol. Biol. Cell. 10**:** 1733-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U., Liao, G.C., Niemann, H., and Rubin, G.M. 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3**:** RESEARCH0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce S.B., Yost, C., Britton, J.S., Loo, L.W., Flynn, E.M., Edgar, B.A., and Eisenman, R.N. 2004. dMyc is required for larval growth and endoreplication in Drosophila. Development 131**:** 2317-2327. [DOI] [PubMed] [Google Scholar]
- Poy M.N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., Macdonald, P.E., Pfeffer, S., Tuschl, T., Rajewsky, N., Rorsman, P., et al. 2004. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432**:** 226-230. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403**:** 901-906. [DOI] [PubMed] [Google Scholar]
- Rong Y.S. and Golic, K.G. 2000. Gene targeting by homologous recombination in Drosophila. Science 288**:** 2013-2018. [DOI] [PubMed] [Google Scholar]
- Rorth P. 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78**:** 113-118. [DOI] [PubMed] [Google Scholar]
- Sempere L.F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., and Ambros, V. 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5**:** R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos A. and Levine, M. 2002a. Dorsal gradient networks in the Drosophila embryo. Dev. Biol. 246**:** 57-67. [DOI] [PubMed] [Google Scholar]
- ____. 2002b. Linear signaling in the Toll-Dorsal pathway of Drosophila: Activated Pelle kinase specifies all threshold outputs of gene expression while the bHLH protein Twist specifies a subset. Development 129**:** 3411-3419. [DOI] [PubMed] [Google Scholar]
- Sullivan K.M., Scott, K., Zuker, C.S., and Rubin, G.M. 2000. The ryanodine receptor is essential for larval development in Drosophila melanogaster. Proc. Natl. Acad. Sci. 97**:** 5942-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. and Pfeifle, C. 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98**:** 81-85. [DOI] [PubMed] [Google Scholar]
- Tepass U. and Hartenstein, V. 1994. The development of cellular junctions in the Drosophila embryo. Dev. Biol. 161**:** 563-596. [DOI] [PubMed] [Google Scholar]
- Thisse B., Stoetzel, C., Gorostiza-Thisse, C., and Perrin-Schmitt, F. 1988. Sequence of the twist gene and nuclear localization of its protein in endomesodermal cells of early Drosophila embryos. EMBO J. 7**:** 2175-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman, W.P., Miska, E., Alvarez-Saavedra, E., Berezikov, E., de Bruijn, E., Horvitz, R.H., Kauppinen, S., and Plasterk, R.H. 2005. MicroRNA expression in zebrafish embryonic development. Science 309**:** 310-311. [DOI] [PubMed] [Google Scholar]
- Xu P., Vernooy, S.Y., Guo, M., and Hay, B.A. 2003. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13**:** 790-795. [DOI] [PubMed] [Google Scholar]
- Yin Z., Xu, X.L., and Frasch, M. 1997. Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development 124**:** 4971-4982. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Samal, E., and Srivastava, D. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436**:** 214-220. [DOI] [PubMed] [Google Scholar]
- Zinke I., Schutz, C.S., Katzenberger, J.D., Bauer, M., and Pankratz, M.J. 2002. Nutrient control of gene expression in Drosophila: Microarray analysis of starvation and sugar-dependent response. EMBO J. 21**:** 6162-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]