SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis (original) (raw)
Abstract
Post-transcriptional gene silencing (PTGS) provides protection in plants against virus infection and can suppress expression of transgenes. Arabidopsis plants carrying mutations at the SDE3 locus are defective in PTGS mediated by a green fluorescent protein transgene. However, PTGS mediated by tobacco rattle virus (TRV) was not affected by sde3. From these results we conclude that SDE3, like the previously described RNA polymerase encoded by SDE1, acts at a stage in the mechanism that is circumvented when PTGS is mediated by TRV. The product of SDE3 is similar to RNA helicase-like proteins including GB110 in mouse and other proteins in Drosophila and humans. These proteins are similar to, but clearly distinct from Upf1p and SMG-2, which are required for nonsense-mediated mRNA decay in yeast and Caenorhabditis elegans and, in the case of SMG-2, for PTGS.
Keywords: double-stranded RNA/nonsense-mediated decay/RNA degradation/RNAi/virus resistance
Introduction
Eukaryotic cells suppress foreign genetic elements, including transgenes, through a process that operates at the RNA level and is referred to here as post-transcriptional gene silencing (PTGS) (Kooter et al., 1999; Plasterk and Ketting, 2000). In higher plants this system provides protection against viruses (Ratcliff et al., 1997, 1999; Mourrain et al., 2000), whereas in Caenorhabditis elegans (Ketting et al., 1999), Drosophila (Jensen et al., 1999) and Chlamydomonas (Wu-Scharf et al., 2000) the targeted elements are transposons. Expression of transgenes is suppressed by PTGS in higher plants (Kooter et al., 1999), Chlamydomonas (Wu-Scharf et al., 2000), Neurospora crassa (Cogoni et al., 1996), C.elegans (Ketting and Plasterk, 2000) and Drosophila (PalBhadra et al., 1997). Presumably, in PTGS of transgenes, the foreign DNA or the corresponding RNA is perceived by the cell as if it were a virus or a transposon.
A remarkable feature of PTGS is its ability to provide nucleotide sequence-specific protection against many different types of foreign genetic element. In effect, it is a type of immune system that operates at the nucleic acid level. However, unlike antibody-mediated immunity, the specificity of the system is not genetically programmed. Instead, it seems that the foreign genetic element is the source of the specificity determinant in PTGS through a process that is likely to involve an RdRP homologue in higher plants (Dalmay et al., 2000b; Mourrain et al., 2000), N.crassa (Cogoni and Macino, 1999a) and C.elegans (Smardon et al., 2000). Other proteins that are involved include an EIF2C homologue in higher plants (Fagard et al., 2000), N.crassa (Catalanotto et al., 2000) and C.elegans (Tabara et al., 1999). Proteins with homology to RNase D/ReqQ are involved in N.crassa (Cogoni and Macino, 1999b) and C.elegans (Ketting et al., 1999).
Double-stranded (ds) RNA is also a likely component of the PTGS mechanism. It is a potent activator of PTGS in plants (Chuang and Meyerowitz, 2000; Smith et al., 2000) and animals (Fire et al., 1998; Wianny and Zernicka-Goetz, 2000). In Drosophila, dsRNA is processed into small 21–23 nucleotide RNAs (Zamore et al., 2000) that are incorporated as guide RNAs into an RNase complex (Hammond et al., 2000). These short RNAs, which are also found in plants exhibiting PTGS (Hamilton and Baulcombe, 1999), would anneal with complementary RNAs and thereby ensure that the RNase specifically targets the RNA species that are homologous to the original dsRNA.
With PTGS mediated by viruses, viroids and inverted repeat transposons, a plausible scenario is that the dsRNA is produced directly as a replication intermediate or by transcription. However, in other examples of PTGS, the foreign genetic element may produce single-stranded (ss) RNA that the RdRP converts to a ds form. Presumably, the ssRNA template of the RdRP is differentiated from native RNA species by a distinctive structural feature or aberration. Currently, the nature of this feature aberration is not known, but, from work in Chlamydomonas (Wu-Scharf et al., 2000), it is possible that either misprocessed or prematurely terminated RNAs are involved.
PTGS in virus-infected plants is targeted against the viral genome and we have exploited this finding with virus vectors carrying fragments of host genes as a means of inactivating host gene expression (Ruiz et al., 1998; Baulcombe, 1999; Burton et al., 2000). The PTGS is targeted against the RNA of the host gene so that the symptoms in the infected plant reflect the function of the encoded protein. This approach complements genetic approaches to assigning gene function (Baulcombe, 1999) but is also useful in the dissection of PTGS. For example, in virus-mediated PTGS of a green fluorescent protein (GFP) transgene we have demonstrated that there is a virus-dependent initiation stage of PTGS. A later stage that accounts for maintenance of PTGS is transgene- rather than virus-dependent (Ruiz et al., 1998) and is associated with methylation of the GFP transgene (Jones et al., 1999). In contrast, the virus-mediated PTGS of phytoene desaturase (PDS) and rubisco small subunit endogenous genes does not exhibit the progression from initiation to maintenance (Ruiz et al., 1998; Jones et al., 1999), does not become virus independent and is not associated with DNA methylation.
In this paper we describe a genetic analysis of PTGS that incorporates the use of virus-mediated gene silencing. We focus on SDE3, which is one of at least four silencing defective (SDE) loci in Arabidopsis encoding proteins required for PTGS (Dalmay et al., 2000b). One of these, SDE1, encodes an RdRP (Dalmay et al., 2000b) and is the same as the SGS2 locus described independently (Mourrain et al., 2000). Here we show that SDE3, like SDE1, is required for PTGS mediated by a GFP transgene but not by a tobacco rattle virus (TRV) vector construct. We also show that SDE3 represents a group of RNA helicase-like proteins in mice, humans and Drosophila. These proteins are similar to, but clearly distinct from, the SMG-2 RNA helicase involved in nonsense-mediated RNA degradation and PTGS in C.elegans (Domeier et al., 2000).
Results
Plant lines
The sde3 locus was identified through its loss of PTGS phenotype in Arabidopsis plants carrying a PVX:GFP amplicon [A] and a 35S:GFP reporter gene [G] (Dalmay et al., 2000b). We refer to plants carrying both loci in the homozygous condition as [GxA]. In wild-type (wt) [GxA] plants, the [A] locus mediated PTGS of the [G] locus whereas, in plants carrying sde mutations, the PTGS was attenuated or absent. In plants carrying the sde1 mutation, there was complete loss of PTGS whereas, in sde3 plants, the PTGS was reduced in the true leaves and flowers but was as strong as in wt plants in the hypocotyl and cotyledons (Dalmay et al., 2000b).
SDE3 affects susceptibility to CMV but not to TRV
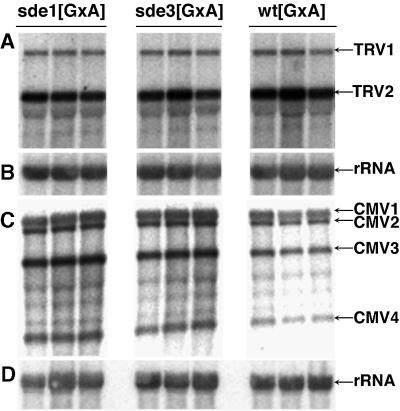
We have shown previously that virus-induced PTGS is a manifestation of an antiviral defence in plants (Ratcliff et al., 1997, 1999). If the sde3 mutation results in loss of this defence mechanism, the plants should exhibit hypersusceptibility to virus infection and viral RNAs should accumulate at a higher level in mutant than in wt plants. To test this possibility we infected plants with TRV and cucumber mosaic virus (CMV). Viral RNA accumulation was monitored by northern analysis. The results in Figure 1 reveal that, with sde3, the mutations had no effect on susceptibility to TRV: the level of viral RNA was the same in sde3 (Figure 1A, tracks 4–6) and wt plants (Figure 1A, tracks 7–9). Figure 1 also confirms our previous finding that sde1 had no effect on TRV accumulation (Figure 1A, tracks 1–3). TRV symptoms were also unaffected by sde3. In contrast, with CMV the virus accumulation measured by quantitative analysis of phosphorimager data was five times higher in the sde1 and sde3 plants than in wt (Figure 1C), and symptoms were more severe in the flowering stem (data not shown).
Fig. 1. SDE3 affects susceptibility to CMV but not to TRV. RNA was isolated from leaves of TRV- (A and B) or CMV- (C and D) infected wild-type (wt[GxA]), sde1[GxA] and sde3[GxA] plants. Northern blot analysis was carried out using 32P-labelled probes corresponding to TRV (A), CMV (C) and rRNA (B and D). The RNA species detected were TRV RNA1 and 2 (TRV1 and TRV2), CMV RNA1, 2, 3 and 4 (CMV1, CMV2, CMV3 and CMV4) and rRNA.
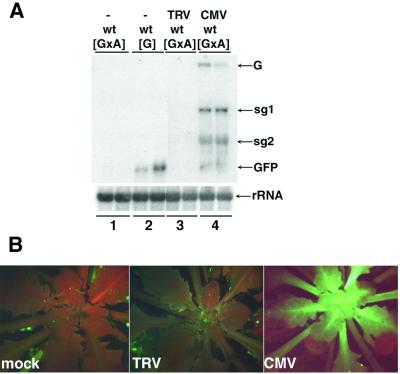
It has been suggested previously that virus-encoded suppressors of PTGS could mask the phenotype of PTGS mutations (Mourrain et al., 2000). If that is the case, any effects of sde1/3 on virus accumulation could have been suppressed by CMV- or TRV-encoded proteins. To investigate this possibility we inoculated TRV and CMV to wt[GxA] plants. We reasoned that the virus-encoded suppressors of PTGS would cause an increase in the levels of the PVX:GFP RNA from locus [A] and of GFP RNA from locus [G] (Brigneti et al., 1998; Voinnet et al., 1999). The increase in these RNAs would lead to enhanced GFP fluorescence in the presence of viral suppressors of PTGS.
These predicted effects of suppressors of PTGS were evident in CMV-infected plants (Figure 2A, tracks 1 and 4, and B). Presumably, the CMV-encoded 2b protein, which is a suppressor of PTGS (Brigneti et al., 1998), was responsible for these effects. In contrast, the TRV-infected plants failed to exhibit GFP fluorescence (Figure 2B) and the levels of PVX:GFP and GFP RNA were as low in TRV-infected [GxA] as in non-infected plants (Figure 2A, tracks 1 and 3). From these results we can rule out the possibility that TRV encodes a strong suppressor of PTGS in Arabidopsis that would mask the effect of mutations on virus-mediated PTGS. Therefore, our finding that sde3 has no effect on TRV accumulation indicates that the encoded protein, like SDE1 (Dalmay et al., 2000b), is not required for TRV-mediated PTGS.
Fig. 2. CMV encodes for a stronger suppressor than TRV. (A) RNA was isolated from leaves of non-inoculated [GxA] (lane 1) or [G] (lane 2) and TRV- or CMV-inoculated [GxA] (lanes 3 and 4, respectively) plants. Northern blot analysis was carried out using 32P-labelled probes corresponding to the full-length GFP RNA sequence (top) or to rRNA (bottom). The RNA species detected were the genomic (G) or subgenomic (sg1 and sg2) RNA of PVX, GFP mRNA (GFP) and rRNA. (B) The images of [GxA] plants were produced under UV light in a dissecting microscope 2 weeks after inoculation with water (mock), TRV or CMV. The red fluorescence is due to chlorophyll.
SDE3 is not required for TRV-mediated gene silencing
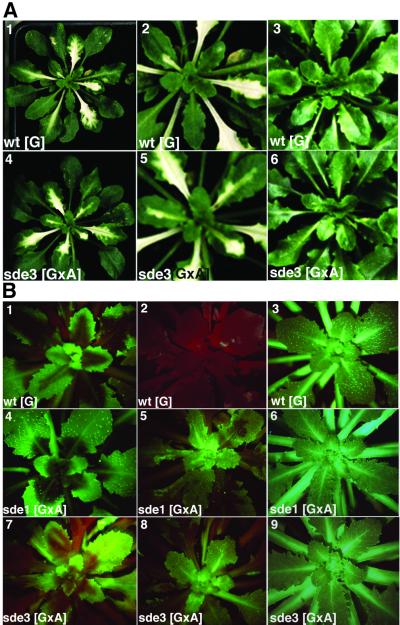
To further investigate the role of SDE3 we infected plants with a TRV vector carrying an insert from the endogenous PDS gene. In wt plants, this virus causes PTGS to be targeted against the endogenous PDS RNA, resulting in low levels of photoprotective carotenoids and photobleaching in the leaves. These symptoms appeared at ∼10 days post-inoculation (d.p.i.) and persisted for 5–7 days (Figure 3A). At later times, the PTGS phenotype was lost and the newly emerging leaves were fully green. This transient PTGS most likely reflects the kinetics of TRV accumulation in infected plants. Initially, in the phase when there is abundant virus accumulation, the PTGS is strong. In the later stages of the infection process, the virus levels are low (Ratcliff et al., 1999).
Fig. 3. SDE3 is not required for TRV-induced gene silencing. (A) Photographs were taken of TRV:PDS-infected wt[G] and sde3[GxA] plants, 3 weeks after inoculation (1 and 4). Panels 2 and 5 show the same plant as 1 and 4, respectively, focusing on the newly emerging leaves. The white areas are a result of photobleaching due to the PTGS of the PDS gene. Panels 3 and 6 show non-infected wt[G] and sde3[GxA]. (B) Plants were infected with TRV:GFP and images were produced under UV light in a dissecting microscope; the red fluorescence is due to chlorophyll. The photographs were taken of TRV:GFP-infected wt[G], sde1[GxA] and sde3[GxA] at 10 (panels 1, 4 and 7) or 20 (panels 2, 5 and 8) d.p.i. Panels 3, 6 and 9 show non-infected wt[G], sde1[GxA] and sde3[GxA].
We predicted that if SDE3 is required for virus-mediated PTGS, the photobleaching symptoms of PTGS would be delayed or more transient than on wt plants. However, in three independent experiments with 10 infected plants, the photobleaching on sde3 plants developed at the same rate and was as persistent as on wt plants (Figure 3A). These results therefore provide confirmation of the susceptibility data (Figure 2), indicating that SDE3 is not required for TRV-mediated PTGS.
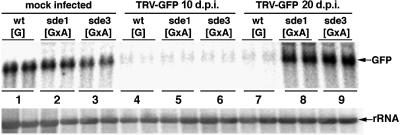
In a second assay for TRV-mediated PTGS we infected plants with TRV:GFP. At 10 d.p.i. on wt[G] plants, this virus induced PTGS of the GFP transgene in the regions around the veins (Figure 3B, 1). By 20 d.p.i., the silencing had spread throughout the plant leaves (Figure 3B, 2) and, unlike the PDS silencing, this effect persisted for the life of the plant. In both the early and late stages of this process, the absence of GFP fluorescence was associated with a reduced steady-state level of GFP RNA (Figure 4, lanes 1, 4 and 7).
Fig. 4. SDE3 is required for persistent silencing of GFP. RNA was isolated from the emerging leaves of mock-inoculated (lanes 1, 2 and 3) or TRV:GFP-inoculated wt[G], sde1[GxA] and sde3[GxA] plants. Samples were taken from the TRV:GFP-inoculated plants at 10 (lanes 4, 5 and 6) or 20 (lanes 7, 8 and 9) d.p.i. Northern blot analysis was carried out using 32P-labelled probes corresponding to the full-length GFP RNA sequence (top) or rRNA (bottom).
When TRV:GFP was inoculated to the sde1 and sde3 mutants of [GxA], the initial stages of PTGS were exactly the same as on the wt[G] lines: GFP expression was lost from the regions around the veins (Figure 3B, 4 and 7) and GFP RNA levels were 10-fold lower in the infected leaves (Figure 4, lanes 5 and 6) than in equivalent tissue of the mock-inoculated plants (Figure 4, lanes 2 and 3). However, the PTGS of GFP in the sde1 and sde3 plants did not persist. By 20 d.p.i., the GFP expression in the newly emerging leaves of the TRV:GFP-infected plants (Figure 3B, 5 and 8) was as extensive as on the non-inoculated plants (Figure 3B, 3, 6 and 9), and the GFP RNA was as abundant as in mock-inoculated plants (Figure 4, lanes 2, 3, 8 and 9).
The TRV:GFP inoculations were also carried out on sde1 and sde3 mutants of line [G] carrying a GFP transgene but not a PVX:GFP amplicon. The aim of these further tests was to investigate the possibility that the PVX:GFP RNA or PVX-encoded proteins could influence the outcome of the TRV-induced silencing of GFP. On both sde1 and sde3 mutants of line [G], the outcome was the same as in the [GxA] background: there was transient loss of GFP expression and GFP RNA but, by 20 d.p.i., the GFP levels were the same as on non-infected plants (data not shown). From these results we have shown that the persistence but not initiation of TRV-induced PTGS of GFP was lost on the sde1 and sde3 mutant plants. The transient TRV-induced PTGS of PDS was unaffected by the mutations.
SDE3 encodes an RNA helicase-like protein
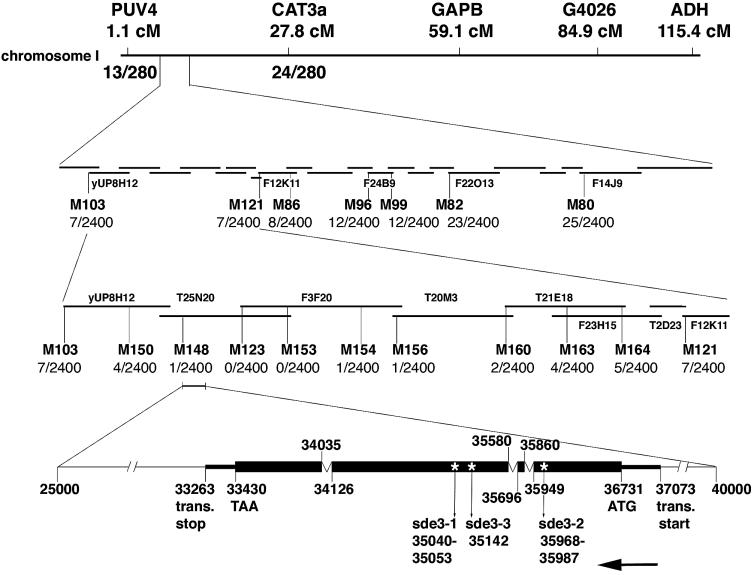
To shed light on the role of SDE3 in the PTGS mechanism we used genetic markers to identify the DNA coding sequence at the SDE3 locus. First we assigned SDE3 to chromosome 1 between PUV4 (1.1 cM) and CAT3a (27.8 cM). We then used a series of markers in the interval of ∼2 Mbp and a mapping population of 1200 plants from a cross of _sde3_[GxA] × wt Landsberg ecotype to assign SDE3 to the interval M148–M154. Within this interval there are 23 predicted open reading frames (ORFs). We were not able to generate a more precise map position of the SDE3 locus due to the low frequency of recombination in this interval. However, using primers designed to PCR amplify each of the predicted ORFs from wt and three mutant plants, we identified only one polymorphic gene.
Sequence analysis of cDNA generated by 5′ and 3′ RACE indicates that this polymorphic SDE3 candidate has three introns. This gene has a 14 bp deletion in sde3-1 and a 20 bp deletion in sde3-2 that is replaced by a 38 bp insertion. The third allele (sde3-3) had a point mutation that introduced an early stop codon (Figure 5).
Fig. 5. Mapping of SDE3. The sde3 phenotype co-segregated with PUV4 and CAT3a markers on chromosome 1. Seven markers were generated throughout this region and the mapping population was increased to 1200 plants. There were seven recombination events between M103 and SDE3 and seven between SDE3 and M121, indicating that SDE3 is located between these two markers. Another nine markers were generated to make a high resolution map around SDE3. There was one recombination event between SDE3 and both M148 and M154, locating SDE3 in a 100 kb region. This sequence is represented in BAC clones T25N20 and F3F20. The bottom part shows the organization of SDE3. The arrow indicates that SDE3 is in a complementary orientation on T25N20 in the database. The numbers indicating the translation start point, the start codon, the exon–intron boundaries, the stop codon, the translation stop point and the positions of the mutation in different mutant alleles are according to the numbering of T25N20.
Transgenes comprising 5 kb of DNA spanning the wt allele of the candidate SDE3 were transformed into [GxA] lines carrying sde3. Three independent transformants were indistinguishable from the wt [GxA] plants with GFP fluorescence restricted to small regions in the growing points of shoots and leaves. None of the emerged leaves of these plants was green fluorescent and the levels of GFP mRNA were as low as in the wt plants. Thus, these complementation data and the occurrence of three independent mutations confirm that this candidate gene is the SDE3 locus.
BLASTP database searches revealed that the 113 363 Da SDE3 has motifs that are conserved in RNA helicase-like proteins (Koonin, 1992). In SDE3, these motifs are typical of a class of RNA helicases that also includes the yeast protein Upf1p (Linder and Daugeron, 2000). However, a mouse protein encoded by gb110 (Mov10) (Mooslehner et al., 1991; Hamann et al., 1993) rather than Upf1p was the closest homologue of SDE3, as identified by BLASTP. All of these SDE3 homologues have RNA helicase motifs that are quite distinct from those of the DEAD, DEAH and Ski2p types of RNA helicase (Linder and Daugeron, 2000).
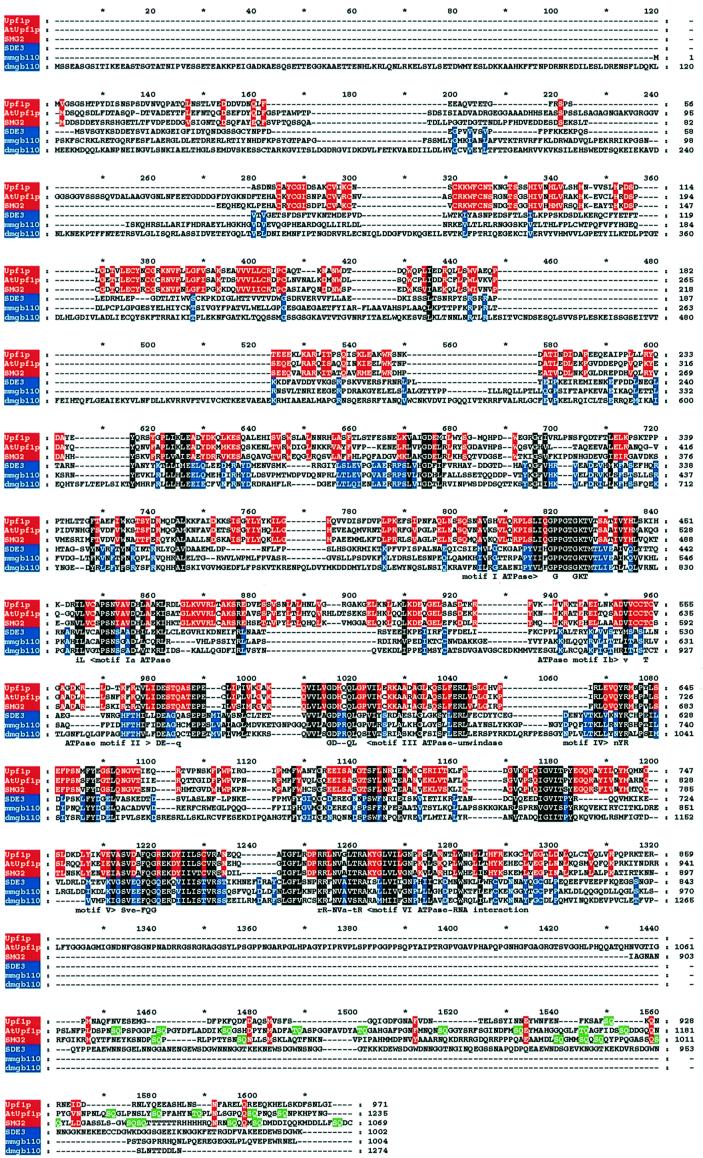
Several features (Figure 6) indicate that there are two groups of SDE3-like helicase. For example, in the N-terminal region of Upf1p and its homologue in C.elegans (SMG-2), there is a conserved cysteine-rich region. In addition, near the C-terminus of these proteins, there are multiple SQ doublets (Leeds et al., 1992; Page et al., 1999). The presence of these domains defines one of the two groups that includes a predicted Arabidopsis protein of unknown function (SWISSPROT accession No. SWALL:BAB10240) but not SDE3. We refer to the SWALL:BAB10240 protein as AtUpf1p because it is the likely functional homologue of Upf1p/SMG-2 in Arabidopsis.
Fig. 6. Sequence alignments of SDE3 and other RNA helicase-like proteins. The sequence of SDE3 was aligned with other RNA helicase-like proteins Upf1p from yeast (accession No. SWALL:P30771), AtUpf1p from Arabidopsis (accession No. SWALL:BAB10240), SMG-2 from C.elegans (accession No. SWALL:O76512) and homologues of a protein in mouse (mmgb110; accession No. SWALL:P23249) and Drosophila (dmgb110; accession No. SWALL:Q9VZP4) using CLUSTALW. These proteins represent the best matches to SDE3 in a database search using BLASTP. The output of CLUSTALW was shaded with GeneDoc; residues in blue are similar in SDE3, mmgb110 and dmgb110; residues in red are similar in Upf1p, AtUpf1p and SMG-2. Residues in white on black are similar in all six sequences. Motifs that have previously been used to define the Upf1p group of RNA helicases (Linder and Daugeron, 2000) are shown beneath the aligned sequences. Residues indicated on a green background are the repeated SQ motifs in the C-terminal domain. It is likely that the predicted AtUpf1p and dmgb110 sequences do not account for introns that were not recognized in the annotation of the genome sequence.
The second group of Upf1p-like helicases includes SDE3 and proteins from mouse, humans and Drosophila (Figure 6 and data not shown). This group is defined by the absence of the cysteine-rich and SQ domains. In addition there are conserved motifs, between 620 and 740 of the aligned sequences (Figure 6), in which the SDE3 group is distinct from Upf1p and its close homologues. Motif searches failed to identify a function of these conserved motifs.
Discussion
A role for SDE3 in synthesis of dsRNA?
We proposed previously that the role of the SDE1 RdRP was to convert aberrant ssRNA of a transgene into a ds form (Dalmay et al., 2000b). The dsRNA would be processed into short 21–23 nucleotide RNAs (Hamilton and Baulcombe, 1999; Zamore et al., 2000) that guide RNase to the targets of PTGS (Hammond et al., 2000). The proposed role of the RdRP in production of dsRNA was based in part on the finding that SDE1 is not required for TRV:PDS-mediated PTGS of PDS (Dalmay et al., 2000b). It seemed likely that the TRV-encoded RdRP could synthesize a ds replication intermediate and thereby compensate in PTGS for the absence of SDE1. As SDE3 is also not required for TRV:PDS-mediated PTGS of PDS (Figure 3A), it is likely that this protein, like SDE1, is involved in production of dsRNA.
The early initiation of TRV-induced PTGS of GFP was not affected by sde1/3 and most likely involves the same virus-mediated mechanism as in TRV:PDS-infected plants. However, the later persistence of TRV-induced PTGS of GFP was affected by sde1/3 (Figures 3 and 4). To explain this difference in early and late stages we propose that the virus-dependent initiation of GFP PTGS progressed into a virus-independent and transgene-dependent maintenance stage, as described previously for PVX-mediated PTGS of GFP in N.benthamiana (Ruiz et al., 1998). Presumably the SDE1/SDE3 proteins would be required at this stage for production of dsRNS
At present we cannot explain why there was no virus-independent maintenance of PDS PTGS. One possibility is that endogenous genes are not able to support the maintenance stage and, consistent with this idea, we showed that virus-mediated PTGS of rubisco does not exhibit the maintenance stage (Jones et al., 1999). Analyses of PTGS induced by a graft-transmissible signal of silencing (Palauqui and Vaucheret, 1998) are also consistent with this idea. Clearly, more examples with transgenes and endogenous genes will be needed to investigate this issue further.
A role for SDE1 and SDE3 in antiviral defence?
As PTGS represents a type of antiviral defence, it was expected that the sde1/3 plants would be hypersusceptible to virus infection. However, in most examples tested, that was not the case; the sde1 and sde3 plants were as susceptible to tobacco mosaic virus, TRV and turnip crinkle virus (TCV) as were the wt plants (Figure 1 and T.Dalmay, unpublished data). Of the viruses tested, only CMV is affected by sde3 or sde1/sgs2 (Figure 1) (Mourrain et al., 2000). To account for the unexpected lack of sde1/_3_-mediated hypersusceptibility we propose that viruses have evolved several strategies for overcoming PTGS; if a virus has overcome PTGS, it would not be affected by sde mutations.
TMV and TCV may manifest one of these strategies. These viruses may have lost the ability to produce the aberrant ssRNA that we have proposed is required for SDE1/_SDE3_-dependent PTGS. Alternatively, or additionally, these viruses may encode suppressors of PTGS (Voinnet et al., 1999). The action of these proteins would suppress PTGS in both wt and mutant plants so that there would be no discernible effect of the mutations.
The counter-defence strategy of TRV does not involve a strong suppressor of PTGS (Figure 2) (Voinnet et al., 1999) and it is possible that this virus does not produce the putative aberrant RNAs required for SDE1/_SDE3_-dependent PTGS. An alternative possibility is that these RNAs are produced but that they are somehow hidden in the cell so that they do not participate in the SDE1/_SDE3_-dependent branch of PTGS.
A third counter-defence strategy, exhibited by CMV, is different from that of the other viruses tested, as illustrated by the effect of sde3 or sgs2/sde1 on accumulation of this virus (Figure 1) (Mourrain et al., 2000). Thus, CMV is affected by the SDE1/SDE3 mechanism despite its ability to produce the 2b protein suppressor of PTGS (Brigneti et al., 1998). Clearly, this virus does produce the RNA species that are required for the SDE1/SDE3 mechanism. Perhaps the suppression of PTGS by the 2b protein is enough to allow accumulation and spread of CMV in the infected plant but not so complete that the sde1/sde3 phenotype is masked.
We predict that viruses other than CMV will accumulate to a lower level on wt than on sde1/sde3 plants. Some of these, like CMV, may be viruses for which Arabidopsis is a recognized host. However, there may be others that are so effectively targeted by the PTGS in the wt plants so that Arabidopsis is not a host. These might be viruses encoding suppressors of PTGS that are effective in their hosts but not in Arabidopsis. If that is the case, PTGS may be significant as a component of a general defence system, referred to as non-host resistance, which accounts for the maxim that ‘most plants are resistant against most viruses’. A role for PTGS in non-host resistance would be confirmed if viruses that do not infect wt Arabidopsis do accumulate and spread on the sde1/sde3 plants.
RNA helicases in PTGS
Several helicase-like proteins have been implicated in PTGS. In addition to SDE3, there are the QDE3/MUT7 RecQ helicases in N.crassa and C.elegans (Cogoni and Macino, 1999b; Ketting et al., 1999) and SMG-2 from C.elegans (Domeier et al., 2000), which is a member of the Upf1p group of RNA helicases (Linder and Daugeron, 2000). MUT6 is a DEAH helicase required for PTGS in Chlamydomonas (Wu-Scharf et al., 2000).
SDE3 differs markedly from QDE3/MUT7, has slight similarity to MUT6 in the helicase motifs and is highly similar to Upf1p/SMG-2 (Figure 6). However, SDE3 lacks a cysteine-rich region in the N-terminal region of Upf1p/SMG-2 (Page et al., 1999), which interacts with Nmd2p (He et al., 1997) to facilitate nonsense-mediated mRNA decay. The repeated SQ motifs in the C-terminal region of SMG-2 and Upf1p are also absent from SDE3 (Figure 6). It has been proposed that these motifs are the substrate of phophatidylinositol 3-kinase-related kinase (Page et al., 1999). Thus, from the absence of important motifs, despite the extensive helicase domain similarity, it is unlikely that SDE3 is the functional homologue of Upf1p and SMG-2. A more likely candidate is the Arabidopsis protein AtUpf1p, which has the N-terminal cysteine-rich region, the Upf1p helicase motifs and the C-terminal SQ repeats (Figure 6).
The absence of Upf1p-like motifs in the N- and C-terminal regions indicates that SDE3 represents a subgroup of the Upf1p-like RNA helicases. The SDE3 subgroup is further characterized by motifs on the N-terminal side of the RNA helicase motifs (Figure 6). Other members of this group are the GB110 protein of mouse and homologues in humans and Drosophila. Presumably, the absence of the cysteine-rich region indicates that this subgroup of proteins does not interact with homologues of Nmd2p and, therefore, that they are available to interact with other as yet unidentified proteins.
It is notable that there are no RNA helicases in the SDE3 subgroup encoded in the genome of C.elegans, although this organism is competent in PTGS. One explanation for this discrepancy is that SDE3-like proteins are regulators rather than essential cofactors of PTGS and are not used in C.elegans. Consistent with that idea, we have reported that sde3 plants exhibit only partial loss of PTGS. PTGS of GFP in the cotyledons is as complete as in the wt plants and, in the true leaves, the PTGS defect is less pronounced than in sde1 and sde2 plants. A second explanation is based on the idea, as discussed above, that SDE3 mediates conversion of ssRNA into a ds form. If that is the case, an SDE3 homologue would not be required in C.elegans because PTGS is normally induced by direct introduction of dsRNA (Fire et al., 1998). It could also be that SMG-2 is a multifunctional protein and, despite the sequence difference in the N-terminal region, carries out the role of SDE3.
It is not known to what extent PTGS in mammals is used as defence against foreign genetic elements. Mouse embryos are competent in PTGS induced by dsRNA (Wianny and Zernicka-Goetz, 2000) but there is currently only limited information about other cell types or the possibility that PTGS can be induced by genetic elements that do not produce dsRNA (Bahramian and Zarbl, 1999). The finding that GB110 is an SDE3 homologue in the mouse genome suggests that PTGS can operate in mammals as in plants. However, we have not been able to identify homologues of SDE1 in mammalian genomes (T.Dalmay, unpublished data) and the possibility remains that SDE3 has multiple roles. The involvement of SMG-2 in RNA interference and nonsense-mediated mRNA decay provides a precedent for a multifunctional RNA helicase involved in PTGS. It would be interesting to test RNA interference in embryos of GB110 mice.
Genetic and biochemical analyses of PTGS
SDE3 is just one of many genes required for PTGS that have been identified from a genetic approach (Plasterk and Ketting, 2000). In some instances, from epistasis analysis and by exploiting viruses, transgenes and transposons as initiators of PTGS, it is possible to produce schemes that assign roles of the encoded proteins to the surveillance and response parts of the mechanism (Plasterk and Ketting, 2000; Voinnet et al., 2000). However, there are indications from the mutation analysis of plants described here and previously (Dalmay et al., 2000b; Mourrain et al., 2000) that a genetic approach may not access all parts of the PTGS mechanism. These projects led to proteins involved in the same stage of the PTGS mechanism and have not so far provided information about the processing of dsRNA or the RNase complex in PTGS. One possible explanation for this bias in the output of the genetic approach is that genes required for other stages of PTGS encode proteins that are essential for normal growth. If that is the case, for a definitive description of PTGS, it will be necessary to develop biochemical approaches. The RNA processing and RNA degrading extracts of Drosophila (Hammond et al., 2000; Zamore et al., 2000) are indicators of the types of system that will be needed.
Materials and methods
Transgenic plants and mutagenesis
The wt[A], wt[G]and wt[GxA] Arabidopsis (C24 ecotype) were described (Dalmay et al., 2000a) as Amp243, GFP142 and GFP142 × Amp243, respectively. The wt[A] contains a 35S-PVX:GFP and wt[G]carries a 35S-GFP transgene. Both lines are homozygous and have a single copy of the respective transgene. The wt[GxA] is the progeny of a cross between the two lines above and it is homozygous for both of the transgenes. The mutagenesis of wt[GxA] and a screen for loss of PTGS plants were described (Dalmay et al., 2000b).
GFP imaging
GFP expression was monitored using an MZ12 dissecting microscope (Leica, Heidelberg, Germany) coupled to an epifluorescence module. Photographs were taken using Kodak Ektachrome Panther (400 ASA) film.
RNA analysis
RNA gel blot analysis was performed as described previously (Mueller et al., 1995). DNA fragments were labelled by random priming incorporation of [32P]dCTP (Amersham). After hybridization, the signal present in the membranes was analysed and quantified using the Fujix Bio-Imaging Analyzer Bas 1000 (Fuji Photo Film Co., Ltd, Fuji, Japan) equipment. PCR-amplified full-length GFP DNA was used for the GFP-specific probe. TRV RNA was detected with a probe made of a _Bst_EII and _Sma_I fragment (5345–6792) of a TRV1 clone (pTR7116) (Hamilton and Baulcombe, 1989), and pCa7 (Ratcliff et al., 2001), which contains the full sequence of RNA2. The CMV probe was pK1, pK2 and pK3, each of which contains the full sequence of RNA 1, 2 and 3, respectively (Boccard and Baulcombe, 1992).
Genetic mapping and DNA sequence analysis
A set of CAPS markers described by Konieczny and Ausubel (1993) was used to detect polymorphism between the Columbia and Landsberg ecotypes and to map sde loci to the 10 chromosome arms. However, several of these markers did not show polymorphism between C24 and Landsberg ecotypes, and in some instances it was necessary to generate alternative markers. The detail of those markers is available on our website (http://www.jic.bbsrc.ac.uk/welcome.htm).
The sde3 mutation was mapped into a 2 Mb region between PUV4 (1.1 cM) and CAT3a (27.8 cM). There were 13 (PUV4) and 24 (CAT3a) recombinants in 140 plants. Seven markers were generated throughout this region and the mapping population was increased to 1200 plants. SDE3 was localized to a 550 kb region between markers M103 and M121 (seven recombinants with both markers). Using another nine markers, SDE3 was finally mapped between M148 and M154, with one recombination between the gene and both markers. This region is ∼100 kb and is represented in BAC clones T25N20 and F3F20. These BACs are sequenced and annotated by the Arabidopsis Sequencing Project (www.Arabidopsis.org). Primers were designed to amplify all 23 predicted ORFs within the genetically defined region. DNA from wt and three different sde3 mutant plants was used to amplify these ORFs. The PCR products were digested with eight restriction enzymes (_Hpa_II, _Rsa_I, _Alu_I, _Hae_III, _Hin_fI, _Hin_cII, _Dde_I and _Sau_3AI) and run in 1.5% agarose gels. Only the PCR products amplifying T25N20.11 showed polymorphism between wt and mutants, after digestion with _Dde_I (sde3-1) and _Dde_I or _Hae_III (sde3-2).
Eight overlapping DNA fragments were generated by PCR from the wt, sde3-1, sde3-2 and sde3-3 plants at the position in BAC T25N20 positions 33 000–37 500 kb and sequenced directly using the Big Dye Terminator Mix (PE Applied Biosystem). The sequencing reactions were resolved on an ABI377 automated sequencer (Applied Biosystem, La Jolla, CA). The regions where mutation was found were sequenced on both strands from three independent PCRs from both the wt and the mutant plants.
The 5′ and 3′ ends of the SDE3 cDNA were determined by RACE using the Marathon cDNA amplification kit (Clontech). RACE products were cloned into pGEM-T plasmid (Promega) and sequences of 10 independent 3′ and 5′ end clones were determined. The cDNA library, obtained by the Marathon cDNA amplification kit (Clontech), was used as a template to PCR amplify eight overlapping fragments that were directly sequenced. The number and location of introns were determined by comparing the sequence data obtained from the genomic DNA with the data obtained from the cDNA.
A 5039 bp region (32 581–37 620 on the BAC T25N20) containing the transcribed region of SDE3 together with 547 bp upstream and 682 bp downstream sequences was PCR amplified from wt Arabidopsis (Columbia ecotype) using the High Fidelity Expand PCR kit (Roche). The PCR product was cloned into pGreen1179 (Hellens et al., 2000) and then transformed into Agrobacterium tumefaciens GV3101. Sequence analysis confirmed that the cloned PCR product was identical to the wt genomic sequence. Flowers of sde3-1[GxA] plants were transformed with the above construct and the seeds were selected on hygromycin-containing medium (50 µg/ml) (Sigma). DNA was extracted from the resistant seedlings and PCR analysis confirmed that the plants carried an SDE3 transgene in an sde3 homozygote background.
In order to obtain sde1 and sde3 plants that contain the 35S-GFP but not the 35S-PVX-GFP transgene, sde1-3[GxA] and sde3-1[GxA] plants were crossed with the [G] line. In the F2 plants, the [G] locus was homozygous while the sde3 mutations and the 35S-PVX-GFP [A] locus segregated. The _sde1_[G] and _sde3_[G] F2 plants were screened for absence of the [A] locus and the presence of the sde1 or sde3 mutations by PCR.
Other protein sequences homologous to SDE3 were identified using the BLASTP program (Altschul et al., 1990). Protein sequences were obtained and collated using the Wisconsin package [Wisconsin Package Version 10.0, Genetics Computer Group (GCG), Madison, WI]. Sequence alignments were produced using CLUSTAL W (Thompson et al., 1994) and were displayed using Genedoc (Nicholas and Nicholas, 1997).
The sequence data of the SDE3 cDNA have been deposited in the DDBJ/EMBL/GenBank database (accession No. AF339908). The sequence of the mutants is available on our website (http://www.jic. bbsrc.ac.uk).
Wild-type and recombinant viruses
The TRV vector carrying the PDS fragment is similar to TRV:GFP described previously (Ratcliff et al., 1999) This vector is described in more detail elsewhere (Ratcliff et al., 2001). The insert in RNA2 of the TRV:PDS vector is the 1770 nucleotides of the Arabidopsis PDS cDNA sequence extending between residues corresponding to 2573 and 4343 of the genomic sequence. The insert was in the sense orientation relative to the TRV ORFs.
Acknowledgments
Acknowledgements
We are grateful to Stephen Rudd for advice on computer analyses, and Louise Jones and Alan Herr for comments on a draft manuscript. Financial support was from the Gatsby Charitable Foundation and the European Community framework IV COMREP project. The work with modified and imported viruses was carried out under licence from the Ministry of Agriculture Fisheries and Food (PHL 24A/2921).
Note added in proof
SDE3 is not similar to DICER, an RNA helicase implicated in RNAi in Drosophila (Bernstein,E. et al. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363--366).
References
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local aligment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bahramian M.B. and Zarbl,H. (1999) Transcriptional and post-transcriptional silencing of rodent α1(I) collagen by a homologous transcriptionally self-silenced transgene. Mol. Cell. Biol., 19, 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D.C. (1999) Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol., 2, 109–113. [DOI] [PubMed] [Google Scholar]
- Boccard F. and Baulcombe,D.C. (1992) Infectious in vitro transcripts from amplified cDNAs of the Y and Kin strains of cucumber mosaic virus. Gene, 114, 223–227. [DOI] [PubMed] [Google Scholar]
- Brigneti G., Voinnet,O., Li,W.X., Ji,L.H., Ding,S.W. and Baulcombe,D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J., 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Burton R.A., Gibeaut,D.M., Bacic,A., Findlay,K., Roberts,K., Hamilton,A., Baulcombe,D.C. and Fincher,G.B. (2000) Virus-induced silencing of a plant cellulose synthase gene. Plant Cell, 12, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C., Azzalin,G., Macino,G. and Cogoni,C. (2000) Gene silencing in worms and fungi. Nature, 404, 245. [DOI] [PubMed] [Google Scholar]
- Chuang C.-H. and Meyerowitz,E.M. (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C. and Macino,G. (1999a) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature, 399, 166–169. [DOI] [PubMed] [Google Scholar]
- Cogoni C. and Macino,G. (1999b) Post-transcriptional gene silencing in Neurospora by a RecQ DNA helicase. Science, 286, 342–344. [DOI] [PubMed] [Google Scholar]
- Cogoni C., Irelan,J.T., Schumacher,M., Schmidhauser,T.J., Selker,E.U. and Macino,G. (1996) Transgene silencing of the Al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA–DNA interactions or DNA methylation. EMBO J., 15, 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Hamilton,A.J., Mueller,E. and Baulcombe,D.C. (2000a) Potato virus X amplicons in Arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell, 12, 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Hamilton,A.J., Rudd,S., Angell,S. and Baulcombe,D.C. (2000b) An RNA-dependent RNA polymerase gene in Arabidopsis is required for post-transcriptional gene silencing mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Domeier M.E., Morse,D.P., Knight,S.W., Portereiko,M., Bass,B.L. and Mango,S.E. (2000) A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science, 289, 1928–1930. [DOI] [PubMed] [Google Scholar]
- Fagard M., Boutet,S., Morel,J.-B., Bellini,C. and Vaucheret,H. (2000) AGO1, QDE-2 and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi and RNA interference in animals. Proc. Natl Acad. Sci. USA, 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Hamann L., Jensen,K. and Harbers,K. (1993) Consecutive inactivation of both alleles of the gb110 gene has no effect on the proliferation and differentiation of mouse embryonic stem cells. Gene, 126, 279–284. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe,D.C. (1999) A novel species of small antisense RNA in post-transcriptional gene silencing. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hamilton W.D.O. and Baulcombe,D.C. (1989) Infectious RNA produced by in vitro transcription of a full-length tobacco rattle virus RNA-1 cDNA. J. Gen. Virol., 70, 963–968. [Google Scholar]
- Hammond S.M., Bernstein,E., Beach,D. and Hannon,G. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cell extracts. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- He F., Brown,A.H. and Jacobson,A. (1997) Upf1p, Nmd2p and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol., 17, 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Edwards,E.A., Leyland,N.R., Bean,S. and Mullineaux,P.M. (2000) pGreen: a versatile and flexible binary Ti Vector for _Agrobacterium_-mediated plant transformation. Plant Mol. Biol., 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Jensen S., Gassama,M.-P. and Heidmann,T. (1999) Taming of transposable elements by homology-dependent gene silencing. Nature Genet., 21, 209–212. [DOI] [PubMed] [Google Scholar]
- Jones L., Hamilton,A.J., Voinnet,O., Thomas,C.L., Maule,A.J. and Baulcombe,D.C. (1999) RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell, 11, 2291–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R.F. and Plasterk,R.H.A. (2000) A genetic link between co-suppression and RNA interference in C. elegans. Nature, 404, 296–298. [DOI] [PubMed] [Google Scholar]
- Ketting R., Haverkamp,T., van Luenen,H. and Plasterk,R. (1999) mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell, 99, 133–141. [DOI] [PubMed] [Google Scholar]
- Konieczny A. and Ausubel,F.M. (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J., 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Koonin E.V. (1992) A new group of putative RNA helicases. Trends Biochem. Sci., 17, 495–497. [DOI] [PubMed] [Google Scholar]
- Kooter J.M., Matzke,M.A. and Meyer,P. (1999) Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci., 4, 340–347. [DOI] [PubMed] [Google Scholar]
- Leeds P., Wood,J.M., Lee,B.-S. and Culbertson,M.R. (1992) Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 2165–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder L. and Daugeron,M.-C. (2000) Are DEAD-box proteins becoming respectable helicases? Nature Struct. Biol., 7, 97–99. [DOI] [PubMed] [Google Scholar]
- Mooslehner K., Muller,U., Karls,U., Hamann,L. and Harbers,K. (1991) Structure and expression of a gene encoding a putative GTP-binding protein identified by provirus integration in a transgenic mouse strain. Mol. Cell. Biol., 11, 886–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P. et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for post-transcriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Mueller E., Gilbert,J.E., Davenport,G., Brigneti,G. and Baulcombe,D.C. (1995) Homology-dependent resistance: transgenic virus resistance in plants related to homology-dependent gene silencing. Plant J., 7, 1001–1013. [Google Scholar]
- Nicholas K.B. and Nicholas,H.B. (1997) GeneDoc: a tool for editing and annotating multiple sequence alignments. http://www.psc.edu/biomed/genedoc
- Page M.F., Carr,B., Anders,K.R., Grimson,A. and Anderson,P. (1999) SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol., 19, 5943–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui J.-C. and Vaucheret,H. (1998) Transgenes are dispensable for the RNA degradation step of cosuppression. Proc. Natl Acad. Sci. USA, 95, 9675–9680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PalBhadra M., Bhadra,U. and Birchler,J.A. (1997) Cosuppression in Drosophila: gene silencing of alcohol dehydrogenase by white-ADH transgenes is Polycomb dependent. Cell, 90, 479–490. [DOI] [PubMed] [Google Scholar]
- Plasterk R.H.A. and Ketting,R.F. (2000) The silence of the genes. Curr. Opin. Genet. Dev., 10, 562–567. [DOI] [PubMed] [Google Scholar]
- Ratcliff F., Harrison,B.D. and Baulcombe,D.C. (1997) A similarity between viral defense and gene silencing in plants. Science, 276, 1558–1560. [DOI] [PubMed] [Google Scholar]
- Ratcliff F., MacFarlane,S. and Baulcombe,D.C. (1999) Gene silencing without DNA: RNA-mediated cross protection between viruses. Plant Cell, 11, 1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F., Martin-Hernandez,A.M. and Baulcombe,D.C. (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J., 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Ruiz M.T., Voinnet,O. and Baulcombe,D.C. (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell, 10, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon A., Spoerke,J.M., Stacey,S.C., Klein,M.E., Mackin,N. and Maine,E.M. (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol., 10, 169–178. [DOI] [PubMed] [Google Scholar]
- Smith N.A., Singh,S.P., Wang,M.B., Stoutjesdijk,P.A., Green,A.G. and Waterhouse,P.M. (2000) Gene expression—total silencing by intron-spliced hairpin RNAs. Nature, 407, 319–320. [DOI] [PubMed] [Google Scholar]
- Tabara H., Sarkissian,M., Kelly,W.G., Fleenor,J., Grishok,A., Timmons,L., Fire,A. and Mello,C.C. (1999) The rde-1 gene, RNA interference and transposon silencing in C. elegans. Cell, 99, 123–132. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Pinto,Y.M. and Baulcombe,D.C. (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses. Proc. Natl Acad. Sci. USA, 96, 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Lederer,C. and Baulcombe,D.C. (2000) A viral movement protein prevents systemic spread of the gene silencing signal in Nicotiana benthamiana. Cell, 103, 157–167. [DOI] [PubMed] [Google Scholar]
- Wianny F. and Zernicka-Goetz,M. (2000) Specific interference with gene function by double-stranded RNA in early mouse development. Nature Cell Biol., 2, 70–75. [DOI] [PubMed] [Google Scholar]
- Wu-Scharf D., Jeong,B., Zhang,C. and Cerutti,H. (2000) Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science, 290, 1159–1162. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]