Sex chromosome evolution: Molecular aspects of Y-chromosome degeneration in Drosophila (original) (raw)
Abstract
Ancient Y-chromosomes of various organisms contain few active genes and an abundance of repetitive DNA. The neo-Y chromosome of Drosophila miranda is in transition from an ordinary autosome to a genetically inert Y-chromosome, while its homolog, the neo-X chromosome, is evolving partial dosage compensation. Here, I compare four large genomic regions located on the neo-sex chromosomes that contain a total of 12 homologous genes. In addition, I investigate the partial coding sequence for 56 more homologous gene pairs from the neo-sex chromosomes. Little modification has occurred on the neo-X chromosome, and genes are highly constrained at the protein level. In contrast, a diverse array of molecular changes is contributing to the observed degeneration of the neo-Y chromosome. In particular, the four large regions surveyed on the neo-Y chromosome harbor several transposable element insertions, large deletions, and a large structural rearrangement. About one-third of all neo-Y-linked genes are nonfunctional, containing either premature stop codons and/or frameshift mutations. Intact genes on the neo-Y are accumulating amino acid and unpreferred codon changes. In addition, both 5′- and 3′-flanking regions of genes and intron sequences are less constrained on the neo-Y relative to the neo-X. Despite heterogeneity in levels of dosage compensation along the neo-X chromosome of D. miranda, the neo-Y chromosome shows surprisingly uniform signs of degeneration.
Sex chromosomes have evolved independently many times in different groups of organisms, including plants, nematodes, Drosophila, butterflies, birds, and mammals (Ohno 1967; Bull 1983). The best-studied Y-chromosomes to date are those of humans and Drosophila melanogaster (Lahn et al. 2001; Carvalho 2002). These highly derived Y-chromosomes retain few functional genes, and consist of a large fraction of repetitive “junk” DNA (Carvalho et al. 2001; Skaletsky et al. 2003). For example, the human Y-chromosome harbors only 27 distinct protein families, and only 16 genes have homologs on the X-chromosome (Skaletsky et al. 2003). About half of the human Y-chromosome is composed of heterochromatin, and the euchromatic portion is a mosaic of X-degenerated, X-transposed, and ampliconic sequences (Skaletsky et al. 2003). Similarly, the D. melanogaster Y-chromosome is entirely heterochromatic, containing a total of only ∼15 genes (Carvalho 2002), while the remaining chromosome consists entirely of highly repetitive and transposable-element (TE)-derived DNA. These Y-chromosomes are ancient (i.e., 50-300 million years old) and are thought to have evolved from ordinary autosomes, but retain few clues about the processes that initiated their degeneration (see Carvalho 2002 for an alternative hypothesis on the origin of the D. melanogaster Y-chromosome). For example, it is unclear whether repetitive DNA began to accumulate on the Y-chromosome only after it already lost most of its genes, or whether it precipitated this degeneration in the first place. To investigate the causes of Y-chromosome degeneration, it is necessary to study systems that are still in the process of degeneration (Charlesworth and Charlesworth 2000).
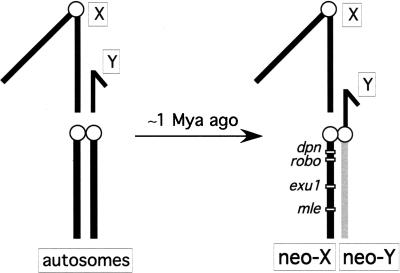
Here, I take advantage of the recently formed neo-sex chromosomes of Drosophila miranda (MacKnight 1939). The neo-Y chromosome of D. miranda was formed about 1.2 million years ago (Mya) (Bachtrog and Charlesworth 2002), by the fusion of an autosome (corresponding to Chromosome 3 in Drosophila pseudoobscura, the closest relative of D. miranda) to the ancestral Y-chromosome (see Fig. 1; MacKnight 1939). The neo-Y is in transition from an ordinary autosome to a degenerate Y-chromosome (MacKnight 1939; Steinemann and Steinemann 1998; Bachtrog 2003a). Interestingly, comparative mapping studies have shown that large parts of the human Y-chromosome are also derived from an autosomal region added to the ancestral Y-chromosome; thus parts of the human Y also represent a degenerate neo-Y chromosome (Waters et al. 2001). In addition, the Y-chromosome of D. pseudoobscura has been proposed to derive from a degenerate neo-Y chromosome (Carvalho and Clark 2005). While the neo-Y chromosomes of humans or D. pseudoobscura are very ancient and almost completely degenerated, the much younger neo-Y of D. miranda still harbors many functional genes. Thus, the neo-Y chromosome of D. miranda serves as an excellent model to study the processes underlying the early stages of sex chromosome evolution.
Figure 1.
Schematic karyotype of the D. miranda neo-sex chromosomes. The neo-sex chromosomes were formed by the fusion of an autosome (Chromosome 3 of D. pseudoobscura) to the Y-chromosome of D. miranda. Shown are the relative locations of the four genomic regions investigated on the neo-X chromosome of D. miranda.
The X- and Y-chromosomes are thought to have evolved from an ordinary pair of autosomes that stopped recombining with each other after acquiring a sex-determining role (Charlesworth 1996; Rice 1996). In the absence of recombination, these originally homologous chromosomes continued to differentiate. Evolutionary theory predicts that deleterious mutations should accumulate faster on a nonrecombining chromosome, whereas beneficial mutations are less likely to become incorporated (Charlesworth and Charlesworth 2000). This will lead to a decline in the level of adaptation on a nonrecombining chromosome. The X-chromosome, which still can recombine in females, maintains most of its ancestral genes, whereas the Y-chromosome degenerates because it is sheltered from recombination (Charlesworth 1996; Rice 1996). Since there is no recombination in Drosophila males, the male-limited neo-Y chromosome of D. miranda completely lacks recombination over its entire length. The decline in the level of adaptation of a proto-Y relative to its homolog favors the evolution of dosage compensation on the X-chromosome (Lucchesi 1978). In fact, the D. miranda neo-X chromosome already shows signs of partial dosage compensation, suggesting that dosage compensation may have an important role early on in the process of sex chromosome evolution (Bone and Kuroda 1996; Marin et al. 1996).
In a previous study, I compared patterns of genome evolution in a 40-kb gene-rich region on the neo-X and neo-Y (Bachtrog 2003a). Genes on the nonrecombining neo-Y chromosome showed various signs of degeneration, including TE insertions, frameshift mutations, and a higher rate of amino acid substitutions, suggesting that deleterious mutations, and particularly TEs, can start to accumulate in the very early stages of Y-chromosome evolution. However, given that only one region of the neo-Y was surveyed, it was unclear how general these observations are, especially since the neo-X chromosome has evolved partial dosage compensation (Bone and Kuroda 1996; Marin et al. 1996). Regions on the neo-Y that are dosage-compensated on the neo-X may be under reduced functional constraint and evolve essentially neutrally; conversely, neo-Y regions that are not yet dosage-compensated may be much more constrained.
Here, I compare patterns of molecular evolution of four large segments containing 12 homologous gene pairs dispersed along the neo-sex chromosomes. In addition, I isolate 56 smaller homologous gene fragments from the neo-sex chromosomes, to address the following questions: What is the molecular nature of the changes causing the degeneration of the neo-Y chromosome? Given heterogeneity in levels of dosage compensation on the neo-X (Marin et al. 1996), how much does the level of Y-chromosome degeneration vary along the chromosome? How large of a role do TEs play in the process of Y-chromosome degeneration? Do most protein-coding genes show signs of degeneration on the neo-Y, or only a subset? What is the fraction of nonfunctional genes on the neo-Y? Do regulatory regions show a similar reduction in levels of adaptation on the neo-Y as protein-coding genes?
Results
To investigate the level of degeneration of the neo-Y chromosome, I isolated and sequenced four large genomic regions located on the neo-sex chromosomes of D. miranda (see Methods). I obtained a total of ∼100 kb of sequence information from both the neo-Y chromosome of D. miranda and the homologous regions located on the neo-X. I confirmed by in situ hybridization that the four genomic regions investigated are, indeed, located on the neo-sex chromosomes of D. miranda, as expected based on chromosome homology in the genus Drosophila (Muller 1940). The relative positions of the four fragments are shown in Figure 1; the mle region is found most distal at position 26D, the exu1 region at 30A, the robo region is at position 33C, and the dpn region is located most proximal at chromosomal position 34B, based on the chromosome map of D. miranda by Das et al. (1982). The genomic regions characterized contain coding sequences for a total of 12 homologous genes (Fig. 2). In addition to the four large contiguous genomic regions above, I isolated 56 homologous gene pairs with an average length of 1.2 kb from the neo-sex chromosomes (see Methods and Supplemental Table 1). These genes were selected randomly with the only requirement of containing at least 650 bp of coding sequence, and are distributed all over the neo-X chromosome. Thus, a total of ∼170 kb of sequence was analyzed from both the neo-X and the neo-Y chromosomes. Comparison with sequence from Chromosome 3 of the published D. pseudoobscura genome (the autosomal homolog of the neo-sex chromosome) allowed me to infer the direction of the evolutionary changes occurring on either the neo-X or the neo-Y, using parsimony criteria. Applying this method reveals that little modification has occurred on the neo-X chromosome, although many deletions, insertions, and single-nucleotide changes have accumulated on the neo-Y.
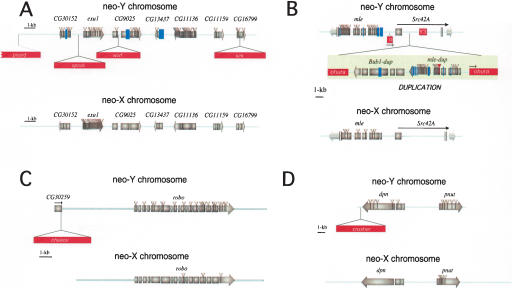
Figure 2.
Organization of the four genomic regions investigated on the neo-sex chromosomes. (A) The exu1 region (Bachtrog 2003a). A total of ∼40 kb of neo-X and ∼45 kb of neo-Y sequence was investigated. (B) The mle genomic region. Here 8 kb of neo-X- and 25 kb of neo-Y-derived sequence were analyzed. (C) The robo genomic region. A total of 6 kb of neo-X and 11 kb of neo-Y sequence was analyzed. (D) The dpn genomic region. Here 24 kb was analyzed from the neo-X chromosome, and 17 kb from the neo-Y. Protein-coding genes are shown as gray bars, and transposable element insertions are displayed as red bars. Amino acid changes are indicated as gray triangles, premature stop codons are displayed as red triangles, and deletions destroying the reading frame of the protein are represented as blue bars.
Transposable element insertions
Seven large DNA insertions were detected on the neo-Y chromosome that are absent from the neo-X (Fig. 2). Sequence analysis revealed that each of these insertions represents a transposable element. Four out of the seven TEs are integrated into intergenic regions, two are found in introns, and one is inserted into the 3′-coding region of the CG30259 gene (Fig. 2). All seven TEs are retrotransposons (i.e., they replicate through an RNA intermediate) and were searched against Repbase (Jurka 2000). Two of the elements, chekov and ohura, belong to the copia family of LTR retrotransposons; crusher and picard are both LTR retrotransposons homologous to diver2; kirk is a non-LTR transposon homologous to Crla; and spock and worf are both non-LTR retrotransposons and were described in detail before (Bachtrog 2003b). As previously shown for spock and worf (Bachtrog 2003b), I used PCR to investigate the frequency of the five new TE insertions in a sample of 12 wild-derived D. miranda strains; all seven TEs are fixed at their specific genomic positions on the neo-Y chromosome (data not shown).
Duplication of the mle and Bub1 gene region
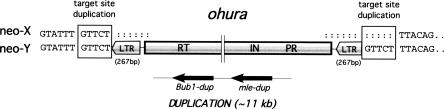
One of the four contiguous regions investigated contains an unusual structural modification on the neo-Y chromosome, consisting of the duplication of the _mle_-Bub1 gene region (Fig. 2B). This duplication is ∼11 kb in size, and appears to be embedded in an LTR retrotransposable element (Figs. 2B and 3). The insertion is contained within an intron of the Src42A gene (Fig. 2B), and is accompanied by a 5-bp target site duplication (Fig. 3). The TE involved in this duplication, ohura, is bounded by two 267-bp-long direct repeats that differ by only two nucleotides (Fig. 3). The duplicated copy of mle is most closely related to the mle gene from the neo-Y and not the neo-X, indicating that the duplication arose from the neo-Y. Both mle copies have several frameshift mutations, some of which are shared, indicating that the gene was nonfunctional before its duplication. The Bub1-dup gene also contains a frameshift mutation. Silent site divergence between the ancestral and the duplicated genomic regions is 0.61%. This is about fivefold lower than average levels of divergence between neo-X- and neo-Y-linked genes (Bachtrog and Charlesworth 2002), suggesting this was a recent duplication relative to the age of the neo-Y chromosome. Although ohura is a retroelement, both copies of mle and the Bub1-dup genes have introns, arguing against retrotransposition as a mechanism generating this duplication. The original copy of mle contains a 715-bp insertion in its 5′-flanking region that is homologous to the first 720 bp of the ohura transposable element 5′ of the mle-dup gene (<1% divergence). The ohura element is located at precisely the same location in the original and duplicate copy of the mle gene (see Fig. 2B). In addition, an ISY3 sequence (a short 713-bp motif that is distributed more than 100 times in the female genome of D. miranda) (see Steinemann and Steinemann 1993) is found in close proximity to the duplicated region (Fig. 2B).
Figure 3.
Insertion site of the ohura transposable element on the neo-Y chromosome of D. miranda. The TE insertion is flanked by a 5-bp target site duplication. The ohura TE is flanked by a 267-bp LTR, and contains three protein domains, a protease (PR), an intergrase (IN), and a reverse transcriptase (RT).
Degeneration of protein-coding genes
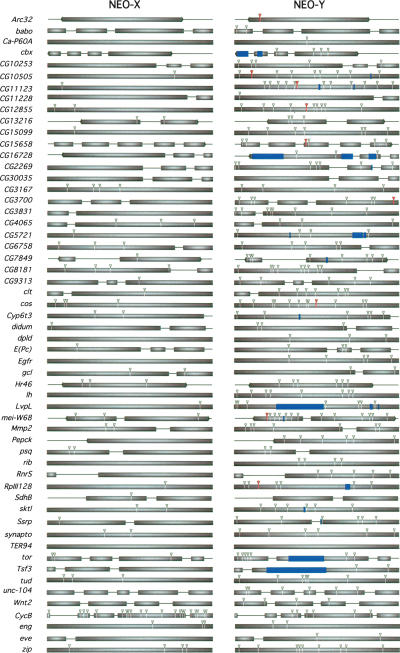
Protein-coding loci on the neo-Y chromosome show various degrees of degeneration. Of the 68 homologous gene pairs studied here, 24 contain premature stop codons and/or frameshift mutations on the neo-Y (Figs. 2 and 4); the remaining genes appear to be translatable into functional proteins. In contrast, none of the homologous neo-X genes contain null mutations. Genes on the neo-Y chromosome show generally higher levels of amino acid substitution than their neo-X homologs (Table 1). The exu1 and CycB genes were previously shown to have undergone adaptive protein evolution on the neo-X (Bachtrog and Charlesworth 2002; Bachtrog 2003a), and were excluded from further analysis. The remaining 66 gene pairs investigated contain a total of 364 amino acid substitutions on the neo-Y chromosome, while the neo-X homologs only have 80 amino acid changes. Thus, amino acid substitutions are almost fivefold more abundant on the neo-Y compared to most neo-X genes (p < 0.01, two-tailed signs test) (see Table 2). This rapid accumulation of amino acid mutations cannot be explained by a higher mutation rate or the neo-Y chromosome, using synonymous substitutions as a neutral standard (p < 0.001, Fisher's exact test).
Figure 4.
Evolution of protein-coding genes on the neo-sex chromosomes. Boxes correspond to exons (with left or right arrows corresponding to the first or last exon of a gene, respectively), and lines correspond to introns or flanking intergenic regions. Gray triangles indicate amino acid changes, blue bars represent frameshift deletions, and red triangles correspond to premature stop codons.
Table 1.
Genes analyzed in this study from the neo-sex chromosome of D. miranda
| Total sites | Synonymous sites | Replacement sites | Noncoding sites | neo-X | neo-Y | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| _K_a (%) | _K_s (%) | _K_a/_K_s | _K_a (%) | _K_s (%) | _K_a/_K_s | |||||
| All genes (n = 68) | 92,498 | 16,809.7 | 61,217.3 | 14,166 | 0.23 | 1.80 | 0.13 | 0.72 | 2.44 | 0.47 |
| All genesa (n = 66) | 88,678 | 16,107.6 | 58,802.4 | 13,486 | 0.17 | 1.76 | 0.12 | 0.72 | 2.46 | 0.47 |
| Functional genes (n = 44) | 60,647 | 11,272.7 | 41,005.3 | 8,174 | 0.26 | 1.74 | 0.17 | 0.59 | 2.55 | 0.35 |
| Nonfunctional genes (n = 24) | 31,851 | 5,537.0 | 20,212.0 | 5,992 | 0.16 | 1.92 | 0.08 | 0.97 | 2.23 | 0.68 |
Table 2.
Observed numbers of nucleotide substitutions along the neo-X and neo-Y chromosomes at different classes of sites (excluding CycB and exu1)
| neo-X | neo-Y | Ratio | |
|---|---|---|---|
| Synonymous substitutions | |||
| Total | 237 | 342 | 1.44* |
| P → U | 108 | 193 | 1.79* |
| U → P | 42 | 42 | 1 |
| Neutral | 87 | 107 | 1.23 |
| Amino acid changes | 80 | 364 | 4.6* |
| Noncoding substitutions | |||
| Total | 144 | 248 | 1.72* |
| Introns | 107 | 174 | 1.63 |
| 5′ and 3′ flanking regions | 37 | 75 | 2.03* |
I also used a maximum-likelihood method to estimate the ratio of amino acid versus synonymous substitutions (i.e., _K_a/_K_s) on the neo-sex chromosomes, as implemented in the PAML software package. A model in which _K_a/K_s was allowed to differ on the neo-X and neo-Y branches fits the data significantly better than a model that assumes the same rate for both branches of the phylogeny (2Δ_L = 60.4, p < 10–6). The average _K_a/_K_s for neo-Y genes is 0.47, about fourfold higher than the average _K_a/_K_s for most neo-X-linked genes (0.12). Thus, the neo-Y chromosome shows a significantly higher rate of protein evolution than the neo-X.
Synonymous substitutions also show a slight (1.5-fold) increase in their rate of accumulation on the neo-Y (Table 2); a total of 237 synonymous substitutions were observed on the neo-X chromosome, but 342 on the neo-Y. This could reflect a higher mutation rate of the male-limited neo-Y chromosome (i.e., male-driven evolution), if synonymous sites are evolving entirely neutrally. Alternatively, synonymous sites might be under weak purifying selection (i.e., codon bias), and the efficiency of this selection is reduced on the neo-Y.
To distinguish between these alternatives, I divided synonymous codons into different categories, with preferred codons (P) corresponding to the selectively favored codon for a specific amino acid, and unpreferred (U) codons corresponding to disfavored codons (Akashi and Schaeffer 1997). Changes from preferred to unpreferred codons (P → U) are presumed to be deleterious, while changes from an unpreferred to a preferred codon (U → P) are presumably beneficial; changes within a class (i.e., P → P or U → U) are assumed to be selectively neutral. Dividing the synonymous substitutions on the neo-X and neo-Y chromosomes in these various categories reveals several interesting features (Table 2). The number of neutral changes is similar on the neo-X and neo-Y chromosomes (87 vs. 107 substitutions). This suggests that there might be a slight (i.e., 1.2-fold), but not statistically significant, elevation in mutation rate on the neo-Y (p > 0.1, two-tailed signs test). This ratio of neutral changes will be used in subsequent analyses as a “neutral standard” to detect differences in patterns of nucleotide substitutions on the neo-X and neo-Y chromosomes. There are significantly more substitutions from P → U codons on both the neo-X and neo-Y chromosomes than there are from U → P (p < 0.01 for both the neo-X and neo-Y, sign test). This excess of unpreferred codon substitutions probably reflects a recent reduction in the amount of codon bias in the _D. miranda_ lineage (Bachtrog 2003c). The numbers of _U_ → _P_ substitutions are identical on the neo-X and neo-Y chromosomes (i.e., 42), and do not statistically differ from the neutral expectation (_p_ > 0.1, Fisher's exact test). However, the neo-Y has significantly more P → U substitutions than the neo-X chromosome (193 vs. 108, p < 0.05 relative to the numbers of neutral changes, using a Fisher's exact test). Thus, there are almost five times more P → U substitutions than U → P on the neo-Y (compared to a 2.5-fold excess of P → U substitutions on the neo-X) (Table 2). This suggests that selection is maintaining some codon bias on the neo-X chromosome (though this selection appears to be weaker than it was in its ancestor). However, unpreferred codons are rapidly accumulating on the neo-Y chromosome, including genes that are still functional on the neo-Y, likely because of the decreased efficiency of natural selection.
Evolution of noncoding, potentially regulatory regions
To characterize changes in noncoding (potentially regulatory) regions, intron sequences and up to 200 bp upstream and downstream of the coding sequence of all genes were aligned. Changes were localized to either the neo-X or neo-Y chromosome, using parsimony criteria. Similar to protein-coding regions, noncoding regions show a significantly higher number of nucleotide changes on the neo-Y compared to the neo-X (248 vs. 144, p < 0.05, Fisher's exact test vs. neutral synonymous changes). Considering intron and flanking regions independently, this effect remains significant for flanking regions (p < 0.05, Fisher's exact test), and goes in the same direction for introns but is not significant (probably reflecting a lack of statistical power; p = 0.08, Fisher's exact test vs. neutral synonymous changes).
The observation of faster evolution on the neo-Y in noncoding flanking regions has two important implications. First, it suggests that intron and intergenic regions are under selective constraint in D. miranda. That is, if these regions would lack any selective constraints, their rate of molecular evolution should be similar on the neo-X and neo-Y chromosomes (or more accurately, it should be similar to the one of neutral synonymous changes). Secondly, the higher rate of substitution in noncoding regions on the neo-Y chromosome can again be understood in terms of a reduction in the effectiveness of natural selection on the neo-Y, causing the accumulation of deleterious regulatory changes on the neo-Y that are eliminated from the neo-X.
Discussion
The role of transposable elements and repetitive DNA
One of the most striking findings on the neo-Y of D. miranda is the abundance of transposable elements (Steinemann and Steinemann 1998; Bachtrog 2003b). Ancient Y-chromosomes, like those of humans or D. melanogaster, consist largely of repetitive and TE-derived DNA. An accumulation of TEs might be expected on these highly degenerated Y-chromosomes because they almost entirely lack functional genes (Charlesworth et al. 1994). Under this model, TE insertions have no fitness consequences, and accumulate on the Y-chromosome neutrally. Alternatively, TEs could accumulate in the early stages of Y-chromosome evolution (i.e., when the Y-chromosome still contains many functional genes) and thus actively contribute to the observed loss of gene function. In the four contiguous neo-Y-linked regions studied here, I find a large number of TE insertions interspersed among functional genes (comprising 1/3 of the surveyed neo-Y sequence). Using in situ hybridization experiments, I and others have shown that there is a striking accumulation of TEs on the neo-Y of D. miranda (Steinemann and Steinemann 1998; Bachtrog 2003b). Together these findings support the notion that TEs are playing an active and early role in the degeneration of the neo-Y.
The accumulation of TEs likely reflects the reduced efficiency of selection at removing these, often deleterious, mutations from the neo-Y. In support of this model, I have shown that each of these TEs is fixed at its individual position in a population sample of D. miranda. This is unusual for TEs in euchromatic regions in Drosophila, which normally segregate at low frequencies at individual insertion sites (Charlesworth and Langley 1989). However, it is not unexpected for the neo-Y chromosome of D. miranda, as the neo-Y probably underwent a recent selective sweep (Bachtrog 2004). The fixation of a single beneficial mutation on the nonrecombining neo-Y would eliminate all variation segregating in the population on the neo-Y, and drag to fixation all TEs with which it is initially associated. Also, the fact that all TEs identified so far on the D. miranda neo-Y are retrotransposons is not unexpected given that analysis of the D. melanogaster genome suggests that 75% of all TEs in this species are retrotransposons (Kaminker et al. 2002).
TEs can have both direct and indirect roles in contributing to Y-chromosome degeneration. Direct effects are apparent as insertions into coding and regulatory regions on the neo-Y (Fig. 2). However, TEs can also indirectly interfere with regular gene expression by producing antisense transcripts of adjacent genes (Kashkush et al. 2003; Puig et al. 2004), or by altering the chromatin structure of the Y-chromosome. While autosomes and X-chromosomes are composed mainly of transcriptional active euchromatin, Y-chromosomes typically contain large quantities of heterochromatin. Heterochromatin has been found to consist largely of TE-derived DNA (The Cold Spring Harbor Laboratory, Washington University Genome Sequencing Center, and PE Biosystems Arabidopsis Sequencing Consortium 2000), and experimental evidence in Arabidopsis shows that heterochromatin can be induced by TE insertions (Lippman et al. 2004). It is interesting to note that portions of the neo-Y of D. miranda are already partially heterochromatic (MacKnight 1939; Steinemann and Steinemann 1998). This may at least in part be caused by the accumulation of TE insertions (Steinemann and Steinemann 1998; Bachtrog 2003b). A change in chromatin structure could, in turn, influence gene expression of a large number of neo-Y-linked loci, similar to the phenomenon of position effect variegation (Schotta et al. 2003). Detailed gene expression analysis should reveal the effect of individual TE insertions on the function of neo-Y-linked genes.
TEs are also playing a role in generating chromosomal rearrangements on the neo-Y chromosome. TEs have been implicated in the creation of chromosomal inversions in Drosophila (Andolfatto et al. 1999; Caceres et al. 1999), Anopheles (Mathiopoulos et al. 1998), and Caenorhabditis (Coghlan and Wolfe 2002). Here, I characterize a large duplication of the mle_-Bub1 gene region that is flanked by a retrotransposable element. An ISY3 sequence is inserted in close proximity to this duplication on the neo-Y. This arrangement parallels another duplication event of the Lcp2 gene identified on the D. miranda neo-Y chromosome that is also associated with both a retrotransposon and an ISY3 element (Steinemann and Steinemann 1993). This suggests that the ISY3 motif and/or the retrotransposable element may be directly involved in generating gene duplications on the neo-Y of D. miranda. The mechanism for the observed rearrangement is unknown, although two possibilities include unequal crossing-over between sister chromatids or duplication of the region resulting from replication accidents. In addition, some of the PCR-isolated genes appear to be duplicated on the neo-Y. For three genes—_zip, CG15099, and _Ssrp_—I obtained a single gene sequence from females, but three gene copies from males (data not shown). This suggests that these genes are also duplicated on the neo-Y chromosome. The human Y-chromosome also contains several gene families created by gene duplications; examples include the DAZ gene cluster (Saxena et al. 1996) or testis transcripts 1 and 2 (Lahn et al. 2001). In addition, the recently formed Y-chromosome of sticklebacks harbors several local duplications (Peichel et al. 2004). Thus, frequent gene duplications appear to be another prominent feature of Y-chromosome evolution.
Reduced constraint at coding DNA and regulatory regions on the neo-Y
Genes on the neo-Y chromosome show a higher rate of amino acid evolution compared to the neo-X (Figs. 2 and 4), and a subset of the neo-Y genes contains large deletions, frameshift mutations, and premature stop codons. In addition, genes on the neo-Y chromosome show an increased rate of accumulation of unpreferred codons. In principal, any changes on the neo-Y can either be detrimental, neutral, or beneficial. The neo-Y chromosome of D. miranda does not recombine; this reduces the efficiency of natural selection and can lead to the accumulation of deleterious mutations that would otherwise be eliminated from a recombining chromosome. Large deletions, frameshift mutations, and premature stop codons are probably deleterious since they tend to destroy gene function (and such changes are not observed on the neo-X lineage). Also, unpreferred codons are a priori assumed to be deleterious, since these codons are less frequently used in highly expressed genes and genes showing high codon bias (Akashi and Schaeffer 1997).
However, the mode of selection acting on amino acid mutations is more difficult to identify. Some genes on the neo-Y are clearly nonfunctional, and any mutation occurring after pseudogenization of the neo-Y copy is probably neutral. If genes are divided into potentially functional genes and those that contain frameshift mutations, the former class of genes are more constrained. The rate of amino acid evolution versus synonymous substitutions of neo-Y loci is significantly lower for functional genes (_K_a/_K_s = 0.35) than for nonfunctional genes (_K_a/_K_s = 0.68; t_-test, p < 0.05) (see Table 1). This suggests that at least some of the amino acid accumulation on the neo-Y chromosome is neutral. However, potentially functional neo-Y genes still have a higher rate of protein evolution compared to their neo-X homologs, using the maximum likelihood approach (2Δ_L = 24.5, p < 10–6).
Amino acid changes at functional genes are less likely to be neutral, and are probably deleterious. This said, some of the amino acid changes observed in intact genes on the neo-Y chromosome could also reflect adaptive changes. For example, genes on a Y-chromosome are male-limited and thus might evolve male-specific functions. Although theory predicts that the overall rate of adaptation should be lower on the neo-Y, this chromosome has apparently experienced recent positive selection (Bachtrog 2004). Thus, at least some genes on the neo-Y chromosome must evolve adaptively.
If positive selection driving protein evolution is limited to a fraction of genes only, there should be significant heterogeneity in the rate of amino acid evolution among genes. This is, indeed, the case for genes investigated on the neo-X chromosome (χ2 = 352.7; p < 0.001). Amino acid substitutions occur more uniformly among neo-Y genes. Of the 68 gene pairs analyzed, four show more amino acid changes on the neo-X (two of which are _CycB_ and _exu1_), eight genes show equal numbers of amino acid substitutions, but 56 genes show more amino acid changes on the neo-Y chromosome. However, there is significant heterogeneity in amino acid evolution even among neo-Y genes (χ2 = 109.0; _p_ < 0.01). If neo-Y genes are divided into genes that contain null mutations and those that are potentially functional, there is no significant heterogeneity within the first class (χ2 = 28.6; _p_ > 0.05) but some within the second class (χ2 = 63.1; p = 0.02). This heterogeneity could be caused by variation in selective constraints among genes, or adaptive amino acid evolution at some genes. In summary, these patterns suggest that positive amino acid evolution might occur but is probably not rampant on the neo-Y and that deleterious mutations accumulate rather uniformly among neo-Y genes. Nonfunctional genes show a significant elevation in rates of amino acid evolution, as expected if these genes evolve essentially neutrally, once inactivated.
A similarly elevated rate of amino acid evolution has also been found for some of the few loci left on the human Y-chromosome that have homologs on the X-chromosome (Wyckoff et al. 2002). Also, plants and birds show an elevated rate of amino acid evolution on their nonrecombining Y- and W-chromosomes, respectively (Fridolfsson and Ellegren 2000; Filatov 2005). Thus, nonrecombining genomes show reduced levels of adaptation in several, evolutionarily independent genomes.
Interestingly, putatively regulatory regions flanking protein-coding genes also show more mutations on the neo-Y compared to the neo-X (Table 2). This observation has two important implications. First, it suggests that these noncoding regions are evolving under selective constraints on the neo-X chromosome of D. miranda, and thus must be of functional importance. In fact, a large portion of eukaryotic genomes is composed of noncoding DNA, and the function of this DNA is currently the subject of intensive research. Consistent with the functionality of noncoding DNA, evolutionary constraint has been detected in D. melanogaster at intergenic DNA upstream and downstream of protein-coding genes (Halligan et al. 2004), and in long introns (Haddrill et al. 2005). Secondly, the reduction in constraint for noncoding regions on the neo-Y chromosome can be understood for reasons analogous to those discussed for protein-coding genes. Purifying selection is less efficient on the neo-Y; thus, slightly deleterious changes in proximal and upstream regulatory regions are more likely to accumulate. In addition, if the protein encoded by a neo-Y-linked gene is maladapted or nonfunctional, the regulatory elements themselves should be under reduced selective constraint and might accumulate (previously deleterious) changes in a neutral manner. Finally, positive selection may drive the divergence of flanking regions on the neo-Y chromosome. An example may be genes on the neo-Y that evolve novel expression patterns in response to being limited to the male sex. Alternatively, positive selection could down-regulate genes on the neo-Y, either because they are maladapted at the protein level or because they are dosage-compensated on the neo-X (Orr and Kim 1998). Again, gene expression analysis should help to distinguish between these hypotheses.
Confounding effects of segregating versus fixed differences on the neo-sex chromosomes
Sampling of a single allele from the neo-X and neo-Y chromosomes, as done in this study, does not allow fixed and segregating mutations to be distinguished. That is, a small fraction of the mutations assigned as a fixed difference occurring on the neo-sex chromosome might, in fact, be segregating in the population. The severe reduction in levels of variability on the neo-Y chromosome implies that nearly all mutations on the neo-Y are fixed within the population (Bachtrog 2004). However, a few of the variants that are assigned as a fixed difference occurring on the neo-X might be polymorphic in the population. In particular, mutations under weak purifying selection will segregate within populations, but selection will ultimately prevent their fixation. Indeed, it has been argued that the observed excess fixation of P → U codons on the neo-X of D. miranda is an artifact of misclassifying segregating polymorphic variants as fixed differences (Bartolome et al. 2005). However, a within-species polymorphism data set for 24 neo-X genes (D. Bachtrog, unpubl.) shows significantly more fixed differences from P → U codon changes occurring on the neo-X lineage, than U → P (27 vs. 7, p < 0.01), supporting the original conclusion that selection for codon bias is, indeed, reduced in D. miranda compared to its ancestor (Bachtrog 2003c).
For all other conclusions drawn from comparisons of the neo-X and neo-Y chromosome, this potential misclassification of fixed sites on the neo-X is, in fact, conservative. Some amino acids, unpreferred codons, or changes at noncoding regions on the neo-X chromosome are probably slightly deleterious and represent a segregating variant on the particular neo-X allele studied that will eventually be eliminated by natural selection. This will cause the excess of mutation accumulation on the neo-Y to be underestimated.
Conclusions
The D. miranda neo-Y chromosome provides several fascinating insights into the early stages of Y-chromosome evolution. In contrast to ancient Y-chromosomes—like those of humans or _D. melanogaster_—this neo-Y chromosome retains most of its ancestral genes. However, about one-third of the genes studied to date are clearly nonfunctional on the neo-Y, containing frameshift mutations or premature stop codons. Most remaining neo-Y genes show more subtle changes, such as an elevated rate of amino acid substitution, an accumulation of unpreferred codons, and less evolutionary constraint in regulatory regions. Transposable elements show a striking accumulation on the neo-Y chromosome and are interspersed among functional genes. This pattern strongly suggests that TEs play an active and early role in the process of Y-chromosome degeneration. Thus, a diverse array of molecular changes, likely representing a wide range of deleterious fitness effects, contributes to the observed degeneration of Y-chromosomes. Despite the heterogeneity observed in levels of dosage compensation on the neo-X, all four contiguous genomic regions investigated in this study show similar patterns and degrees of degeneration. Likewise, the many protein-coding genes investigated show surprisingly uniform levels of degeneration on the neo-Y. This may indicate that dosage compensation does not have a huge impact in the early stages of Y-chromosome degeneration. It will be of great interest to measure expression profiles of neo-sex-linked genes to directly relate the amount of molecular degeneration to levels of gene expression and dosage compensation.
Methods
Cloning of the neo-X- and the neo-Y-linked regions of D. miranda
A genomic library was constructed from D. miranda males (strain 0101.3) using the Lambda Fix II Library kit (Stratagene). High molecular weight genomic DNA from D. miranda males was partially digested with Sau3A and ligated to the vector following the instructions of the manufacturer. This genomic library was screened for the neo-Y- and neo-X-linked copy of exuperantia1, male-less lethal, roundabout, and deadpan. Library screening and standard molecular techniques were carried out according to Sambrook et al. (1989) and are described in Bachtrog (2003b). DNA was prepared from the isolated phage and subcloned into the pZero2.1 vector (Invitrogen). A combination of long PCR (Roche) using allele-specific primers and library screening was carried out to obtain the sequences investigated. Both strands were sequenced using the ABI Prism BigDye Chemistry Version 3 (Perkin-Elmer) on an ABI 377 automated sequencer. Sequences were edited with the Sequencher 4.1 software. A total of ∼100 kb was isolated and sequenced from the neo-X and the neo-Y chromosomes. About 17 kb of this represents coding sequence; the rest consists of intron or intergenic DNA. The gene arrangements in the neo-X regions analyzed are identical to those in D. melanogaster, despite the large evolutionary distance between the pseudoobscura and the melanogaster groups (Richards et al. 2005).
PCR isolation and sequencing of neo-sex-linked protein-coding gene fragments
Genomic DNA was extracted from single males and females from an inbred D. miranda line (strain 0101.3). Primer pairs for >75 neo-sex-linked genes were designed using the D. pseudoobscura genome sequence. PCR products were amplified as ∼1.2-kb fragments from both male and female genomic DNA. PCR products were used as sequencing templates after polyethylene glycol (PEG 8000) purification. Gene-specific primers and the original PCR primers were used for sequencing with the BigDye 3.0 cycle sequencing kit. Each fragment was sequenced at least once on both strands. PCR products from female-genomic DNA were usually completely homozygous (i.e., they only contain the neo-X copy). A subset of the primer pairs that successfully amplified in D. miranda females yielded a heterozygous product using male genomic DNA (i.e., I amplified both the neo-X and neo-Y alleles). To identify the neo-X and neo-Y alleles, the amplified PCR product was cloned with the TOPO TA cloning kit (Invitrogen), and at least 10 clones (and sometimes up to 20 clones) were analyzed per gene, to ensure sufficient sampling of the neo-Y copy to allow the identification and exclusion of recombinant clones and PCR errors. This combination of direct sequencing and cloning of PCR products allowed me to obtain sequence information from 52 neo-X- and neo-Y-linked genes. Primer pairs for PCR amplification are given in Supplemental Table 2. I also added four previously studied genes from the neo-sex chromosomes (zip, CycB, eng, and eve) (see Bachtrog and Charlesworth 2002; Bachtrog 2003c).
Genome queries, TE characterization, and evolutionary analysis
The D. miranda sequences were searched against the D. pseudoobscura genome using BLAST (http://flybase.bio.indiana.edu/blast/). The sequences were aligned using CLUSTAL, and annotated using the homologous D. pseudoobscura genes. Transposable elements were searched against Repbase (http://www.girinst.org/Censor_Server.html), and annotated by comparing the genomic DNA sequence to the protein sequence using Wise2 (http://www.ebi.ac.uk/Wise2/). _K_a and _K_s were calculated with a maximum-likelihood method that accounts for unequal transition and transversion rates and unequal base and codon frequencies. A likelihood ratio test was used to test different models of evolution by application of the codeml program within the PAML software package. A model assuming the same _K_a/_K_s ratio for the neo-X and neo-Y branches was compared with a model that allowed the _K_a/_K_s ratio to differ. Twice the difference in log-likelihood between models is assumed to be distributed approximately as χ2, with the degrees of freedom equal to the differences in the number of parameters.
Supplementary Material
[Supplemental Research Data]
Acknowledgments
D.B. is supported by the Austrian Academy of Science, and this research was supported in part by a BBSRC grant to Brian Charlesworth. I am grateful to Peter Andolfatto for comments on the manuscript.
E-mail dbachtrog@ucsd.edu; fax (858) 534-7108.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3543605\. Article published online before print in September 2005.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. DQ145178-DQ145281.]
References
- Akashi, H. and Schaeffer, S.W. 1997. Natural selection and the frequency distributions of “silent” DNA polymorphism in Drosophila. Genetics 146**:** 295-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P., Wall, J., and Kreitman, M. 1999. Unusual haplotype structure at the proximal breakpoint of In(2L)t in a natural population of Drosophila melanogaster. Genetics 153**:** 1297-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog, D. 2003a. Adaptation shapes patterns of genome evolution in sexual and asexual genomes in Drosophila. Nat. Genet. 34**:** 215-219. [DOI] [PubMed] [Google Scholar]
- ———. 2003b. Accumulation of spock and worf, two novel non-LTR retrotransposons on the neo-Y chromosome of Drosophila miranda. Mol. Biol. Evol. 20**:** 173-181. [DOI] [PubMed] [Google Scholar]
- ———. 2003c. Protein evolution and codon usage bias on the neo-sex chromosomes of Drosophila miranda. Genetics 165**:** 1221-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2004. Evidence that positive selection drives Y-chromosome degeneration in Drosophila miranda. Nat. Genet. 36**:** 518-522. [DOI] [PubMed] [Google Scholar]
- Bachtrog, D. and Charlesworth, B. 2002. Reduced adaptation of a non-recombining neo-Y chromosome. Nature 416**:** 323-326. [DOI] [PubMed] [Google Scholar]
- Bartolome, C., Maside, X., Yi, S., Grant, A., and Charlesworth, B. 2005. Patterns of selection on synonymous and nonsynonymous variants in Drosophila miranda. Genetics 169**:** 1495-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone, J.R. and Kuroda, M.I. 1996. Dosage compensation regulatory proteins and the evolution of sex chromosomes in Drosophila. Genetics 144**:** 705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull, J.J. 1983. Evolution of sex determining mechanisms. Benjamin Cummings, Menlo Park, CA.
- Caceres, M., Ranz, J.M., Barbadilla, A., Long, M., and Ruiz, A. 1999. Generation of a widespread Drosophila inversion by a transposable element. Science 16**:** 415-418. [DOI] [PubMed] [Google Scholar]
- Carvalho, A.B. 2002. Origin and evolution of the Drosophila Y chromosome. Curr. Opin. Genet. Dev. 12**:** 664-668. [DOI] [PubMed] [Google Scholar]
- Carvalho, A. and Clark, A. 2005. Y Chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science 307**:** 108-110. [DOI] [PubMed] [Google Scholar]
- Carvalho, A.B., Dobo, B.A., Vibranovski, M.D., and Clark, A.G. 2001. Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. 98**:** 13225-13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B. 1996. The evolution of chromosomal sex determination and dosage compensation. Curr. Biol. 6**:** 149-162. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B. and Charlesworth, D. 2000. The degeneration of Y chromosomes. Philos. Trans. R Soc. Lond. B Biol. Sci. 355**:** 1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B. and Langley, C.H. 1989. The population genetics of Drosophila transposable elements. Annu. Rev. Genet. 23**:** 251-287. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., Sniegowski, P., and Stephan, W. 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371**:** 215-220. [DOI] [PubMed] [Google Scholar]
- Coghlan, A. and Wolfe, K. 2002. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 12**:** 857-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cold Spring Harbor Laboratory, Washington University Genome Sequencing Center, and PE Biosystems Arabidopsis Sequencing Consortium. 2000. The complete sequence of a heterochromatic island from a higher eukaryote. Cell 100**:** 377-386. [PubMed] [Google Scholar]
- Das, M., Mutsuddi, D., Duttagupta, A.K., and Mukherjee, A.S. 1982. Segmental heterogeneity in replication and transcription of the X2 chromosome of Drosophila miranda and conservativeness in the evolution of dosage compensation. Chromosoma 87**:** 373-388. [Google Scholar]
- Filatov, D. 2005. Substitution rates in a new Silene latifolia sex-linked gene, SlssX/Y. Mol. Biol. Evol. 22**:** 402-408. [DOI] [PubMed] [Google Scholar]
- Fridolfsson, A.K. and Ellegren, H. 2000. Molecular evolution of the avian CHD1 genes on the Z and W sex chromosomes. Genetics 155**:** 1903-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill, P.R., Halligan, D., Charlesworth, B., and Andolfatto, P. 2005. Patterns of intron sequence evolution depend upon length and GC content. Genome Biol. 6**:** R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan, D., Eyre-Walker, A., Andolfatto, P., and Keightley, P. 2004. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 14**:** 273-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka, J. 2000. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 9**:** 418-420. [DOI] [PubMed] [Google Scholar]
- Kaminker, J.S., Bergman, C.M., Kronmiller, B., Carlson, J., Svirskas, R., Patel, S., Frise, E., Wheeler, D.A., Lewis, S.E., Rubin, G.M., et al. 2002. The transposable elements of the Drosophila melanogaster euchromatin: A genomics perspective. Genome Biol. 3**:** research0084.1-0084.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush, K., Feldman, M., and Levy, A. 2003. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33**:** 102-106. [DOI] [PubMed] [Google Scholar]
- Lahn, B.T., Pearson, N.M., and Jegalian, K. 2001. The human Y chromosome, in the light of evolution. Nat. Rev. Genet. 2**:** 207-216. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., Gendrel, A., Black, M., Vaughn, M., Dedhia, N., McCombie, W., Lavine, K., Mittal, V., May, B., Kasschau, K., et al. 2004. Role of transposable elements in heterochromatin and epigenetic control. Nature 430**:** 471-476. [DOI] [PubMed] [Google Scholar]
- Lucchesi, J.C. 1978. Gene dosage compensation and the evolution of sex chromosomes. Science 202**:** 711-716. [DOI] [PubMed] [Google Scholar]
- MacKnight, R.H. 1939. The sex-determining mechanism of Drosophila miranda. Genetics 24**:** 180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin, I., Franke, A., Bashaw, G.J., and Baker, B.S. 1996. The dosage compensation system of Drosophila is co-opted by newly evolved X chromosomes. Nature 383**:** 160-163. [DOI] [PubMed] [Google Scholar]
- Mathiopoulos, K., della Torre, A., Predazzi, V., Petrarca, V., and Coluzzi, M. 1998. Cloning of inversion breakpoints in the Anopheles gambiae complex traces a transposable element at the inversion junction. Proc. Natl. Acad. Sci. 95**:** 12444-12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H.J. 1940. Bearings of the Drosophila work on systematics. In The new systematics (ed. J.S. Huxley), pp. 185-268. Oxford University Press, London.
- Ohno, S. 1967. Sex chromosomes and sex linked genes. Springer Verlag, Berlin.
- Orr, H.A. and Kim, Y. 1998. An adaptive hypothesis for the evolution of the Y chromosome. Genetics 150**:** 1693-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel, C., Ross, J., Matson, C., Dickson, M., Grimwood, J., Schmutz, J., Myers, R., Mori, S., Schluter, D., and Kingsley, D. 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 14**:** 1416-1424. [DOI] [PubMed] [Google Scholar]
- Puig, M., Cáceres, M., and Ruiz, A. 2004. Silencing of a gene adjacent to the breakpoint of a widespread Drosophila inversion by a transposon-induced antisense RNA. Proc. Natl. Acad. Sci. 101**:** 9013-9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, W.R. 1996. Evolution of the Y sex chromosome in animals. BioScience 46**:** 331-343. [Google Scholar]
- Richards, S., Liu, Y., Bettencourt, B.R., Hradecky, P., Letovsky, S., Nielsen, R., Thornton, K., Hubisz, M.J., Chen, R., Meisel, R.P., et al. 2005. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and _cis_-element evolution. Genome Res. 15**:** 1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Saxena, R., Brown, L., Hawkins, T., Alagappan, R., Skaletsky, H., Reeve, M., Reijo, R., Rozen, S., Dinulos, M., Disteche, C., et al. 1996. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat. Genet. 14**:** 292-299. [DOI] [PubMed] [Google Scholar]
- Schotta, G., Ebert, A., Dorn, R., and Reuter, G. 2003. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell Dev. Biol. 14**:** 67-75. [DOI] [PubMed] [Google Scholar]
- Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P.J., Cordum, H.S., Hillier, L., Brown, L.G., Repping, S., Pyntikova, T., Ali, J., Bieri, T., et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423**:** 825-837. [DOI] [PubMed] [Google Scholar]
- Steinemann, M. and Steinemann, S. 1993. A duplication including the Y allele of Lcp2 and the TRIM retrotransposon at the Lcp locus on the degenerating neo-Y chromosome of Drosophila miranda: Molecular structure and mechanisms by which it may have arisen. Genetics 134**:** 497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1998. Enigma of Y chromosome degeneration: neo-Y and neo-X chromosomes of Drosophila miranda: A model for sex chromosome evolution. Genetica 102-103: 409-420. [PubMed] [Google Scholar]
- Waters, P.D., Duffy, B., Frost, C.J., Delbridge, M.L., and Graves, J.A. 2001. The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80-130 million years ago. Cytogenet. Cell Genet. 92**:** 74-79. [DOI] [PubMed] [Google Scholar]
- Wyckoff, G.J., Li, J., and Wu, C.I. 2002. Molecular evolution of functional genes on the mammalian Y chromosome. Mol. Biol. Evol. 19**:** 1633-1636. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://flybase.bio.indiana.edu/blast/; FlyBase.
- http://www.ebi.ac.uk/Wise2/; Wise2.
- http://www.girinst.org/Censor_Server.html; Repbase.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Research Data]