Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing (original) (raw)
Abstract
Su(var)3–9 is a dominant modifier of heterochromatin-induced gene silencing. Like its mammalian and Schizosaccharomyces pombe homologues, Su(var) 3–9 encodes a histone methyltransferase (HMTase), which selectively methylates histone H3 at lysine 9 (H3-K9). In Su(var)3–9 null mutants, H3-K9 methylation at chromocentre heterochromatin is strongly reduced, indicating that SU(VAR)3–9 is the major heterochromatin-specific HMTase in Drosophila. SU (VAR)3–9 interacts with the heterochromatin-associated HP1 protein and with another silencing factor, SU(VAR)3–7. Notably, SU(VAR)3–9–HP1 interaction is interdependent and governs distinct localization patterns of both proteins. In Su(var)3–9 null mutants, concentration of HP1 at the chromocentre is nearly lost without affecting HP1 accumulation at the fourth chromosome. By contrast, in HP1 null mutants SU(VAR)3–9 is no longer restricted at heterochromatin but broadly dispersed across the chromosomes. Despite this interdependence, Su(var)3–9 dominates the PEV modifier effects of HP1 and Su(var)3–7 and is also epistatic to the Y chromosome effect on PEV. Finally, the human SUV39H1 gene is able to partially rescue Su(var)3–9 silencing defects. Together, these data indicate a central role for the SU(VAR)3–9 HMTase in heterochromatin-induced gene silencing in Drosophila.
Keywords: gene silencing/H3-K9 methylation/heterochromatin/SU(VAR)3–9
Introduction
Heritable changes in gene expression between successive generations (imprinting) and during ontogenesis (gene silencing) are controlled by higher order chromatin structure. For an analysis of molecular processes connected with gene silencing the phenomenon of position-effect variegation (PEV) in Drosophila is of special interest. In PEV, euchromatic regions become subjected to transcriptional silencing after their placement into a heterochromatic neighbourhood. Nuclease digestion studies (Wallrath and Elgin, 1995) and DNA methylase accessibility assays (Boivin and Dura, 1998) demonstrate a significant change in the higher order chromatin structure of regions subjected to heterochromatin-induced silencing. Spreading of the heterochromatic structure beyond the breakpoint, _trans_-interactions between heterochromatic regions and topological changes within the interphase nucleus all appear to be involved in PEV-mediated gene silencing (Weiler and Wakimoto, 1995; Henikoff, 1997).
Genetic dissection of these processes has been performed by isolation of a large number of dominant modifiers of PEV mutations (Reuter and Wolff, 1981; Sinclair et al., 1983). Altogether, more than 150 modifier genes have been identified and mapped. Molecular analysis has proved that several of these genes encode structural or regulatory components of chromatin (Reuter and Spierer, 1992; Wallrath, 1998). Genetic analysis revealed dosage-dependent effects for PEV modifier genes. All dominant Su(var) and E(var) loci studied to date display haplo-dependent effects, but only a subset are connected with triplo opposite effects (Locke et al., 1988; Wustmann et al., 1989). Three genes with a haplo-suppressor and triplo-enhancer effect have been molecularly characterized and shown to encode proteins enriched in heterochromatic regions. Su(var)2–5 encodes the heterochromatin protein HP1 (James and Elgin, 1986; Eissenberg et al., 1990), which interacts with several other chromatin proteins and therefore might function as a linker module in heterochromatin protein complexes (Wang et al., 2000). Su(var)3–7 encodes a protein that contains seven widely spaced zinc fingers, interacts with HP1 and has the potential to bind DNA (Cleard et al., 1997; Delattre et al., 2000). Association of SU(VAR)3–9 with heterochromatic regions in Drosophila was recently demonstrated (Schotta and Reuter, 2000). The SU(VAR)3–9 protein contains the chromo and SET domains, which are two evolutionarily conserved modules also found in several other chromatin proteins (Jenuwein et al., 1998; Jones et al., 2000). Recently, a site-specific in vitro histone H3-K9 methyltransferase activity of the SET domain of mammalian SUV39H1 has been demonstrated (Rea et al., 2000). A similar function was shown for the fission yeast homologue Clr4p (Nakayama et al., 2001) and for Drosophila SU(VAR)3–9 (Czermin et al., 2001). This function of SU(VAR)3–9 proteins was suggested to impart heterochromatin association of SU(VAR)3–9 protein complexes as an essential prerequisite for gene silencing (Lachner et al., 2001; Nakayama et al., 2001).
Here we show that SU(VAR)3–9 controls heterochromatin-dependent gene silencing by histone H3-K9 methylation within chromocentre heterochromatin. SU(VAR)3–9 associates with heterochromatin through direct interactions with HP1. Localization of SU(VAR)3–9 and HP1 to heterochromatin are interdependent. In HP1 null mutants SU(VAR)3–9 not only binds to heterochromatin but also disperses across euchromatic regions and causes ectopic H3-K9 methylation. HP1 in Su(var)3–9 null mutants is nearly lost from chromocentre heterochromatin. Despite this interdependence, Su(var)3–9 mutations dominate the PEV modifier effect of extra copies of Su(var)2–5 and Su(var)3–7. Thus, the heterochromatin specific H3-K9 histone methyltransferase (HMTase) activity of SU(VAR)3–9 plays a central role in heterochromatin association of SU(VAR)3–9–HP1 complexes and their function in gene silencing.
Results
SU(VAR)3–9 associates with chromocentre heterochromatin
After fixation heterochromatic regions usually appear fuzzy in structure and are composed of numerous attenuated strands. In contrast, confocal microscopy of unfixed nuclei expressing enhanced green fluorescent protein (EGFP) fusions of heterochromatin-associated proteins demonstrates that chromocentre heterochromatin represents a rather solid and large structure (Schotta and Reuter, 2000). In our studies these techniques were used to analyse in vivo distribution of wild-type and mutant SU(VAR)3–9 proteins in wild-type and null mutant background.
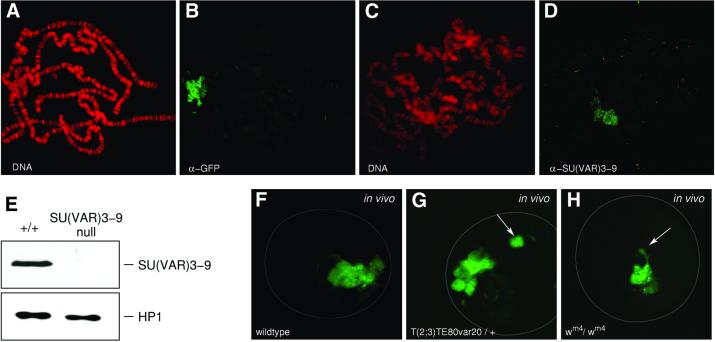
Chromosomal distribution of SU(VAR)3–9 has been studied in transgenic lines expressing an EGFP fusion of the full-length cDNA [SU(VAR)3–9–EGFP]. The transgenes used rescue Su(var)3–9 mutations (data not shown). In unfixed polytene nuclei the fusion protein is detected by GFP fluorescence in chromocentre heterochromatin and along the fourth chromosome (Schotta and Reuter, 2000). Immunostaining of larval salivary gland polytene chromosomes using an anti-GFP antibody furthermore revealed association of SU(VAR)3–9–EGFP with telomeres and a few euchromatic bands (Figure 1A and B). With a SU(VAR)3–9-specific polyclonal antibody (Figure 1E), association of the endogenous SU(VAR)3–9 protein with chromocentre heterochromatin and the fourth chromosome is seen (Figure 1C and D). However, only very weak signals are detected for endogenous SU(VAR)3–9 at telomeres and the few euchromatic sites, which could be due to higher concentrations of the fusion protein in transgenic lines or to stronger affinity of the GFP antibody.
Fig. 1. Distribution of the SU(VAR)3–9 protein in salivary gland polytene chromosomes. (A and B) Immunostaining of polytene chromosomes from a pP{GS[ry+, hs (Su(var)3–9 cDNA)–EGFP]} transgenic line with a rabbit polyclonal α-GFP antibody. (C and D) Heterochromatin association of endogenous SU(VAR)3–9 after staining with a rabbit polyclonal α-SU(VAR)3–9 antibody. DNA staining with propidium iodide (A and C) and immunostaining of the same nuclei, respectively (B and D). (E) Western blot of nuclear extracts from wild-type and Su(var)3–9 null mutant embryos. Blot probed with α-SU(VAR)3–9 antibody (upper panel) and α-HP1 antibody (lower panel). (F–H) Confocal sections of salivary gland nuclei showing in vivo distribution of SU(VAR)3–9–EGFP in wild-type (F), T(2;3)TE80var20/+; pP{GS[ry+, hs (Su(var)3–9 cDNA)–EGFP]} (G) and _w_m4/_w_m4; pP{GS[ry+, hs (Su(var)3–9 cDNA)–EGFP]} (H) larvae. The chromosomal rearrangements relocate regions of heterochromatin to distal euchromatin (arrows).
To determine whether SU(VAR)3–9–EGFP binds to different blocks of heterochromatin, we studied 21 PEV rearrangements with breakpoints in Xh, 2Lh, 2Rh, 3Lh and 3Rh (Materials and methods). For each rearrangement, unfixed salivary gland nuclei heterozygous for the P{GS[ry+, hs (Su(var)3–9 cDNA)–EGFP]} transgene were analysed by confocal microscopy. The rearrangements relocate blocks of pericentric heterochromatin to distal euchromatic regions. In all 19 autosomal rearrangements studied, an additional SU(VAR)3–9–EGFP site was detected (Figure 1G). In nuclei homozygous for the X chromosomal inversions _w_m4 and _w_m51b the rearranged heterochromatic regions remain associated with chromocentre heterochromatin and threads of heterochromatic material stained with SU(VAR)3–9–EGFP are detected (Figure 1H).
SU(VAR)3–9 causes histone H3-K9 methylation in chromocentre heterochromatin
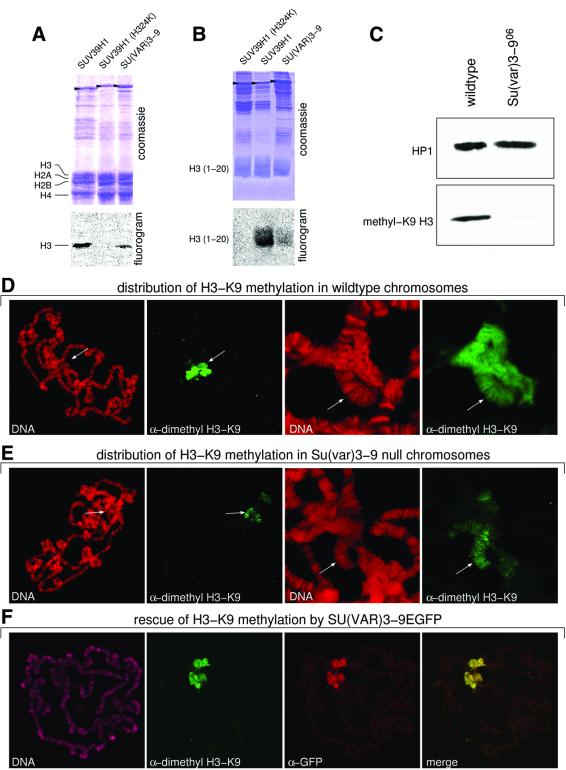
Drosophila SU(VAR)3–9, like its human orthologue SUV39H1 (Rea et al., 2000), specifically causes H3-K9 methylation. HMTase activity of SU(VAR)3–9 was shown after immunoaffinity purification of SU(VAR)3–9 from Drosophila embryo extracts (Czermin et al., 2001) and is confirmed by in vitro assays with the recombinant SU(VAR)3–9 protein (Figure 2A and B). Western analysis of nuclear extracts of wild-type and homozygous _Su(var)3–9_06 mutant embryos with an dimethyl H3-K9 specific antibody revealed strong reduction of H3-K9 methylation in the Su(var)3–9 null mutant embryos (Figure 2C). The small amount of K9-methylated H3 histones that is detected in null mutant embryos suggests that SU(VAR)3–9 is the main but not the only H3-K9 HMTase in Drosophila. Immunocytological analysis of wild-type salivary gland chromosomes with the same antibody shows strong staining of chromocentral heterochromatin and the fourth chromosome (Figure 2D). In larvae homozygous for the _Su(var)3–9_06 null mutant, staining of chromocentre heterochromatin is strongly reduced, whereas staining of the fourth chromosome appears not to be affected (Figure 2E). This demon strates that SU(VAR)3–9-dependent H3-K9 methylation is mainly limited to chromocentre heterochromatin. Histone H3-K9 methylation in chromocentre heterochromatin is re-established in _Su(var)3–9_06 null mutant larvae that carry the pP{GS[ry+, hs(Su(var)3–9 cDNA)–EGFP]} transgene after a 5 min heat-shock treatment (Figure 2F). These results indicate that suppression of PEV by _Su(var)3–9_06 and reduction of H3-K9 methylation within chromocentre heterochromatin are correlated.
Fig. 2. Histone H3-K9 methylation in heterochromatin is controlled by SU(VAR)3–9. (A and B) H3-K9 HMTase activity of recombinant SU(VAR)3–9 protein for histone H3 (A) and N-terminal peptide (1–20) of histone H3 (B) compared with wild-type and mutant (H324L) human SUV39H1. Arrowheads mark the recombinant proteins. (C) Western analysis of nuclear extracts from wild-type and Su(var)3–9 null mutant [_Su(var)3–9_06/_Su(var)3–9_06] embryos with an α-dimethyl H3-K9 antibody. (D) Immunostaining of wild-type larval salivary gland chromosomes with α-dimethyl H3-K9 antibody. Whole chromosome set and enlargement of chromocentre region. Arrows denote the fourth chromosome. (E) Immuno staining of SU(VAR)3–9-deficient [_Su(var)3–9_06/_Su(var)3–9_06] larvae with α-dimethyl H3-K9 antibody. Arrows in enlarged chromocentre denotes the fourth chromosome. (F) Reconstitution of H3-K9 methylation in chromocentre heterochromatin by expression of SU(VAR)3–9–EGFP in Su(var)3–9 null mutant larvae. DNA staining with propidium iodide (D and E) and TOTO-3 (F).
The N-terminus of SU(VAR)3–9 interacts with the chromo-shadow domain of HP1 and the C-terminal region of SU(VAR)3–7
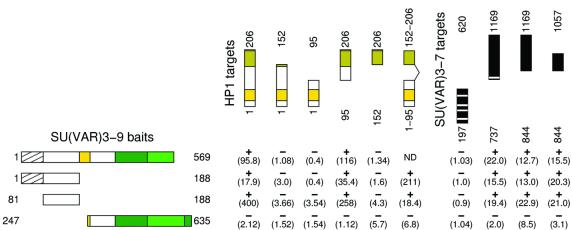
Putative SU(VAR)3–9 interacting proteins were isolated in yeast two-hybrid screens using a cDNA library of 21-h-old Drosophila embryos. The Su(var)3–9 bait construct contains amino acids 1–569, because the full-length Su(var)3–9 cDNA (635 amino acids) causes strong activation of the lacZ and LEU2 reporter genes. Of 350 000 yeast transformants scored, 2500 showed activation in the assay on XGal medium but only ∼300 clones caused galactose-dependent activation of the lacZ reporter. Of these clones, 86 also activated LEU2 and 23 clones could be confirmed as positives after retransformation into EGY48 cells containing the SU(VAR)3–9 bait. In 12 cases HP1 cDNAs were identified by sequence analysis. The remaining clones are currently being subjected to further analysis.
Domains of interaction between SU(VAR)3–9 and HP1 were identified in two-hybrid tests with a series of SU(VAR)3–9 and HP1 bait and prey constructs (Figure 3). The SU(VAR)3–9 N-terminus (amino acids 81–188), excluding the overlapping 81 amino acid region with eIF2γ (Krauss and Reuter, 2000), interacts with the HP1 chromo-shadow domain.
Fig. 3. Yeast two-hybrid analysis of SU(VAR)3–9 interaction with HP1 and SU(VAR)3–7. Two-hybrid interaction tests of SU(VAR)3–9 baits with HP1 and SU(VAR)3–7 target constructs. LacZ activities are shown in parentheses. SU(VAR)3–9 regions are indicated, shaded for the overlapping 81 amino acid region with eIF2γ: yellow for the chromodomain, dark green for the SAC domain and green for the SET domain. In HP1 the chromodomain is indicated in yellow and the chromo-shadow domain in amber. Zinc fingers in SU(VAR)3–7 are indicated by white stripes.
The Drosophila SU(VAR)3–7 heterochromatin protein has been shown to interact with HP1 (Delattre et al., 2000). SU(VAR)3–9 bait constructs and SU(VAR)3–7 prey constructs were tested for interaction. In SU(VAR)3–9 the same region (amino acids 81–188) found to interact with HP1 also showed interaction with SU(VAR)3–7, indicating that this region of SU(VAR)3–9 is an interaction target for both HP1 and SU(VAR)3–7. The region in SU(VAR)3–7 that interacts with SU(VAR)3–9 is situated C-terminal to the zinc fingers (Figure 3).
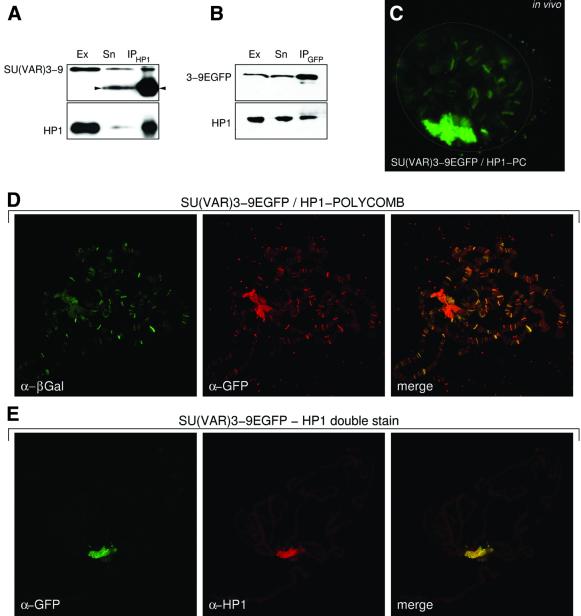
To determine whether there was evidence for the interaction of SU(VAR)3–9 and HP1 in a protein complex in vivo, nuclear extracts of 0–12 h wild-type or SU(VAR)3–9–EGFP transgenic embryos were immunoprecipitated with an HP1 or a GFP antibody, respectively (Figure 4A and B). Fractions retained either by the HP1 (Figure 4A) or GFP (Figure 4B) antibody and the supernatants were analysed by western blotting with HP1 and either SU(VAR)3–9 or GFP-specific antibodies. The antibodies detect the corresponding proteins in the anti-HP1 or the anti-GFP precipitate.
Fig. 4. In vivo and cytological analysis of interaction between SU(VAR)3–9 and HP1. (A and B) Immunoprecipitation of SU(VAR)3–9–EGFP (A) and HP1 (B) from heat-shocked transgenic and untreated wild-type 0–12 h embryos, respectively. Blots were probed with (A) α-SU(VAR)3–9, and (B) α-GFP (upper panels) and α-HP1 (lower panels). Ex, nuclear extract; Sn, supernatant; IP, immunoprecipitate. Arrowheads mark the signal of the antibody used for co-immunoprecipitation. (C) In vivo relocation of SU(VAR)3–9–EGFP by an HP1/PC chimeric protein. GFP fluorescence in a confocal section of a salivary gland nucleus. (D) Immunocytological analysis of transheterozygous larvae expressing SU(VAR)3–9–EGFP and the HP1/PC chimeric protein with α-GFP for SU(VAR)3–9–EGFP and α-β-gal for HP1/PC. (E) Double immunostaining for SU(VAR)3–9–EGFP with a mouse monoclonal α-GFP antibody and with a rabbit polyclonal α-HP1 antibody.
In vivo interaction of SU(VAR)3–9 with HP1 was also tested with a chimeric HP1/PC protein. This chimeric HP1 protein contains the chromodomain of the POLYCOMB (PC) protein and relocates endogenous HP1 to PC-binding sites (Platero et al., 1996). Consequently, HP1-interacting proteins should also be relocated to PC binding sites. We constructed genotypes expressing both the HP1/PC chimeric and the SU(VAR)3–9–EGFP fusion protein under heat-shock control. In salivary gland nuclei of these larvae, SU(VAR)3–9–EGFP decorates many euchromatic sites (Figure 4C). These sites correspond to binding sites of the HP1/PC chimeric protein as shown by double staining with GFP antibody for SU(VAR) 3–9–EGFP and β-galactosidase (β-gal) antibody, which detects the β-gal-tagged HP1/PC protein (Figure 4D). Co-localization of both SU(VAR)3–9 and HP1 represents further support for a direct physical interaction (Figure 4E).
Multiple control of SU(VAR)3–9 heterochromatin association
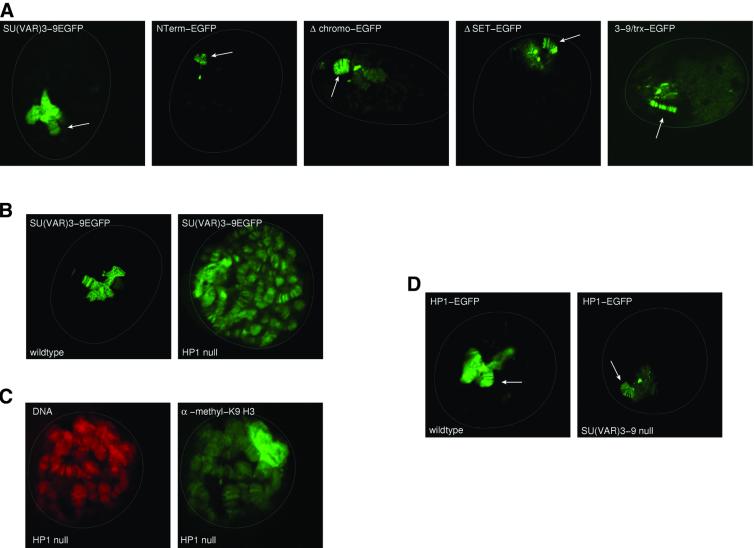
We studied different regions of SU(VAR)3–9 for their possible function in control of heterochromatin association by in vivo analysis of EGFP-tagged protein variants in larvae deficient for the endogenous protein. The N-terminus of SU(VAR)3–9, which interacts with HP1 and SU(VAR)3–7, might also play an important role in its heterochromatin association. An EGFP fusion construct including amino acids 1–188 (NTerm–EGFP) was transformed into Drosophila and its in vivo chromosomal distribution was studied in salivary gland nuclei of _Su(var)3–9_06 homozygous larvae. In vivo analysis of unfixed polytene nuclei showed small amounts of the NTerm–EGFP protein in chromocentre heterochromatin, weak GFP fluorescence in nucleoplasm, but almost normal association with the fourth chromosome (Figure 5A), suggesting that the N-terminus of SU(VAR)3–9 is not sufficient for normal association of the protein.
Fig. 5. Control of SU(VAR)3–9 and HP1 heterochromatin association. (A) Confocal sections of unfixed larval salivary gland nuclei expressing full-length SU(VAR)3–9–EGFP, SU(VAR)3–9NTerm–EGFP, SU(VAR)3–9Δchromo–EGFP, SU(VAR)3–9ΔSET–EGFP and SU(VAR)3–9TRX–EGFP protein variants in SU(VAR)3–9-deficient larvae. Arrows point to the fourth chromosome. (B) In vivo distribution of SU(VAR)3–9–EGFP in wild-type and HP1 deficient [Df(2L)TE128 × 11/_Su(var)2–5_04] salivary gland nuclei. (C) α-dimethyl H3-K9 antibody staining of an HP1-deficient salivary gland nucleus. DNA staining with propidium iodide. (D) In vivo analysis of HP1–EGFP heterochromatin association in wild-type and SU(VAR)3–9-deficient [_Su(var)3–9_06/_Su(var)3–9_06] nuclei of third instar larval salivary glands. Arrows point to the fourth chromosome.
In order to test for a possible role of the chromodomain in heterochromatin association, we constructed transgenic lines expressing an EGFP-tagged SU(VAR)3–9 with a chromodomain deletion (Δchromo–EGFP). This fusion protein also shows strongly reduced in vivo staining of chromocentre heterochromatin but normal association with the fourth chromosome, demonstrating that the chromodomain is also essential for normal heterochromatin association of SU(VAR)3–9.
The effects of SET domain mutations on SU(VAR)3–9 chromosomal distribution were studied with different EGFP fusion constructs. We introduced SU(VAR) 3–9–EGFP proteins where the SET domain was either deleted or replaced with the SET domain of the TRITHORAX (TRX) protein. ΔSET deletes the complete SET domain (amino acids 484–635) but not the cysteine-rich region. In SU(VAR)3–9TRX–EGFP, amino acids 484–635 were replaced with the SET domain of the TRX protein (amino acids 3586–3726). All the SET domain mutant proteins differ from wild-type SU(VAR)3–9 in their in vivo distribution within chromocentre heterochromatin. The mutant proteins only bind to the middle of chromocentre heterochromatin (Figure 5A), but association with the fourth chromosome is not affected. NTerm–EGFP, Δchromo–EGFP and the mutant SET– EGFP fusion proteins interact in vivo with HP1 and associate with numerous euchromatic sites when co-expressed with an HP1/PC chimeric protein (data not shown). Our data show that the N-terminus, the chromodomain and the SET domain are all essential for normal association of SU(VAR)3–9 with the chromocentre heterochromatin. In contrast, the N-terminus is sufficient for association of SU(VAR)3–9 with the fourth chromosome.
Heterochromatin association of SU(VAR)3–9 and HP1 are interdependent
The role of HP1 in heterochromatin association of SU(VAR)3–9 was studied in HP1-deficient third instar larvae expressing SU(VAR)3–9–EGFP. HP1-deficient larvae were constructed by combining a deletion of Su(var)2–5 with a null mutant allele (see Materials and methods). These trans-heterozygotes survive up to third larval instar and in salivary gland nuclei of these larvae no HP1 staining is detected (Fanti et al., 1998). In these larvae, SU(VAR)3–9–EGFP still remains within heterochromatin and associates with the fourth chromosome, but in addition becomes associated with euchromatic regions along all chromosomes (Figure 5B). This shows that HP1 is essential for maintaining restricted binding of SU(VAR)3–9 to heterochromatin. Immunocytological analysis of HP1 null nuclei demonstrates that ectopic binding of SU(VAR)3–9–EGFP causes H3-K9 methylation along euchromatic regions (Figure 5C).
When we analysed the distribution of HP1–EGFP in homozygous loss-of-function _Su(var)3–9_06 larvae, its association with chromocentre heterochromatin was dramatically reduced (Figure 5D). In contrast to wild type, in SU(VAR)3–9-deficient larvae the HP1–EGFP protein only remains concentrated in the middle of the chromocentre. GFP background staining in the nucleus is seen by UV microscopy (data not shown), indicating that the unbound HP1 might become dispersed. Association of HP1–EGFP with the fourth chromosome is not affected in Su(var)3–9 homozygous mutant larvae, indicating that different regulatory processes are involved in control of HP1 binding.
Su(var)3–9 dominates the PEV modifier effects of Su(var)2–5, Su(var)3–7 and of Y chromosome aneuploidy
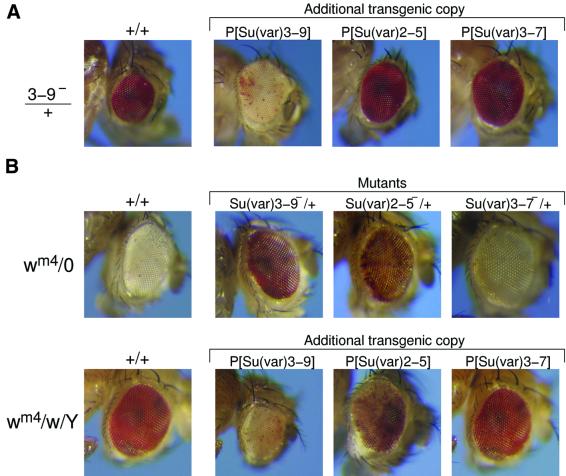
Su(var)3–9, Su(var)3–7 and Su(var)2–5 display a haplo-suppressor/triplo-enhancer dosage-dependent effect on PEV. Additional genomic copies of all three genes cause strong enhancement of white variegation in _w_m4 and genetic interactions between the genes were tested by combining Su(var)3–9 null mutants with additional genomic copies of Su(var)2–5 and Su(var)3–7. In all these combinations the suppressor effect of Su(var)3–9 null mutations dominates the triplo-dependent enhancer effects of Su(var)2–5 and Su(var)3–7 (Figure 6A).
Fig. 6. Genetic interactions between Su(var)3–9, Su(var)2–5 and Su(var)3–7 and the modifier effect of Y chromosome aneuploidy in _w_m4 PEV. (A) Heterozygotes for the loss-of-function mutant alleles _Su(var)3–9_06, _Su(var)2–5_05 and _Df(3R)AceD1_[Su(var)3–7 null] were combined with transgenic constructs introducing extra genomic copies of Su(var)3–9, Su(var)2–5 or Su(var)3–7, respectively. Resultant white variegation in offspring _w_m4 female is shown. (B) Genetic interaction to the PEV modifier effect of Y chromosome aneuploidy. First row shows white variegation in _w_m4/O males of +/+ (control) and of heterozygotes of the loss-of-function _Su(var)3–9_06, _Su(var)2–5_05 and _Df(3R)AceHD1_[Su(var)3–7 null] alleles, respectively. Second row shows white variegation in _w_m4/w/Y females of +/+ (control) and of heterozygotes with an additional genomic copy of Su(var)3–9, Su(var)2–5 and Su(var)3–7, respectively.
Modification of variegation by Y chromosome aneuploidy is a general diagnostic feature of PEV in Drosophila. Addition of Y chromosome suppresses PEV, whereas its loss enhances PEV (Figure 6B). In _w_m4/XY, _wm_2 (XXY) females, strong suppression of white variegation results in a wild-type red eye phenotype. An extra dose of the Su(var)3–9 gene dominates the Y chromosome suppressor effect. An extra Su(var)2–5 copy shows a co-operative effect and results in a variegated eye phenotype, whereas the suppressor effect of the extra Y chromosome is epistatic to the enhancer effect of extra Su(var)3–7 copies. In a complementary fashion Su(var)3–9 mutations dominate the strong enhancer effect caused by loss of the Y chromosome, as shown in _w_m4/0 males (Figure 6B). The _Su(var)2–5_05 null mutation causes a variegated phenotype, whereas the Y-dependent enhancer effect dominates the suppressor effect of the Su(var)3–7 deletion Df(3R)Ace HD1. These genetic interactions indicate a central functional role of SU(VAR)3–9 in heterochromatin-induced white gene silencing in _w_m4. In agreement with the genetic data, SU(VAR)3–9 appears to be associated with the heterochromatic breakpoints in all the 21 PEV rearrangements studied.
Human SUV39H1 partially rescues Su(var)3–9 silencing defects in Drosophila
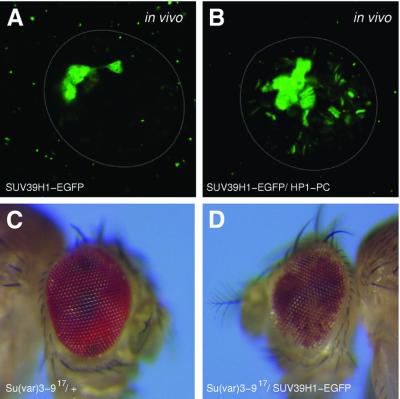
SU(VAR)3–9 is structurally conserved from fission yeast (Ivanova et al., 1998) to human (Aagaard et al., 1999). Functional conservation of human SUV39H1 was tested in transgenic Drosophila lines that express SUV39H1–EGFP under the control of the hsp70 heat-shock promoter. SUV39H1–EGFP is heterochromatin-associated in salivary gland nuclei (Figure 7A). In vivo interaction of SUV39H1 with HP1 is indicated by its association with euchromatic sites if co-expressed with the HP1/PC chimeric protein (Figure 7B). This is also supported by yeast two-hybrid data, where we found interaction between Drosophila HP1 and human SUV39H1 (data not shown). SUV39H1 enhances white gene silencing in _w_m4 (Aagaard et al., 1999). After remobilization of the SUV39H1–EGFP a derivative transgenic line was isolated that shows partial rescue of the dominant suppressor effect of Su(var)3–9 mutations (Figure 7D). These data suggest strong functional conservation between Drosophila and human SU(VAR)3–9 proteins.
Fig. 7. Expression of human SUV39H1 in Drosophila. (A) Association of SUV39H1–EGFP with chromocentre heterochromatin and the fourth chromosome. In vivo confocal section of a salivary gland nucleus. (B) Relocation of SUV39H1–EGFP to euchromatic sites in transgenic lines with HP1/PC chimeric protein. (C and D) SUV39H1 rescues the dominant suppressor effect of Su(var)3–9 mutations: (C) control _w_m4/Y; _Su(var)3–9_06/+ and (D) _w_m4/Y; _Su(var)3–9_06/pP{GS[ry+, hs cDNA SUV39H1–EGFP]} males.
Discussion
Gene silencing in PEV and heterochromatin-specific H3-K9 methylation by SU(VAR)3–9
Genetic dissection of PEV in Drosophila resulted in identification of new functions causally connected with control of higher order chromatin structure in heterochromatin (Reuter and Spierer, 1992). The heterochromatin-associated proteins encoded by Su(var)2–5, Su(var)3–7 and Su(var)3–9 have been intensively studied. Recent demonstration of in vitro histone H3 methyltransferase activity of SUV39H1 (Rea et al., 2000), the human homologue of Drosophila SU(VAR)3–9, has provided new insights into the molecular mechanisms connecting changes in higher order chromatin structure and gene silencing with the histone code of epigenetic programming (Jenuwein and Allis, 2001). We have shown that in Drosophila, histone H3-K9 methylation is strongly enriched in chromocentre heterochromatin and the fourth chromosome. Immunocytological studies revealed that SU(VAR)3–9 preferentially causes H3-K9 methylation within chromocentre heterochromatin. Although these results suggest a significant role of H3-K9 methylation in altering chromatin structure and gene activity during development, further studies are required to understand how integral components of higher order chromatin complexes in heterochromatin are assembled and their function is regulated.
Heterochromatin association of SU(VAR)3–9 and HP1 is interdependent
SU(VAR)3–9 and HP1 represent evolutionarily conserved components of heterochromatin protein complexes. Interaction between the two proteins has previously been suggested for the mammalian homologues (Aagaard et al., 1999). In Drosophila, the N-terminus of SU(VAR)3–9 and the chromo-shadow domain region of HP1 constitute the sites of their interaction. Ectopic association of SU(VAR)3–9–EGFP along euchromatic regions in HP1-deficient salivary gland nuclei and strongly reduced binding of HP1–EGFP to chromocentre heterochromatin in SU(VAR)3–9-deficient nuclei suggest that interaction between both proteins is essential for their association with chromocentre heterochromatin. Recently, it was shown that H3-K9 methylation creates chromodomain-dependent binding sites of HP1 (Bannister et al., 2001; Lachner et al., 2001). Strong reduction of HP1–EGFP heterochromatin binding in SU(VAR)3–9 null mutants might reflect a requirement of HP1 binding to methylated H3-K9 for heterochromatin-association of SU(VAR)3–9–HP1 complexes. These results suggest a multipstep control for heterochromatin association of SU(VAR)3–9–HP1 complexes. After primary association of SU(VAR)3–9 with heterochromatin, consecutive H3-K9 methylation by SU(VAR)3–9 would create binding sites of HP1, which finally results in stable association of SU(VAR)3–9–HP1 complexes with heterochromatin. These processes are likely to be controlled by several other as yet unknown factors. In these processes the chromodomain as well as the SET domain of SU(VAR)3–9 might be directly involved. Fusion proteins deleting either the chromodomain or the SET domain only show restricted binding to heterochromatin.
Although SU(VAR)3–9 associates with the fourth chromosome, H3-K9 methylation in the fourth chromosome is not changed in Su(var)3–9 null mutants, suggesting that H3-K9 methylation in this chromosome is controlled by a different HMTase activity. In contrast to SU(VAR)3–9 association with chromocentre heterochromatin, which depends on the chromodomain and the SET domain, for its binding to the fourth chromosome the N-terminus is sufficient. A special chromatin structure of the fourth chromosome is also indicated by identification of POF, a chromosome four-specific protein (Larsson et al., 2001). Different requirements of SU(VAR)3–9 and HP1 association with the fourth chromosome and chromocentre heterochromatin suggest occurrence of heterochromatin protein complexes of different composition, as well as differential control of their assembly.
Functional domains of SU(VAR)3–9 and the control of gene silencing
Structure–function analysis with transgenic SU(VAR) 3–9–EGFP protein variants revealed new aspects of their role in heterochromatin localization of SU(VAR)3–9. In contrast to studies with human SUV39H1 (Melcher et al., 2000) we analysed in vivo heterochromatin association of SU(VAR)3–9–EGFP protein variants in nuclei deficient for the endogenous SU(VAR)3–9 protein. The N-terminus of SU(VAR)3–9 (amino acids 81–188), which contains the interaction domain to HP1 and SU(VAR)3–7, is involved in heterochromatin association of the protein. However, association of the truncated protein is restricted to the fourth chromosome and the central region of chromocentre heterochromatin. Deletion of the chromodomain in SU(VAR)3–9 also affects its normal chromosomal distribution and reduces binding to chromocentre heterochromatin, but not with the fourth chromosome. In contrast, deletion or point mutations of the chromodomain result in ectopic distribution of human SUV39H1 in HeLa cells (Melcher et al., 2000). These findings might indicate functional differences between the SU(VAR)3–9 and SUV39H1 chromodomain. In both SU(VAR)3–9 and SUV39H1, the N-terminus contains the interaction surface for HP1 and HP1β, respectively. In human cells, overexpression of SUV39H1 results in ectopic chromosomal distribution (Melcher et al., 2000). In contrast, even after strong overexpression of Su(var)3–9 no comparable effects were observed in Drosophila.
Deletion of the SET domain or an exchange of the SU(VAR)3–9 SET domain with the SET domain of the TRX protein strongly affects heterochromatin distribution of the proteins. The proteins become concentrated within the middle of chromocentre heterochromatin, but again show normal association with the fourth chromosome. This suggests that the SET domain of SU(VAR)3–9 is directly involved in the control of SU(VAR)3–9 association with chromocentre heterochromatin. In Drosophila, aberrant heterochromatin distribution of SU(VAR)3–9 proteins with SET domain mutations could be causally connected with suppression of heterochromatin-induced gene silencing. Comparable results have been obtained for clr4, the Schizosaccharomyces pombe homologue of Su(var)3–9, where mutations in the SET domain show defects in silencing and mating-type switching (Ivanova et al., 1998). However, in S.pombe swi6, the homologue of Su(var)2–5 was shown to represent the main dosage-dependent component of gene silencing at the mat2/3 locus, whereas only subtle effects of clr4 are reported (Nakayama et al., 2000). These functional differences observed for SU(VAR)3–9 and HP1 orthologues in fission yeast, Drosophila and mammals might reflect considerable functional and/or structural differences of the silencing complexes in these organisms.
Aberrant heterochromatin distribution of SU(VAR)3–9 SET domain mutant proteins suggests involvement of other factors in a functional control of the SET domain. These factors might also affect its HMTase activity. Mutations in genes encoding these putative regulatory genes should be genetically epistatic to the triplo-dependent enhancer effect of Su(var)3–9. Proteins like the SET domain-binding factor Sbf1 (Firestein et al., 2000), which has been shown to be involved in regulation of the phosphorylation state of the SET domain, might also play a central role. Identification of PEV enhancer mutations like _ptn_D that cause ectopic binding of SU(VAR)3–9 and HP1 to many euchromatic sites (Kuhfittig et al., 2001) indicates the existence of different positive as well as negative control mechanisms for chromosomal distribution of heterochromatin protein complexes. Further studies of modifiers of PEV mutations will contribute substantially to our understanding of the complex regulatory processes involved in the control of higher order chromatin structure and heterochromatin-induced gene silencing.
Materials and methods
PEV modifier mutations, transgenic lines and PEV rearrangements
Su(var)3–9 mutations were identified by their strong dominant suppressor effect on white variegation in _In(1)w_m4 (hereafter denoted as _w_m4). In our studies the X-ray-induced _Su(var)3–9_06 and the ethylmethanesulfonate (EMS)-induced _Su(var)3–9_17 alleles were used. Owing to an insertion of DNA in _Su(var)3–9_06 and an 8 bp deletion in _Su(var)3–9_17 resulting in a frame-shift at codon 93, both alleles represent amorphic mutations (Tschiersch et al., 1994). Su(var)2–5 mutations are described elsewhere (Wustmann et al., 1989; Eissenberg et al., 1990, 1992; Platero et al., 1996). Deficiency Df(3R)AceHD1 uncovers the Su(var)3–7 gene (Reuter et al., 1990), and this deletion was used to study the loss-of-function effect of Su(var)3–7 on white variegation in _w_m4.
Transgenic lines were produced by P element-mediated germline transformation (Rubin and Spradling, 1982). The genomic copy of Su(var)3–9 (P[(ry+) Su(var)3–9 11 kb]) is described in Tschiersch et al. (1994). The different P[(ry+) hs (Su(var)3–9 cDNA)] constructs were remobilized in crosses with TM3, _ry_RK Sb e P[(ry+) Δ2–3] (99B) containing a stable P transposase source (Reuter et al., 1993), and derivative lines were assayed for Su(var)3–9 mutant rescue.
The effects of Y aneuploidy on white variegation in _w_m4 were studied by crosses of C(1)M4, _y_2/0 females and _FM7_·Y, _wm_2/0 males. C(1)M4 is a compound X with _w_m4 + AB-FM7, _y w_m4 – _y_– _w_a _v_Of _sc_8 (Flybase, August 21, 2001). The effect of Su(var) mutations was studied in _w_m4/0; +/+, _w_m4/0; _Su(var)2–5_05/+, _w_m4/0; _Su(var)3–9_17/+, or _w_m4/0; Df(3R)AceHD1[Su(var)3–7_–_]/+ males. The enhancer effect of additional genomic copies of Su(var)3–9, Su(var)2–5 and Su(var)3–7 was tested in _w_m4/_FM7_·Y, _w m_2 and C(1)M4, _y_2/Y females.
EGFP fusion constructs
SU(VAR)3–9–EGFP constructs are based on pP{GS[(ry+) hsEGFP3′]} (Schotta and Reuter, 2000). Parts of Su(var)3–9 cDNA (Tschiersch et al., 1994) were amplified by PCR and cloned into this vector. The following constructs were made: NTerm (amino acids 1–206) with primer pair NTerm forward (5′-acagggcccgatatcgagatttgatgccg-3′) and NTerm back (5′-acagcggccgccgttatttttcgaccgcttggag-3′), Δchromo (missing amino acids 207–267) by inserting a PCR fragment containing amino acids 268–635 amplified with primer chromo– forward (5′-acagcggccgcgagcggcatcagcagctctac-3′) and c3-9 end into the NTerm construct and ΔSET (missing amino acids 484–653) with primer pair c3-9 start and SET– back (5′-acagcggccgcgagcggcatcagcagctctac-3′).
SUV39H1–EGFP was constructed by amplification of the SUV39H1 open reading frame with primer pair SUV39H1 forward (5′-acagcggccgcaacatggcggaaaatttaaaaggctg-3′) and SUV39H1 reverse (5′-acagcggccgccgaagaggtatttgcggcagg-3′) using a cDNA as template followed by cloning into the pP{GS[(ry+) hsEGFP3′]} vector.
HP1–EGFP was constructed by amplification of a full-length HP1 cDNA isolated in the yeast two-hybrid screen with SU(VAR)3–9 with primer pair HP1 cDNA forward (5′-atagcggccgcatgggcaagaaaatcgacaacc-3′) and HP1 cDNA reverse (5′-atagcggccgccatcttcattatcagagtac-3′), followed by cloning into pP{GS[(ry+) hsEGFP3′]}.
Analysis of SU(VAR)3–9 heterochromatin distribution
Nineteen autosomal PEV rearrangements with breakpoints in 2L, 2R, 3L or 3R heterochromatin were balanced over CyO GFP P(w+m hsp70: GAL4) P(w+m UAS:GFP) (Rudolph et al., 1999) and heterozygous males crossed to homozygous pP{GS[(ry+) hs (Su(var)3–9 cDNA)–EGFP]} females. Third instar larvae without the strong GFP fluorescence of CyO GFP identify the genotype with the PEV rearrangement and pP{GS[(ry+) hs Su(var)3–9 cDNA)–EGFP]}. New white mottled rearrangements were isolated after X-ray irradiation of the TE62, TE80, TE196 and TE301 elements (Ising et al., 1981). The nine new white mottled rearrangements have heterochromatic breakpoints in 2Lh [_In(2L)TE62var7_], in 2Rh [In(2LR)TE62var25, In(2LR)TE80var24 and _In(2LR)TE196var2_], in 3Lh [T(2;3)TE62var20 and _T(2;3)TE301var1_] and in 3Rh [T(2;3)TE62var18, T(2;3)TE80var35 and _T(2;3)TE80var40_]. The other PEV rearrangements studied are _In(2LR)bw_v1, _In(2LR)bw_v32g, In(2LR)Rev and _In(2L)lt_m12 with a break in 2Lh, _In(2R)bw_34k, _In(2R)bw_vDe1 and _In(2R)bw_vDe2 in 2Rh, _T(2;3)bw_v5 and _T(2;3)bw_vDe4 in 3Lh, and _T(2;3)bw_vDe3 in 3Rh (Flybase, 2001). For _In(2L)lt_m12, _In(2R)b_wvDe1 and _In(2R)bw_vDe2, the exact breakpoints were localized within heterochromatic blocks h37, h43 and h44/45, respectively (Dimitri, 1991). _In(1)w_m4 and _In(1)w_m51b have breakpoints in the X chromosomal h28 and h30 blocks of heterochromatin, respectively. Homozygous pP{GS[ry+, hs (Su(var)3–9 cDNA)–EGFP]} females were crossed to _In(1)w_m4/Y and _In(1)w_m51b/Y males, respectively. SU(VAR)3–9–EGFP distribution was analysed in third instar female larvae.
SU(VAR)3–9 null larvae expressing HP1–EGFP were of the genotype CyO/HP1–EGFP; _Su(var)3–9_06 e ro/_Su(var)3–9_06 e ro. HP1 null mutant Df(2L)TE128X11/_Su(var)2–5_04; SU(VAR)3–9–EGFP larvae were generated by a cross of CyO GFP/Df(2L)TE128X11; SU(VAR)3–9–EGFP × CyO GFP/_Su(var)2–5_04; SU(VAR)3–9–EGFP. The Df(2L)TE128X11 chromosome uncovers the Su(var)2–5 locus (Wustmann et al., 1989). In vivo analysis of EGFP fusion proteins was performed as described previously (Schotta and Reuter, 2000).
Immunostaining of polytene chromosomes
Preparation of polytene chromosomes was performed as described previously (Silver and Elgin, 1978) with the following modifications: salivary glands of third instar larvae were dissected in 0.7% NaCl, fixed for 10 min and squashed in 45% acetic acid/2% formaldehyde. Chromosomes were incubated with mouse monoclonal α-GFP (Babco; 1:50/5% dry milk), rabbit polyclonal α-GFP (Molecular Probes; 1:50/5% dry milk), mouse monoclonal α-β-gal (Sigma; 1:50/5% dry milk), rabbit polyclonal α-HP1 (gift from J.Eissenberg; 1:70/5% dry milk) or rabbit polyclonal α-SU(VAR)3–9 (1:25/5% dry milk) antibodies at 4°C overnight, followed by incubation with secondary FITC- or TR-conjugated antibodies (Jackson Immunoresearch Laboratories Inc.; 1:50/5% dry milk) for 2 h at 37°C.
For immunfluorescence analysis with the α-dimethyl H3-K9 (Upstate; 1:50/5% dry milk) and polyclonal SU(VAR)3–9 (1:20/5% dry milk) antibodies polytene chromosomes were prepared as described previously (Alfageme et al., 1980) with the following modifications: after dissection in gland medium (Cohen and Gotchel, 1971) the glands were fixed for 2 min in phosphate-buffered saline (PBS) containing 2% formaldehyde, followed by incubation for 3 min and squashing in 2% formaldehyde/45% acetic acid. The remaining steps took place as described above.
SU(VAR)3–9 specific rabbit antisera were raised against purified bacterial GST–SU(VAR)3–9 (amino acids 275–480) antigen and purified by pre-incubation with embryo extracts of Su(var)3–9 null mutant embryos. DNA was stained with propidium iodide or TOTO-3 (Molecular Probes). Preparations were mounted in Vectashield medium and examined with confocal laser scanning microscopy.
Immunostaining of whole salivary glands
Salivary glands from third instar larvae were dissected in gland medium (Cohen and Gotchel, 1971) and fixed for 15 min in PBS containing 3.7% formaldehyde/1% Triton X-100. The glands were washed in PBS/0.1% Triton X-100 and permeabilized for 1 h with PBS/1% Triton X-100. Incubation overnight at 4°C with α-dimethyl H3-K9 antibody (Upstate; 1:100 in PBS/0.1% Triton X-100) was followed by incubation with secondary FITC-conjugated antibody (Jackson Immunoresearch Laboratories Inc.; 1:100 in PBS/0.1% Triton X-100) for 2 h at 37°C. DNA was stained with propidium iodide (Molecular Probes). Preparations were examined with confocal laser scanning microscopy.
Yeast two-hybrid screens, constructs and interaction tests
Plasmid transformation in yeast strain EGY48 (Gyuris et al., 1993) was performed as described previously (Gietz et al., 1992). Expression of the target protein was induced on 2% galactose medium. Activation of the chromosomal LEU2 gene was tested on Ura–, His–, Trp–, Leu– medium, whereas activation of the lacZ marker on the pSH18–34 plasmid was tested on 2% XGal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), Ura–, His–, Trp– medium. Expression of bait proteins was confirmed by western blot analysis with a lexA antibody (Santa Cruz Biotechnology). Tests for activation of lacZ and LEU2 reporter genes were performed for all bait constructs. β-gal activity in units/min was calculated with 1000 × OD420/(OD600 × V × t), where V is the volume of the culture, t is the reaction time in minutes, OD420 is the optical density of _o_-nitrophenole and OD600 is the optical density of the culture (Samson et al., 1989).
SU(VAR)3–9 interacting preys were isolated from the embryonic Drosophila MATCHMAKER LexA cDNA library (Clontech). The truncated Su(var)3–9 cDNA (amino acids 1–569) bait construct had to be used in the two-hybrid screens because of strong activation of lacZ and LEU2 reporter genes by the full-length protein. This Su(var)3–9 bait construct was also used for screens with a human HeLa MATCHMAKER LexA cDNA library (Clontech).
SU(VAR)3–9 bait and target plasmid constructs were generated by PCR-mediated cloning of different regions from a full-length Su(var)3–9 cDNA (Tschiersch et al., 1994) into vectors pEG202 and pJG4-5 (kindly provided by R.Brent). Primers for amplification were _Eco_90 (5′-atcgaattcatggccacggctgaagcc-3′), _Eco_330 (5′-ttggaattcgcagaacgcttgagcg-3′), _Eco_800 (5′-ggataccatgaattcgagaacacctggga-3′), _Xho_1790 (5′-gaacacctcgagattaggatcgcaagagtgatt-3′) and _Bam_2000 (5′-ttcggatccattcaaaagaggacctttc-3′). The SU(VAR)3–9 constructs were generated with the following primer pairs: 1–569 with _Eco_90 and _Xho_1790, 81–635 with _Eco_330 and _Bam_2000, and 247–635 with _Eco_800 and _Bam_2000. The bait constructs 1–188 and 81–188 were prepared by a _Sal_I self-ligation of clones 1–569 and 81–635, respectively. HP1 and SU(VAR)3–7 bait and target constructs are described in Delattre et al. (2000), and were kindly provided by P.Spierer.
In vitro histone methyltransferase assay
The generation and purification of GST fusion proteins and histone methyltransferase assays were performed as described previously (Rea et al., 2000). GST constructs used were human SUV39H1 (H342K) amino acids 82–412, human SUV39H1 amino acids 82–412 (Rea et al., 2000) and Drosophila SU(VAR)3–9 amino acids 305–635.
Co-immunprecipitation of SU(VAR)3–9–EGFP and HP1
Nuclei of wild-type embryos and heat-shocked embryos expressing SU(VAR)3–9–EGFP were prepared as described previously (Elgin and Hood, 1973) and resuspended in immunoprecipitation (IP) buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM NaF, 10 mM β-glycerophosphate). After sonication and centrifugation protein concentration of extracts was determined using standard Bradford assays. Equal protein amounts were used for overnight incubation at 4°C with 1 µg polyclonal α-GFP antibody (Molecular Probes) or 1 µg polyclonal HP1 antibody (kindly provided by J.Eissenberg) followed by incubation with 20 µl protein G beads (Roche) for 3 h at 4°C. Beads were washed three times with IP buffer and proteins were eluted by boiling in SDS loading buffer. Nuclear extracts, supernatants from overnight incubation and immunoprecipitated proteins were loaded on two separate gels, and western blots were probed with α-GFP, α-SU(VAR)3–9 and α-HP1 antibodies.
Western analysis of H3-K9 methylation in Drosophila embryo extracts
Nuclear extracts of wild-type and Su(var)3–9 null embryos (0–12 h) were prepared as described above. The same protein amounts (20 µg) were loaded on two different gels and western blots were probed with an α-HP1 and an α-dimethyl H3-K9 antibody (Upstate), respectively.
Acknowledgments
Acknowledgements
We thank Drs B.Wakimoto and A.Hilliker for helpful comments on the manuscript. We are grateful to Dr P.Spierer for bait clones of SU(VAR)3–7 and Dr J.Eissenberg for HP1 antibodies and Drosophila strains. Yeast strains and all basic yeast plasmids for the two-hybrid system were generously provided by Dr R.Brent. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) to G.R.
References
- Aagaard L. et al. (1999) Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J., 18, 1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfageme C.R., Rudkin,G.T. and Cohen,L.H. (1980) Isolation, properties and cellular distribution of D1, a chromosomal protein of Drosophila. Chromosoma, 78, 1–31. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Zegermann,P., Patridge,J.F., Miska,E.A., Thomas,J.O., Allshire,T.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Boivin A. and Dura,J.M. (1998) In vivo chromatin accessibility correlates with gene silencing in Drosophila. Genetics, 150, 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleard F., Delattre,M. and Spierer,P. (1997) SU(VAR)3–7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation. EMBO J., 16, 5280–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen L.H. and Gotchel,B.V. (1971) Histones of polytene and nonpolytene nuclei of Drosophila melanogaster. J. Biol. Chem., 246, 1841–1848. [PubMed] [Google Scholar]
- Czermin B., Schotta,G., Hulsmann,B.B., Brehm,A., Becker,P.B., Reuter,G. and Imhof,A. (2001) Physical and functional association of SU(VAR)3–9 and HDAC1 in Drosophila. EMBO rep., 2, 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M., Spierer,A., Tonka,C.-H. and Spierer,P. (2000) The genomic silencing of position-effect variegation in Drosophila melanogaster: Interaction between the heterochromatin-associated proteins Su(var)3–7 and HP1. J. Cell Sci., 113, 4253–4261. [DOI] [PubMed] [Google Scholar]
- Dimitri P. (1991) Cytogenetic analysis of the second chromosome heterochromatin of Drosophila melanogaster. Genetics, 127, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J.C., James,T., Foster-Hartnett,D., Hartnett,T., Ngan,V. and Elgin,S.C.R. (1990) Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 87, 9923–9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J.C., Morris,G.D., Reuter,G. and Hartnett,T. (1992) The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics, 131, 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S.C.R. and Hood,L.E. (1973) Chromosomal proteins of Drosophila embryos. Biochemistry, 12, 4984–4991. [DOI] [PubMed] [Google Scholar]
- Fanti L., Giovinazzo,G., Berloco,M. and Pimpinelli,S. (1998) The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell, 2, 527–538. [DOI] [PubMed] [Google Scholar]
- Firestein R., Cui,X., Huie,P. and Cleary,M.L. (2000) Set domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3–9. Mol. Cell. Biol., 20, 4900–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flybase (2001) The Drosophila database. Available online at http://morgan/harvard.edu and http://www.ebi.ac.uk/flybase/
- Gietz D., Jean,A.S., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Henikoff S. (1997) Nuclear organization and gene expression: homologous pairing and long-range interactions. Curr. Opin. Cell Biol., 9, 388–395. [DOI] [PubMed] [Google Scholar]
- Ising G. and Block,K. (1981) Derivation-dependent distribution of insertion sites for a Drosophila transposon. Cold Spring Harb. Symp. Quant. Biol., 45, 527–544. [DOI] [PubMed] [Google Scholar]
- Ivanova A.V., Bonaduce,M.J., Ivanov,S.V. and Klar,A.J.S. (1998) The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nature Genet., 19, 192–195. [DOI] [PubMed] [Google Scholar]
- James T.C. and Elgin,S.C.R. (1986) Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol., 6, 3862–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Laible,G., Dorn,R. and Reuter,G. (1998) SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell. Mol. Life Sci., 54, 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.O., Cowell,I.G. and Singh,P.B. (2000) Mammalian chromo domain proteins: their role in genome organisation and expression. BioEssays, 22, 124–137. [DOI] [PubMed] [Google Scholar]
- Krauss V. and Reuter,G. (2000) Two genes become one: The genes encoding heterochromatin protein SU(VAR)3–9 and translation initiation factor subunit eIF-2γ are joined to a dicistronic unit in holometabolic insects. Genetics, 156, 1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhfittig S., Szabad,J., Schotta,G., Hoffmann,J., Máthé,E. and Reuter,G. (2001) _pitkin_D, a novel gain-of-function enhancer of position-effect variegation affects chromatin regulation during oogenesis and early embryogenesis in Drosophila. Genetics, 157, 1227–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Larsson J., Chen,J.D., Rasheva,V., Rasumson-Lestander,Å. and Pirrotta,V. (2001) Painting of fourth, a chromosome-specific protein in Drosophila. Proc. Natl Acad. Sci. USA, 98, 6273–6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J., Kotarski,M. and Tartof,K. (1988) Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics, 120, 181–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher M., Schmid,M., Aagaard,L., Selenko,P., Laible,G. and Jenuwein,T. (2000) Structure–function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation and mitotic progression. Mol. Cell. Biol., 20, 3728–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Klar,A.J.S. and Grewal,I.S. (2000) A chromo domain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meisosis. Cell, 101, 307–317. [DOI] [PubMed] [Google Scholar]
- Nakayama J., Rice,J.D., Strahl,B.D., Allis,C.D. and Grewal,S.I.S. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science, 292, 110–113. [DOI] [PubMed] [Google Scholar]
- Platero J.S., Sharp,E.J., Adler,P.N. and Eissenberg,J.C. (1996) In vivo assay for protein–protein interactions using Drosophila chromosomes. Chromosoma, 104, 393–404. [DOI] [PubMed] [Google Scholar]
- Rea S. et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature, 406, 593–599. [DOI] [PubMed] [Google Scholar]
- Reuter G. and Wolff,I. (1981) Isolation of dominant suppressor mutations for position-effect variegation. Mol. Gen. Genet., 182, 516–519. [DOI] [PubMed] [Google Scholar]
- Reuter G. and Spierer,P. (1992) Position effect variegation and chromatin proteins. BioEssays, 14, 605–612. [DOI] [PubMed] [Google Scholar]
- Reuter G., Guiarre,M., Farah,J., Gausz,J., Spierer,A. and Spierer,P. (1990) Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature, 344, 219–223. [DOI] [PubMed] [Google Scholar]
- Reuter G., Hoffman,G., Dorn,R. and Saumweber,H. (1993) Construction and characterization of TM3 balancer carrying P[(ry+)Δ2–3] as a stable transposase source. Drosoph. Inf. Serv., 72, 78–79. [Google Scholar]
- Rubin G.M. and Spradling,A.C. (1982) Genetic transformation of Drosophila with transposable element vectors. Science, 218, 348–353. [DOI] [PubMed] [Google Scholar]
- Rudolph T., Lu,B., Westphal,T., Szidonya,J., Eissenberg,J.C. and Reuter,G. (1999) New type of CyO and TM3 green balancers. Drosoph. Inf. Serv., 82, 99–100. [Google Scholar]
- Samson M.-L., Jackson-Grusby,L. and Brent,R. (1989) Gene activation and DNA binding by Drosophila Ubx and abd-A proteins. Cell, 57, 1045–1052. [DOI] [PubMed] [Google Scholar]
- Schotta G. and Reuter,G. (2000) Controlled expression of tagged proteins in Drosophila using a new modular P-element vector system. Mol. Gen. Genet., 262, 916–920. [DOI] [PubMed] [Google Scholar]
- Silver L.M. and Elgin,S.C.R. (1978) Production and characterization of antisera against three individual NHC proteins; a case of a generally distributed NHC protein. Chromosoma, 68, 101–104. [DOI] [PubMed] [Google Scholar]
- Sinclair D.A.R., Mottus,R.C. and Grigliatti,T.A. (1983) Genes which suppress position effect variegation in Drosophila melanogaster are clustered. Mol. Gen. Genet., 191, 326–333. [Google Scholar]
- Tschiersch B., Hofmann,A., Krauss,V., Dorn,R., Korge,G. and Reuter,G. (1994) The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3_–_9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J., 13, 3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath L. (1998) Unfolding the mysteries of heterochromatin. Curr. Opin. Genet. Dev., 8, 147–153. [DOI] [PubMed] [Google Scholar]
- Wallrath L. and Elgin,S.C.R. (1995) Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev., 9, 1263–1277. [DOI] [PubMed] [Google Scholar]
- Wang G., Ma,A., Chow,C.-M., Horsley,D., Brown,N.R., Cowell,I.G. and Singh,P.B. (2000) Conservation of heterochromatin protein 1 function. Mol. Cell. Biol., 20, 6970–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler K.S. and Wakimoto,B.T. (1995) Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet., 29, 577–605. [DOI] [PubMed] [Google Scholar]
- Wustmann G., Szidonya,J., Taubert,H. and Reuter,G. (1989) The genetics of position-effect variegation modifying loci in Drosophila melanogaster. Mol. Gen. Genet., 217, 520–527. [DOI] [PubMed] [Google Scholar]