The root of the angiosperms revisited (original) (raw)
Abstract
Most recent phylogenetic analyses of basal angiosperms have converged on the placement of Amborella as sister to all other extant angiosperms. However, certain recent studies suggest that Amborella and Nymphaeales (water lilies) form a clade sister to all remaining angiosperms or that Nymphaeales alone are the sister to the remaining angiosperms. We report here (i) maximum parsimony, maximum likelihood, and Bayesian phylogenetic analyses of 11 genes (>15,000 bp per taxon) for 16 taxa, (ii) maximum parsimony analysis for a subset of these genes for 104 taxa, and (iii) tests of alternative rootings with the nonparametric bootstrap and the likelihood ratio test with the parametric bootstrap. In addition, we use simulation analyses to examine the amount of bias that may be present in our methods of phylogeny estimation. Amborella continues to receive strong bootstrap support as the sister to all other extant angiosperms, and three of four tests reject alternative hypotheses of the angiosperm root. Although we cannot conclusively choose between Amborella vs. Amborella + Nymphaeales as sister to all other angiosperms, most analyses favor the former rooting.
Within the past few years, phylogenetic relationships among the major lineages of angiosperms have largely been clarified (1–5). Understanding the major branching patterns in the angiosperms is critical for character reconstruction in the earliest angiosperms and for understanding subsequent patterns of diversification. A particularly noteworthy development has been the general agreement, based on maximum parsimony (MP) analyses of DNA sequences from all three genomes (nuclear, chloroplast, and mitochondrial), on the placement of the root of the angiosperms—the monotypic Amborella is sister to all other extant flowering plants, followed by Nymphaeales (water lilies), and a clade of Austrobaileyaceae, Trimeniaceae, Illiciaceae, and Schisandraceae as successive sisters to the remaining lineages of flowering plants (1–3, 6). When multiple gene sequences were combined, high internal support, as measured by the bootstrap or jackknife, was obtained for these basal branches, with values above 90% attained in some analyses (1–3, 6). However, despite considerable support for Amborella as sister to all other angiosperms, some reservations regarding this placement have been expressed. Parkinson et al. (2) and Qiu et al. (7) could not reject, using the Kishino–Hasegawa (KH) test (8), the hypothesis that Nymphaeales are sister to all other flowering plants or that Amborella and Nymphaeales form a clade sister to all remaining angiosperms. In addition, a maximum likelihood (ML) analysis of basal angiosperms with a subset of the taxa analyzed in larger parsimony analyses found a clade of Amborella + Nymphaeales to be the sister to all other flowering plants (9). Similarly, Mathews and Donoghue (10) could not reject alternative rootings at Amborella + Nymphaeales or at Nymphaeales alone with Templeton's significantly less parsimonious test (SLPT) in an expanded analysis of phytochrome A (phyA) and phytochrome C (phyC) sequences. Recently, Barkman et al. (11) conducted an analysis of two “noise-reduced” multigene data sets for basal angiosperms, a 6-gene, 35-taxon data set and a 9-gene, 15-taxon data set. “Noise,” defined as potentially problematic characters or taxa, was reduced with the program rasa [Relative Apparent Synapomorphy Analysis (12)]. The results of Barkman et al. (11) depended on the method of phylogenetic analysis used. After noise reduction, weighted parsimony, neighbor-joining (NJ), and ML indicated Amborella + Nymphaeales as sister to all other angiosperms with the Amborella + Nymphaeales sister-group relationship receiving bootstrap support as high as 96% in the NJ analysis of the 6-gene, noise-reduced data set and 86% in the NJ analysis of the 9-gene, noise-reduced data set. Equally weighted parsimony placed Amborella alone as sister to all other taxa. Graham and Olmstead (13), using data from 17 chloroplast genes, also found Amborella as sister to all remaining angiosperms when both Cabomba and Nymphaea of Nymphaeales were included; however, when Nymphaea was removed, Cabomba and Amborella were successive sisters to all remaining angiosperms.
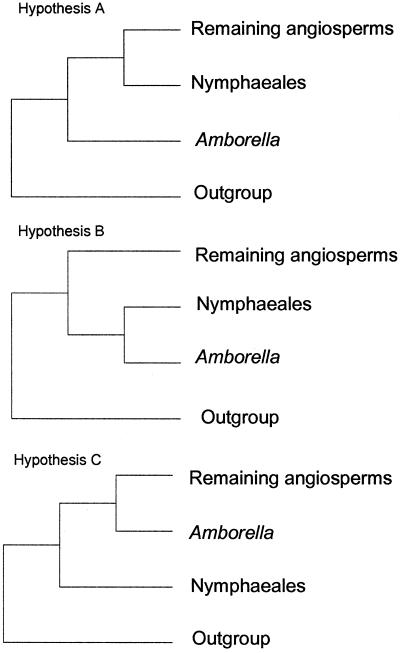
Recent analyses therefore suggest three alternative hypotheses for the root of the angiosperms (Fig. 1). The first (hypothesis A) states that the root of the angiosperms is placed between Amborella and the rest of the angiosperms. The second (hypothesis B) states that the root is between Amborella + Nymphaeales and the rest of the angiosperms. The third (hypothesis C) states that Nymphaeales are sister to Amborella + all other angiosperms. Of these three hypotheses, several studies have provided strong support for A (1–3) as measured by the nonparametric bootstrap (typically referred to simply as the bootstrap) (14), whereas Barkman et al. (11) obtained strong support for hypothesis B, that is, Amborella + Nymphaeales are sister to all other angiosperms.
Figure 1.
The three phylogenetic hypotheses regarding the placement of the root of the angiosperms. (Hypothesis A) Amborella is sister to all other extant angiosperms. (Hypothesis B) Amborella + Nymphaeales are sister to all remaining angiosperms. (Hypothesis C) Nymphaeales are sister to all remaining angiosperms.
To explore the rooting of the angiosperms further and the placement of Amborella relative to other basal angiosperms, we constructed and analyzed two DNA data sets. The first data set includes 16 taxa sequenced for 11 genes (18S rDNA, 26S rDNA, phyA, phyC, mtSSU, cox1, rps2, atpA, matR, rbcL, and atpB), a total of 15,772 bp per taxon. Because the taxon sampling of the individual data sets did not match exactly, data for closely related taxa were sometimes merged to form a single “composite exemplar” sequence. Because several recent empirical and simulation studies have demonstrated the importance of the addition of both taxa and characters in resolving difficult phylogenetic problems (15–18), we constructed a second data set for 104 taxa. For this data set we added taxa that were sequenced for at least 5 of the 11 genes. A table of exemplars, genes, and voucher and GenBank information is available from M.J.Z.
With the 16-taxon data set, we explored the position of Amborella and the root of the angiosperms with MP, ML, and Bayesian inference. This data set also served as a basis for simulation analyses by using the parametric bootstrap with which we tested the three hypotheses regarding the root of the angiosperms. We also used the 16-taxon data set to analyze different data partitions by using ML to locate phylogenetic signal that supports each of the three hypotheses of the angiosperm root. The seven partitions examined were nuclear genes, chloroplast genes, mitochondrial genes, ribosomal genes (18S rDNA, 26S rDNA, and mtSSU), DNA sequences of protein-coding genes (phyA, phyC, cox1, rps2, atpA, matR, rbcL, and atpB), a data set with only first and second codon positions of protein-coding genes, and a data set containing only third codon positions of protein-coding genes.
We used two methods to investigate the degree to which alternative phylogenetic hypotheses were supported by the data, the nonparametric bootstrap (14) and the likelihood-ratio test with the parametric bootstrap (19–22). The nonparametric bootstrap is routinely used as a method of assessing support for phylogenetic hypotheses. The likelihood ratio test with the parametric bootstrap is used to obtain an expected distribution of tree score differences, against which the observed difference can be compared; the advantages of using this approach for testing alternative phylogenetic hypotheses have been reviewed (23). To test hypotheses A–C, data sets were simulated, using estimated parameter values, assuming that either hypothesis B or C is true (see Methods). By comparing the difference in tree scores between hypothesis A and hypotheses B and C with simulated data, we tested whether the observed difference in tree scores could be expected if either hypothesis B or C is true. This test was applied with both ML and MP analyses.
Methods
Phylogenetic Analyses.
All MP and ML analyses and hypothesis tests were conducted with PAUP*4.03b (24). For the 16-taxon data set, all taxa were sequenced for mtSSU, rbcL, atpB, 18S rDNA, and 26S rDNA. An rps2 sequence was not available for Trochodendron; a cox1 sequence was not available for Pinus; a phyA/phyC sequence was not available for Platanus, Pinus, or Ginkgo; a matR sequence was not available for Acorus; and atpA was lacking for Amborella. Pinus was sequenced for five genes but was not sequenced for cox1, mtSSU, and rps2, and sequences from Abies were used instead for these genes. Pinus and Sarcandra were not sequenced for 26S rDNA, and instead sequences from Larix and Chloranthus were used, respectively. The 104-taxon data set had no concatenated “composite exemplar” sequences, and missing data were coded as such (i.e., as ?). Gaps and known mitochondrial editing sites were excluded from all analyses; however, inclusion of gaps scored as missing did not affect the results (data not shown).
MP analyses were performed on both the 16-taxon and 104-taxon data sets with 100 random taxon additions, holding 5 trees per replicate, with tree bisection-reconnection (TBR) branch-swapping and saving multiple parsimonious trees (MULPARS on). Two hundred nonparametric bootstrap replicates were performed with simple taxon addition and TBR branch-swapping, with MULPARS turned off.
ML analyses assumed the Hasegawa–Kishino–Yano (HKY) + Γ or the general-time-reversible + Ι + Γ model of molecular evolution. ML analyses for the 16-taxon data set used transition/transversion ratio, rate heterogeneity, and base frequencies estimated from trees obtained from MP analyses. We also analyzed the 16-taxon data set with HKY + Γ model parameter values estimated from two random trees. We obtained the same ML tree regardless of the tree used to estimate the parameter values and base frequencies. ML analyses used heuristic searches with starting trees obtained by NJ followed by TBR branch-swapping. ML nonparametric bootstrap analyses for the 16-taxon data set used 100 heuristic searches with starting trees obtained with NJ based on p distances followed by TBR and nearest-neighbor interchange branch-swapping, saving all optimal trees.
The Bayesian phylogenetic analyses were conducted with mrbayes version 1.10 (25). We used uniform prior probabilities and estimated base frequencies and the parameters for the HKY + Γ model of molecular evolution. We ran four chains of the Markov chain Monte Carlo, sampling 1 tree every 100 generations for 1,000,000 generations starting with a random tree. Stationarity was reached at about generation 24,000. Thus, the first 24,000 generations were the “burn in” of the chain, and inferences about the phylogeny were based on those trees sampled after generation 24,000.
To conduct the parametric bootstrap we first built constraint trees for hypotheses B and C with MACCLADE 3.04 (26). We then performed MP analyses with the 16-taxon data set, as above, enforcing the constraints imposed by hypotheses B and C, respectively. Each of these constrained analyses found a single MP tree from which we estimated, using ML, parameter values for the HKY + Γ model of molecular evolution and base frequencies. We then performed ML analyses enforcing the constraint topology imposed by either hypothesis B or C. We used the ML trees found for hypotheses B and C to reestimate parameter values for the HKY + Γ model and base frequencies to simulate 100 data sets, using seq-gen (27), with the size of the data set identical to the 16-taxon data set (i.e., 16 taxa, 15,772 bp), for each of the hypotheses, B and C. Each of the 100 data sets for each hypothesis (B and C) was analyzed by means of ML with the HKY + Γ model of molecular evolution with a heuristic search strategy with starting trees obtained by NJ and subtree pruning-regrafting branch-swapping. Parameter values for the model were identical to those used to simulate the data. The −ln tree scores and trees were saved for each of the simulated data sets. We then performed ML analysis on each of the 100 simulated data sets for each hypothesis, enforcing a topological constraint of Amborella as sister to the rest of the angiosperms and saved all −ln tree scores and trees. These analyses of the simulated data allowed us to calculate the null distribution of the likelihood-ratio test statistic, δ = (logL1 − logL0), where logL1 is the −ln tree score of the unconstrained analysis by using data simulated under the assumption that hypothesis B or C is true, and logL0 is the −ln tree score of the tree(s) obtained from the constrained analysis, with Amborella sister to the rest of the angiosperms and the data simulated assuming that either hypothesis B or C is true. We then tested whether δ calculated from analysis of the real 16-taxon data set fell within this null distribution of δ.
The incongruence length difference test (28) was performed with 100 replicates and a heuristic search strategy that used simple taxon addition, holding 5 trees at each step, and TBR branch-swapping.
For simulation analyses of error rates and bias with MP, 500 replicate data sets equal in size to the data analyzed in the 16-taxon total-evidence analysis were generated for each of the three hypotheses of the root of the angiosperms. The data were simulated assuming an HKY + Γ model or a general-time-reversible + Ι + Γ model of molecular evolution with seq-gen (27). Each replicate was analyzed with 100 random taxon additions, TBR branch-swapping, holding 5 trees per taxon addition, and MULPARS. For analyses with ML, we used the same methods as for MP except that we used 100 replicate data sets for each hypothesis, starting trees obtained by means of NJ, and subtree pruning-regrafting branch-swapping.
Results and Discussion
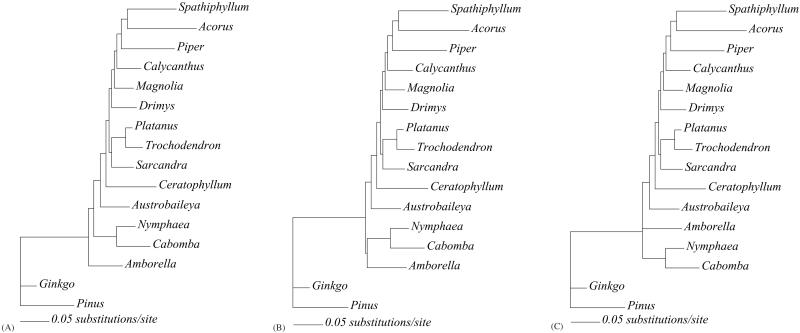
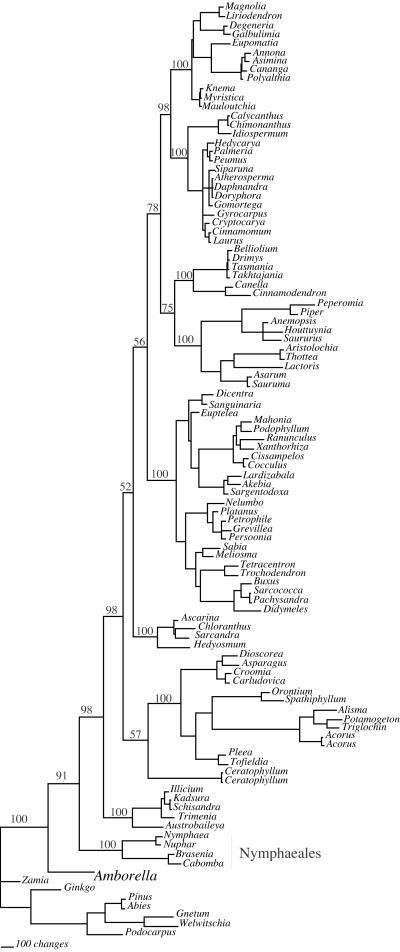
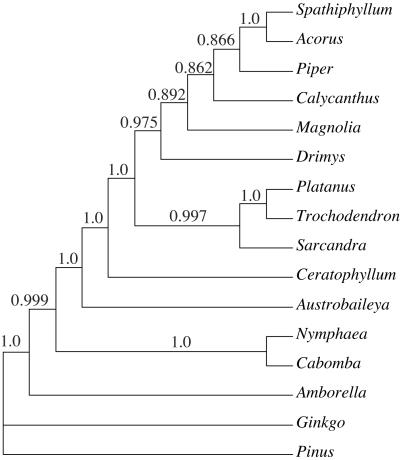
MP bootstrap analyses of both the 16-taxon and 104-taxon data sets show high support (96% and 91%, respectively) for the placement of Amborella as sister to all other extant angiosperms (Figs. 2A and 3). ML analysis of the 16-taxon data set, with either the HKY + Γ or the general-time-reversible + Ι + Γ model of molecular evolution, also recovered Amborella as sister to all other extant angiosperms with fairly high bootstrap support (88% with 200 bootstrap replicates and nearest-neighbor interchange branch-swapping and 90% with 100 replicates by using TBR branch-swapping for the HKY + Γ model). The results of the Bayesian analysis indicate a posterior probability of 0.999 for the placement of Amborella as sister to the rest of the angiosperms (Fig. 4). Thus, as in most recent analyses (1–3, 5), we find strong support for hypothesis A by using MP and fairly strong support for A with ML; only the analyses by Barkman et al. (11) provided strong support for an alternative hypothesis, B.
Figure 2.
Trees showing branch lengths for each of the three hypotheses. Branch lengths estimated with ML assuming an HKY + Γ model of sequence evolution with the total-evidence data set for the three phylogenetic hypotheses outlined in Fig. 1. (A) Tree found in MP and ML analyses of total-evidence data set. (B) ML tree constraining Amborella + Nymphaeales. (C) ML tree constrained to hypothesis C.
Figure 3.
Strict consensus of 16 MP trees obtained in analysis of the 104-taxon data set. Tree length = 23,864; consistency index = 0.394; retention index = 0.518. Bootstrap values (obtained by using 200 replicate searches with TBR with MULPARS turned off) for major clades of angiosperms are indicated above the branches.
Figure 4.
50% majority rule tree derived from those trees sampled after “burn in.” Posterior probabilities are indicated above the branches.
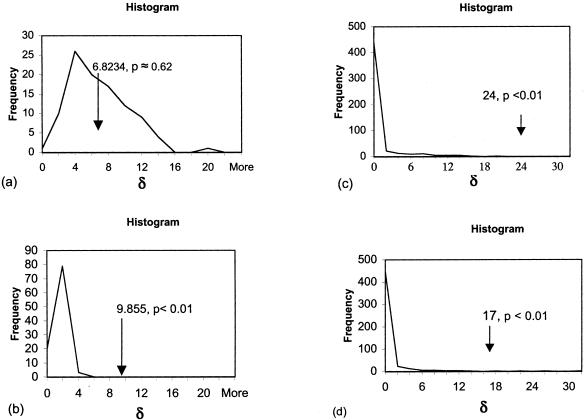
Based on the likelihood-ratio test with the parametric bootstrap and ML as the tree estimation method for the 16-taxon data set, hypothesis A is not significantly different, in terms of tree scores, from hypothesis B, but hypothesis A is significantly different from hypothesis C (Fig. 5). Based on this test and MP as the tree estimation method for the 16-taxon data set, hypothesis A is significantly different, in terms of tree length, from hypotheses B and C (Fig. 5).
Figure 5.
Results of the likelihood ratio test with the parametric bootstrap, showing the distribution of δ (the likelihood ratio test statistic), which is the difference between the optimal trees supporting hypothesis B or C and hypothesis A for simulated data assuming that either hypothesis B or C is correct. The observed value for the actual data (represented by the arrow) is shown in relation to the distribution. If the observed value is much greater than the values found in the distribution, the null hypothesis is rejected. (a) With ML, the difference between optimal trees supporting hypothesis B compared with hypothesis A. The observed value is not significantly different from the values in the distribution (p ≈ 0.62). (b) With ML, the difference between optimal trees supporting hypothesis C compared with hypothesis A. The observed value is significantly different from the values in the distribution (p < 0.01). (c) With MP, the difference between optimal trees supporting hypothesis B compared with hypothesis A. The observed value is significantly different from the values in the distribution (p < 0.01). (d) With MP, the difference between optimal trees supporting hypothesis C compared with hypothesis A. The observed value is significantly different from the values in the distribution (p < 0.01).
We examined different data partitions with ML in an effort to locate phylogenetic signal that may underlie each of the three phylogenetic hypotheses (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org). Of the data partitions examined, the nuclear genes, the ribosomal DNA data, and first and second codon positions recovered hypothesis A (88, 92, and 38% bootstrap support, respectively). The data partitions of chloroplast and protein-coding genes recovered hypothesis B (62% for monophyly of Amborella + Nymphaeales/78% support for monophyly of the remaining angiosperms, and 69%/86% bootstrap support, respectively). Analysis of the mitochondrial data set and the third codon positions also recovered hypothesis B, but with bootstrap support <50%. Thus, although some partitions support hypothesis B rather that A, this conflicting signal is not strongly supported. Moreover, incongruence length difference tests of nuclear, chloroplast, and mitochondrial partitions indicate that these partitions are homogeneous with respect to each other (P = 0.21). However, the ribosomal and protein partitions are significantly heterogeneous (P = 0.04). In MP analyses of the 16-taxon data set, six of the seven partitions recovered Amborella as sister to the remaining angiosperms (data not shown).
We performed MP and ML analyses with Nymphaea removed from the total-evidence data matrix, an approach similar to Graham and Olmstead (13). MP recovered Cabomba sister to the rest of the angiosperms (76% bootstrap support), a result consistent with hypothesis C, whereas ML analysis recovered Amborella sister to the rest of the angiosperms with 80% bootstrap support. We also constructed a data set with the same nine genes used by Barkman et al. (11); however, we did not use rasa on this data set. Both MP and ML recovered Amborella sister to the rest of the angiosperms, with 93% and 54% bootstrap support, respectively.
Several issues pertaining to either character or taxon sampling may lead to the recovery of alternative hypotheses A or B. For example, the addition to our data set of atpA for Amborella might result in the recovery of hypothesis B instead of A, as in the analysis by Barkman et al. (11). Furthermore, different character sets supported alternative hypotheses. Analyses of only protein-coding genes recovered hypothesis B with ML and the HKY + Γ model of sequence evolution, whereas analysis of ribosomal genes recovered hypothesis A. Moreover, omission of certain characters or taxa based on the assumption that they contribute more noise than signal, as by Barkman et al., might result in recovery of alternative hypotheses. Saturated sites, such as some third codon positions, are a potential source of noise. However, recent studies challenge the assumption that third codon positions simply add noise to analyses; instead, they may be important for phylogenetic resolution at even the broadest levels (29–32). Our ML analysis of first and second codon positions in the 16-taxon data set recovered hypothesis A, whereas analysis of all three codon positions recovered hypothesis B. Finally, taxon sampling alone may influence the rooting. For example, the results by Graham and Olmstead (13) depended on their sampling of Nymphaeales. Even the most thorough sampling of extant plants may undersample historical lineages crucial for accurate reconstruction of basal relationships.
To explore the placement of the angiosperm root further, we performed a simulation analysis to calculate error rates and to determine the amount of bias that may be present in our method of tree estimation (MP or ML), following the method of Huelsenbeck (33), Maddison et al. (34), Wiens and Hollingsworth (35), and Sanderson et al. (36). We simulated replicate data sets for each of the three hypotheses, assuming an HKY + Γ or general-time-reversible + Ι + Γ model of molecular evolution, and recorded the frequency of recovering the tree used to simulate the data by using MP and ML (Tables 1 and 2). We would expect, for example, given tree B as the correct tree, that analyses of data simulated under that assumption would recover tree B with a high probability. In our MP analyses, tree (hypothesis) A was recovered 100% of the time when it was the underlying tree. However, even when tree B or C was the underlying tree, we also recovered tree A with high probability (0.843 and 0.862, respectively). This analysis indicates bias in our tree reconstruction with MP.
Table 1.
Error rates estimated with MP and simulated data for each of the three tree hypotheses
| Tree A | Tree B | Tree C | |
|---|---|---|---|
| HKY + Γ model | |||
| Tree A* | 1.000 | 0.000 | 0.000 |
| Tree B* | 0.843 | 0.072 | 0.084 |
| Tree C* | 0.862 | 0.000 | 0.138 |
| GTR + I + Γ model | |||
| Tree A* | 1.000 | 0.000 | 0.000 |
| Tree B* | 0.815 | 0.13 | 0.03 |
| Tree C* | 0.847 | 0.000 | 0.15 |
Table 2.
Error rates estimated with ML and simulated data for each of the three tree hypotheses
| Tree A | Tree B | Tree C | |
|---|---|---|---|
| Tree A* | 1.000 | 0.000 | 0.000 |
| Tree B* | 0.01 | 0.98 | 0.01 |
| Tree C*† | 0.19 | 0.33 | 0.31 |
With ML, tree A was recovered in all replicate searches when it was the underlying tree; tree B was recovered in 98% of the replicates in which it was the correct tree. However, when tree C was the correct tree, we recovered it only 31% of the time. Tree B was reconstructed in 33% of the replicates, and tree A was recovered in 19% of the replicates. The remaining 17% of the replicates were equivocal regarding the relationships among Amborella, Nymphaeales, and the rest of the angiosperms. Tree C is especially difficult to recover, even when it is the underlying tree, because of the short branch length connecting Nymphaeales to the rest of the angiosperms (Fig. 2); with ML, the results are equivocal, but MP shows a clear bias toward reconstructing tree A.
To test whether the placement of the angiosperm root near Amborella and Nymphaeales was simply the result of bias in tree reconstruction methods, we simulated data from two additional trees and analyzed these data sets with MP. The first tree had Magnolia as sister to the rest of the angiosperms, and the second tree had Amborella and Nymphaeales placed in separate parts of the tree with neither sister to the rest of the angiosperms. In this second tree, Amborella was sister to a clade containing Calycanthus, Piper, Spathiphyllum, and Acorus, and the Nymphaeales were sister to the eudicots. In MP analyses of these data sets, Ceratophyllum was reconstructed as sister to the rest of the angiosperms approximately 60% of the time, Amborella was never reconstructed as the sister group to the other angiosperms, and analyses of only 2 of 500 data sets recovered Nymphaeales as sister to the rest of the angiosperms. These analyses demonstrate that placement of Amborella or Nymphaeales as sister to the other angiosperms is not an artifact of the tree reconstruction method.
Morphology does not provide many additional characters as independent evidence regarding the three hypotheses. One character that might unambiguously support a direct link between Amborella and Nymphaeales is the absence of ethereal oil cells in parenchymatous tissue (37–39). However, based on the absence of such cells outside of angiosperms, this condition is likely to be ancestral, with ethereal oil cells evolving later. Another potential synapomorphy is the lack of vessels, but again this may be ancestral in the angiosperms, and this condition in Nymphaeales can reasonably be interpreted in several ways (1, 40). In general, it appears that the hypotheses depicted in Fig. 1 imply similar directions of character evolution in the earliest angiosperms, only slightly shifting our interpretation, and then with little confidence.
In our analyses, Amborella continued to receive strong nonparametric bootstrap support as sister to all other extant angiosperms with MP. Analyses with ML revealed the same topology, but with slightly lower support. In addition, Bayesian phylogeny estimation indicated Amborella sister to the rest of the angiosperms with a posterior probability of 0.999. Alternative topologies were statistically rejected in three of four tests, and hypothesis C was not supported in any of the analyses of the 16-taxon or 104-taxon data sets or their partitions. However, ML analyses of protein-coding genes as well as mitochondrial and chloroplast gene partitions recovered Amborella + Nymphaeales as sister to all remaining angiosperms. Therefore, despite the strong total-evidence support for hypothesis A, the Amborella + Nymphaeales hypothesis cannot be rejected by all analyses. Thus, inferences of character evolution in the angiosperms perhaps should consider both hypotheses A and B.
Improved inferences of ancestral character-state reconstructions for the earliest angiosperms will require additional data, not from molecular phylogenetics, but from fossil discoveries and reanalyses of existing fossil data of flowering plants and groups that may be the sister to the angiosperms (41–47). Future analyses must focus on the integration of molecular and fossil data sets as well as further morphological and functional characterization of extant flowering plants (48, 49). Furthermore, it may be desirable to focus more attention on resolving relationships among lineages of monocots and eudicots.
Supplementary Material
Supporting Table
Acknowledgments
We thank Jack Sullivan for helpful suggestions regarding the parametric bootstrap and likelihood ratio tests, Jim Doyle for valuable discussion of morphological characters, and Mark Chase for helpful comments on the manuscript. This research was supported in part by National Science Foundation Grant DEB-9707868 and by a joint Fulbright Distinguished Professorship (to D.E.S. and P.S.S.).
Abbreviations
MP
maximum parsimony
ML
maximum likelihood
NJ
neighbor-joining
TBR
tree bisection-reconnection
HKY
Hasegawa-Kishino-Yano
References
- 1.Mathews S, Donoghue M J. Science. 1999;286:947–950. doi: 10.1126/science.286.5441.947. [DOI] [PubMed] [Google Scholar]
- 2.Parkinson C L, Adams K L, Palmer J D. Curr Biol. 1999;9:1485–1488. doi: 10.1016/s0960-9822(00)80119-0. [DOI] [PubMed] [Google Scholar]
- 3.Qiu Y-L, Lee J, Bernasconi-Quadroni F, Soltis D E, Soltis P S, Zanis M, Zimmer E A, Chen Z, Savolainen V, Chase M W. Nature (London) 1999;402:404–407. doi: 10.1038/46536. [DOI] [PubMed] [Google Scholar]
- 4.Soltis P S, Soltis D E, Chase M W. Nature (London) 1999;402:402–404. doi: 10.1038/46528. [DOI] [PubMed] [Google Scholar]
- 5.Soltis D E, Soltis P S, Chase M W, Mort M E, Albach D C, Zanis M, Savolainen V, Hahn W H, Hoot S B, Fay M F, et al. Bot J Linn Soc. 2000;133:381–461. [Google Scholar]
- 6.Qiu Y-L, Lee J, Whitlock B A, Bernasconi-Quadroni F, Dombrovska O. Mol Biol Evol. 2001;18:1745–1753. doi: 10.1093/oxfordjournals.molbev.a003962. [DOI] [PubMed] [Google Scholar]
- 7.Qiu Y-L, Lee J, Bernasconi-Quadroni F, Soltis D E, Soltis P S, Zanis M, Zimmer E A, Chen Z, Savolainen V, Chase M W. Int J Plant Sci. 2000;161:S3–S27. [Google Scholar]
- 8.Kishino H, Hasegawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 9.Soltis P S, Soltis D E, Zanis M J, Kim S. Int J Plant Sci. 2000;161:S97–S107. [Google Scholar]
- 10.Mathews S, Donoghue M J. Int J Plant Sci. 2000;161, Suppl. 6:S41–S55. [Google Scholar]
- 11.Barkman T J, Ghenery G, McNeal J R, Lyons-Weiler J, de Pamphilis C W. Proc Natl Acad Sci USA. 2000;97:13166–13171. doi: 10.1073/pnas.220427497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyons-Weiler J, Hoelzer G A, Tausch R J. Mol Biol Evol. 1996;13:749–757. doi: 10.1093/oxfordjournals.molbev.a025635. [DOI] [PubMed] [Google Scholar]
- 13.Graham S W, Olmstead R G. Am J Bot. 2000;87:1712–1730. [PubMed] [Google Scholar]
- 14.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 15.Graybeal A. Syst Biol. 1998;47:9–17. doi: 10.1080/106351598260996. [DOI] [PubMed] [Google Scholar]
- 16.Hillis D M. Nature (London) 1996;383:130–131. doi: 10.1038/383130a0. [DOI] [PubMed] [Google Scholar]
- 17.Rannala B, Huelsenbeck J P, Yang Z, Nielsen R. Syst Biol. 1998;47:702–710. doi: 10.1080/106351598260680. [DOI] [PubMed] [Google Scholar]
- 18.Soltis D E, Soltis P S, Mort M E, Chase M W, Savolainen V, Hoot S B, Morton C M. Syst Biol. 1998;47:32–42. doi: 10.1080/106351598261012. [DOI] [PubMed] [Google Scholar]
- 19.Goldman N, Anderson J P, Rodrigo A G. Syst Biol. 2000;49:652–670. doi: 10.1080/106351500750049752. [DOI] [PubMed] [Google Scholar]
- 20.Hillis D M, Mable B K, Moritz C. In: Molecular Systematics. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 21.Huelsenbeck J P, Hillis D M, Nielsen R. Syst Biol. 1996;45:546–558. [Google Scholar]
- 22.Swofford D L, Olsen G J, Waddell P J, Hillis D M. In: Molecular Systematics. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 23.Huelsenbeck J P, Hillis D M. In: Molecular Zoology: Advances, Strategies, and Protocols. Ferraries J D, Palumbi S R, editors. New York: Wiley; 1996. pp. 19–45. [Google Scholar]
- 24.Swofford D L. In: Phylogenetic Analysis of DNA Sequences. Miyamato M M, Cracraft J, editors. New York: Oxford Univ. Press; 1991. pp. 295–333. [Google Scholar]
- 25.Huelsenbeck J P. MRBAYES 1.10: Bayesian Inference of Phylogeny. Rochester, NY: Univ. of Rochester; 2000. [Google Scholar]
- 26.Maddison W P, Maddison D R. MACCLADE 3.04: Analysis of Phylogeny and Character Evolution. Sunderland, MA.: Sinauer; 1992. [DOI] [PubMed] [Google Scholar]
- 27.Rambaut A, Grassly N C. Comput Appl Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- 28.Mickevich M F, Farris J S. Syst Zool. 1981;30:351–370. [Google Scholar]
- 29.Källersjö M, Albert V A, Farris J S. Cladistics. 1999;15:91–93. [Google Scholar]
- 30.Lewis L A, Mishler B D, Vilgalys R. Mol Phylogenet Evol. 1997;7:377–393. doi: 10.1006/mpev.1996.0395. [DOI] [PubMed] [Google Scholar]
- 31.Olmstead R G, Reeves R A, Yen A C. In: Molecular Systematics of Plants II: DNA Sequencing. Soltis P S, Soltis D E, Doyle J J, editors. Boston: Kluwer; 1998. pp. 164–187. [Google Scholar]
- 32.Sennblad B, Bremer B. Syst Biol. 2000;49:101–113. doi: 10.1080/10635150050207410. [DOI] [PubMed] [Google Scholar]
- 33.Huelsenbeck J P. Syst Biol. 1998;47:519–537. [PubMed] [Google Scholar]
- 34.Maddison D R, Baker M D, Ober K A. Syst Entomol. 1999;24:103–138. [Google Scholar]
- 35.Wiens J J, Hollingsworth B D. Syst Biol. 2000;49:143–159. doi: 10.1080/10635150050207447. [DOI] [PubMed] [Google Scholar]
- 36.Sanderson M J, Wojciechowski M F, Hu J-M, Sher Khan T, Brady S G. Mol Biol Evol. 2000;17:782–797. doi: 10.1093/oxfordjournals.molbev.a026357. [DOI] [PubMed] [Google Scholar]
- 37.Money L L, Bailey I W, Swamy B G L. J Arnold Arbor. 1950;31:372–403. [Google Scholar]
- 38.Qiu Y-L, Chase M W, Les D H, Parks C L. Ann Mo Bot Gard. 1993;80:587–606. [Google Scholar]
- 39.Carlquist S, Schneider E L. Pac Sci. 2001;55:305–312. [Google Scholar]
- 40.Schneider E L, Carlquist S. Nord J Bot. 1996;16:637–641. [Google Scholar]
- 41.Donoghue M J, Doyle J A. Curr Biol. 2000;10:R106–R109. doi: 10.1016/s0960-9822(00)00304-3. [DOI] [PubMed] [Google Scholar]
- 42.Friis E M, Pedersen K R, Crane P R. Plant Syst Evol Suppl. 1994;8:31–49. [Google Scholar]
- 43.Sun G, Dilcher D L, Zheng S, Zhou Z. Science. 1998;282:1692–1695. doi: 10.1126/science.282.5394.1692. [DOI] [PubMed] [Google Scholar]
- 44.Friis E M, Pedersen K R, Crane P R. Int J Plant Sci. 2000;161:S169–S182. doi: 10.1086/314248. [DOI] [PubMed] [Google Scholar]
- 45.Mohr B A R, Friis E M. Int J Plant Sci. 2000;161:S155–S167. doi: 10.1086/314248. [DOI] [PubMed] [Google Scholar]
- 46.Swisher C C, III, Wang Y-Q, Wang X-L, Xu X, Wang Y. Nature (London) 1999;400:58–61. [Google Scholar]
- 47.Friis E M, Pedersen K R, Crane P R. Nature (London) 2001;410:357–360. doi: 10.1038/35066557. [DOI] [PubMed] [Google Scholar]
- 48.Feild T S, Zweiniecki M A, Brodribb T, Jaffré T, Donoghue M J, Holbrook N M. Int J Plant Sci. 2000;161:705–712. [Google Scholar]
- 49.Endress P K. Int J Plant Sci. 2001;162:1111–1140. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Table