Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes (original) (raw)
Abstract
SWI/SNF complexes are ATP-dependent chromatin remodelling enzymes that have been implicated in the regulation of gene expression in yeast and higher eukaryotes. BRG1, a catalytic subunit in the mammalian SWI/SNF complex, is required for transcriptional activation by the estrogen receptor, but the mechanisms by which the complex is recruited to estrogen target genes are unknown. Here, we have identified an interaction between the estrogen receptor and BAF57, a subunit present only in mammalian SWI/SNF complexes, which is stimulated by estrogen and requires both a functional hormone-binding domain and the DNA-binding region of the receptor. We also found an additional interaction between the p160 family of coactivators and BAF57 and demonstrate that the ability of p160 coactivators to potentiate transcription by the estrogen receptor is dependent on BAF57 in transfected cells. Moreover, chromatin immunoprecipitation assays demonstrated that BAF57 is recruited to the estrogen-responsive promoter, pS2, in a ligand-dependent manner. These results suggest that one of the mechanisms for recruiting SWI/SNF complexes to estrogen target genes is by means of BAF57.
Keywords: BAF57/chromatin remodelling/estrogen receptor/p160 coactivators/SWI/SNF
Introduction
Estrogen receptor α (ERα), a member of the nuclear receptor (NR) superfamily (Mangelsdorf et al., 1995; Di Croce et al., 1999b) is a ligand-inducible transcrip tion factor that regulates many physiological processes (Nilsson et al., 2001) in response to its natural ligand, 17β-estradiol (E2). Upon ligand binding, ERα binds to specific DNA sequences (EREs) present in the promoters of estrogen-dependent genes, triggering the recruitment to the promoter of many cofactors that must overcome the barrier to transcription formed by the packaging of DNA into dense chromatin. These cofactors reorganize chromatin templates and recruit basal transcription factors and RNA polymerase II. Two distinct types of chromatin remodelling activities have been well characterized. Histone acetyltransferases (HATs), which acetylate lysine residues in core histones, are proposed to unfold chromatin structure, thereby facilitating the binding of transcriptional regulators to promoters (Roth et al., 2001). The second class consists of ATP-dependent chromatin remodelling complexes (Dilworth and Chambon, 2001; Varga-Weisz, 2001), which use the energy of ATP hydrolysis to locally disrupt the association of histones with DNA. In addition, histones undergo other post-translational modifications including phosphorylation (Lee and Archer, 1998), and methylation of arginine residues by protein methyltransferases (Chen et al., 1999; Wang et al., 2001; Bauer et al., 2002), which are also implicated in gene activation by NRs.
Several lines of evidence have demonstrated the key role of ATP-dependent chromatin remodelling enzymes in the transcriptional activation by NRs. Initially, transient transfection studies showed that the co-expression of BRM/SNFα and BRG1/SNFβ, the catalytic subunits of the human SWI/SNF complex, were able to potentiate the transcriptional activation by ER and other NRs (Muchardt and Yaniv, 1993; Chiba et al., 1994). More recently, chromatin-dependent in vitro purified transcription systems have revealed that members of two different families of ATP-dependent chromatin complexes, SWI/SNF and ISWI, are selectively required for the ligand-dependent transactivation for different NRs (Di Croce et al., 1999a; Dilworth et al., 2000; Lemon et al., 2001).
All these chromatin remodelling activities are recruited to the ERE-containing genes in a highly regulated manner by mechanisms involving complex protein–protein interactions that are not as yet fully elucidated. Biochemical and genetic studies have identified a large number of proteins, generically named coactivators, that are recruited directly to the ligand-activated NRs, which are capable of potentiating their activity (McKenna et al., 1999). Among them are the p160 family of coactivators, SRC1, TIF2/GRIP1 and RAC3/AIB1/ACTR/pCIP (Leo and Chen, 2000), which are encoded by three distinct genes. These highly homologous proteins exhibit a common domain structure and it has been suggested that they act, at least in part, by serving as adapter molecules that recruit chromatin remodelling activities to the hormone-responsive promoters. The p160 coactivators contain a central receptor-interacting domain (RID), with several LXXLL motifs, responsible for the interaction with the ligand-bound NRs (Heery et al., 1997; Torchia et al., 1997), and two conserved C-terminal activation domains, AD1 and AD2. AD1 is responsible for the recruitment of CBP/p300 (Chakravarti et al., 1996; Kamei et al., 1996), which possess HAT activity, and AD2 interacts with arginine methyltransferases (Chen et al., 1999). In addition, the p160 coactivators have a highly conserved N-terminal basic helix–loop–helix/Per-Arnt-Sim (bHLH–PAS) domain (Crews and Fan, 1999), a protein dimerization motif that is the most conserved region between the three members of the family. It has been reported that this domain is involved in the recruitment of p160 coactivators by other transcription factors (Belandia and Parker, 2000; Chen et al., 2000; Wu et al., 2001), but the role of this highly conserved domain in the formation of the preinitiation complex by activated NRs remains elusive.
To investigate the function of the conserved bHLH–PAS domain in SRC1, we performed a yeast two-hybrid screen in an attempt to identify proteins that could directly interact with this domain. Here we described a novel interaction between the p160 coactivators and BAF57 (SMARCE1), a core subunit of the mammalian SWI/SNF family of ATP-dependent chromatin remodelling complexes. We also demonstrate that BAF57 is capable of interacting directly with ER in a ligand-regulated manner, and present evidence suggesting that BAF57 is required to target SWI/SNF complexes to estrogen-responsive promoters and enable p160 coactivators to potentiate transcriptional activity by ER.
Results
Interaction between SRC1 and BAF57
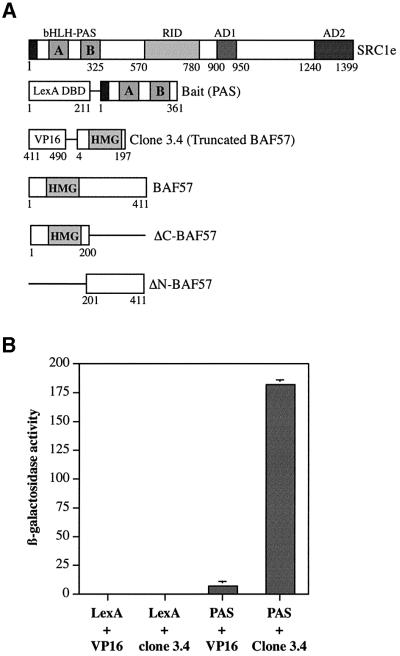
The domain organization of SRC1e that is conserved in the p160 family of coactivators is shown in Figure 1A. In an attempt to identify proteins that interact with the highly conserved bHLH–PAS domain of SRC1, we used the N-terminal region of SRC1 as bait in a yeast two-hybrid screening system. Yeast transformants containing the LexA-DNA-binding domain (DBD) fused to the bHLH– PAS domain (SRC1 amino acids 1–361, Figure 1A), and mouse proteins fused to the VP16 activation domain were selected according to their ability to grow in medium lacking histidine. Positive transformants were identified and tested for β-galactosidase activity. Sequence analysis and database searches revealed that four of the stronger interacting clones encoded an identical polypeptide, comprising the HMG DBD of BAF57 (amino acids 4–197, Figure 1A), a subunit of the mammalian SWI2/SNF2 complex (Wang et al., 1998). Clone 3.4 was chosen for further analysis. To test the specificity of the interaction, truncated BAF57 fused to the VP16 AD, or the isolated VP16 AD, were re-transformed into yeast expressing the LexA-DBD fused to the bHLH–PAS domain or the isolated LexA-DBD. The interactions were studied by determining the levels of lacZ reporter expression in yeast extracts using β-galactosidase assays (Figure 1B). This experiment showed a strong in vivo interaction between the N-terminal region of BAF57 and the SRC1 bHLH–PAS domain.
Fig. 1. BAF57 interacts with the SRC1 bHLH–PAS domain in yeast cells. (A) Schematic representation of SRC1e, the LexA chimera used as bait in the two-hybrid screening, the truncated VP16-tagged BAF57 clone, full-length BAF57 and the two BAF57 deletion mutants used in this study. Numbers refer to amino acids in the full-length proteins. The bHLH and the PAS homology region (containing two imperfect repetitions named PAS A and PAS B regions), the nuclear RID, and the activation domains 1 and 2 (AD1 and AD2, respectively) in SRC1e and the HMG domain in BAF57 are indicated. (B) The L40a yeast strain expressing either LexA-DBD or LexA-DBD fused to the SRC1 bHLH–PAS domain (PAS) was transformed with either the empty pASV3 plasmid or pASV3 expressing clone 3.4 fused to the VP16 activation domain. β-galactosidase activity in each yeast extract was measured in duplicate. Data represent the mean + SD of two independent transformants.
We then used GST pull-down assays to investigate the interaction between BAF57 and p160 coactivators in vitro. We confirmed that the SRC1 PAS domain (amino acids 1–361), used as bait in the two-hybrid screening, was able to bind to the truncated BAF57 protein encoded by clone 3.4 fused to GST (Figure 2A). The N-terminal portion of BAF57 was also capable of interacting with the full-length SRC1e, RAC3 (Figure 2A) and TIF2 (data not shown). Thus, all three members of the p160 family of coactivators interact in vitro with the N-terminal region of BAF57. When the full-length BAF57 protein fused to GST was used in the assay, we detected a similar interaction between full-length BAF57 and all full-length p160 coactivators (Figure 2B). The C-terminal region of BAF57 (amino acids 201–411), fused to GST, showed no interaction at all with the SRC1 bHLH–PAS domain or full-length SRC1 (data not shown), indicating that there are no additional SRC1-interacting domains in BAF57. In order to investigate whether the interaction between BAF57 and SRC1e was restricted to the bHLH–PAS domain, we tested the interaction between truncated SRC1e mutants, lacking the bHLH–PAS domain (ΔPAS– SRC1e) and full-length BAF57 fused to GST. In vitro translated ΔPAS–SRC1e did not interact with the GST–BAF57 fusion protein (Figure 2B), suggesting that the bHLH–PAS region is the only domain in SRC1e responsible for the interaction with BAF57. In a control experiment, ΔPAS–SRC1e was able to bind to the hormone-binding domain of ERα in GST pull-down assays (data not shown), indicating that its function is preserved in the deletion mutant. Finally, in the converse experiment, in vitro translated full-length BAF57 was able to bind to the N-terminal region of SRC1 (amino acids 1–450; Figure 2C).
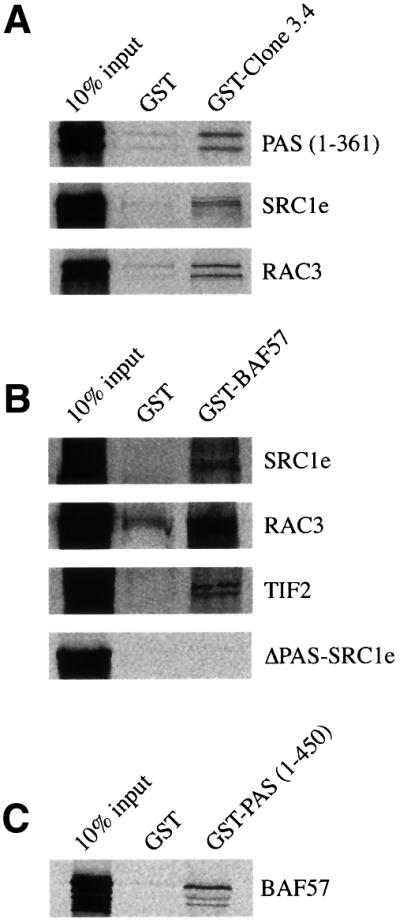
Fig. 2. In vitro interaction of BAF57 with p160 coactivators. (A) Binding of GST fusion proteins of truncated BAF57 encoded by clone 3.4 to the SRC1 bHLH–PAS domain, full-length SRC1 and RAC3. GST fusion of amino acids 4–197 of BAF57, coupled to Sepharose beads was incubated with in vitro translated [35S]methionine-labelled SRC1 bHLH–PAS domain (amino acids 1–361), full-length SRC1e or RAC3. After extensive washing, samples were boiled and separated on 10% SDS–PAGE. Gels were fixed and dried, and the labelled proteins were detected by fluorography. (B) Binding of GST–BAF57 to SRC1e, RAC3, TIF2 and ΔPAS-SRC1e. GST fusion proteins of full-length BAF57 were incubated with 35S-labelled full-length SRC1e, RAC3, TIF2 or ΔPAS-SRC1e as described above. (C) Binding of GST–bHLH–PAS to BAF57. GST fusion proteins of the first 450 amino acids of SRC1 were incubated with 35S-labelled full-length BAF57 as described above. In each panel, the input lane represents 10% of the total volume of lysate used in each reaction.
Effect of BAF57 expression on transcriptional activation by ER
Previous studies showed that Brahma-related gene 1 (BRG1), the ATPase subunit of the SWI2/SNF complex, was able to potentiate transcriptional activation by ER and other NRs (Muchardt and Yaniv, 1993; Chiba et al., 1994), and recently it has been shown, using a transient transfection system in a BRG1-deficient cell line, that transcriptional activation by ER requires functional BRG1 (DiRenzo et al., 2000). The observation that BAF57 can interact with p160 coactivators prompted us to investigate its ability to modulate the transcriptional activity of ER in transfected cells.
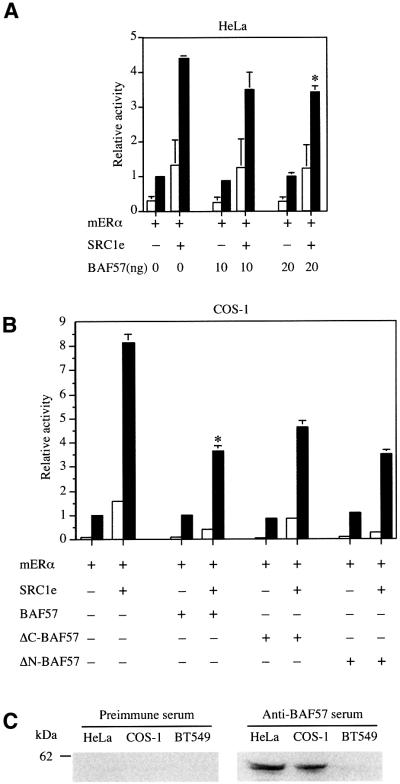
In our initial experiments, BAF57 was unable to stimulate transcriptional activity of ERα. Indeed, the co-expression of BAF57 in HeLa cells partially inhibited the ability of SRC1e to potentiate transcriptional activity of ERα on a 2XERE-pS2-luciferase reporter (Figure 3A). In similar experiments using COS-1 cells, the reduction in reporter gene transcription was more apparent, reaching a maximum of 50% inhibition in the presence of 20 ng of BAF57 (Figure 3B). However, HeLa cells express high levels of BAF57 protein (Wang et al., 1998), and we confirmed the expression of BAF57 in our HeLa and COS-1 cells by western blotting using a rabbit polyclonal antibody generated against BAF57 (Materials and methods; Figure 3C). The repression observed may reflect recruitment of overexpressed BAF57 protein alone rather than intact SWI/SNF complexes, thereby interfering with the action of the endogenous coactivator complex. Alternatively, it is conceivable that the SWI/SNF complex is functioning as a corepressor (Underhill et al., 2000), and that BAF57 is potentiating this activity, but this seems unlikely because the recruitment of SWI/SNF complexes by NRs has been most frequently associated with gene activation (Dilworth and Chambon, 2001). Since the expression of deletion mutants of BAF57 caused a similar repression to that of the full-length protein (Figure 3B), we conclude that BAF57 was behaving in a dominant-negative manner, interfering with the recruitment of endogenous SWI/SNF coactivator complexes.
Fig. 3. Effect of BAF57 expression on transcriptional activation by ER in cell lines expressing endogenous BAF57. (A) HeLa cells were transiently transfected with expression vectors for mERα and SRC1e, the 2XERE-pS2-luciferase reporter, different amounts of BAF57 expression vector and an internal control vector (pRL-CMV, providing constitutive expression of Renilla luciferase). (B) COS-1 cells were transiently transfected with expression vectors for mERα and SRC1e, the 2XERE-pS2-luciferase reporter, 20 ng of full-length BAF57, ΔC-BAF57 or ΔN-BAF57 expression vectors and pRL-CMV as an internal control. In each case, after transfection, cells were washed and incubated with vehicle (white bars) or 17β-estradiol (black bars) at 10–8 M for 24 h. Subsequently, cell lysates were assayed using a dual luciferase reporter system. Normalized values are expressed relative to the activity of mERα alone in the presence of 10–8 M E2. The results shown represent the average of at least two independent experiments assayed in quadruplicate + SD. The asterisks represent statistical analysis, which shows that the results observed were significant (p < 0.05 for HeLa cells and p < 0.001 for COS-1 cells). (C) Western blotting showing expression levels of BAF57 proteins in HeLa, COS-1 and BT549 cell lines.
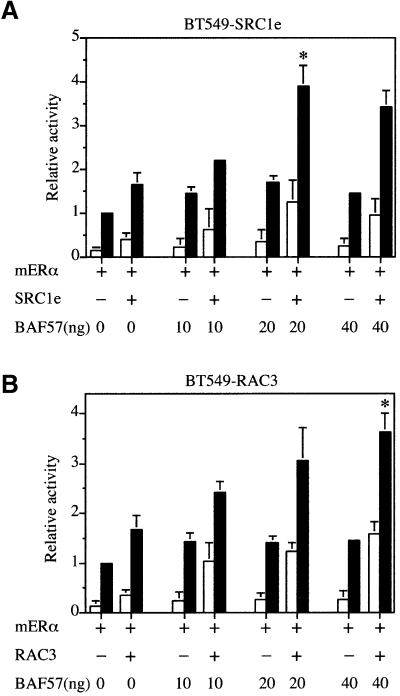
Recently, a breast ductal carcinoma cell line (BT549) lacking BAF57 protein has been described, which nevertheless retains the expression of the remaining SWI2/SNF2 subunits (Decristofaro et al., 2001). We confirmed the lack of BAF57 expression in BT549 cells by western blotting (Figure 3C). These cells provide an opportunity to analyse the importance of BAF57 to transcriptional activation by ER. Interestingly, the ability of SRC1e to potentiate transcriptional activation of ER in BT549 cells was markedly impaired relative to HeLa and COS-1 cells (compare Figures 3A, B and 4A). However, expression of exogenous BAF57 increased the activity of SRC1e as a coactivator in a dose-dependent manner (Figure 4A). Similarly, both RAC3 (Figure 4B) and TIF2 (data not shown) exhibit very little activity as coactivators in BT549 cells, but could be rescued by expressing increasing amounts of BAF57. Taken together, the data imply that BAF57 is required for p160 family members to show full coactivation potential in transiently transfected cells.
Fig. 4. BAF57 is required for ER coactivation by p160 proteins. (A) The BAF57-deficient breast ductal carcinoma cell line, BT549, was transiently transfected with expression vectors for mERα and SRC1e, the 2XERE-pS2-luciferase reporter, different amounts of BAF57 expression vector and pRL-CMV as an internal control. (B) BT549 cells were transiently transfected with expression vectors for mERα and RAC3, the 2XERE-pS2-luciferase reporter, different amounts of BAF57 expression vector and pRL-CMV as an internal control. Data are presented as described in Figure 3. The results shown represent the average of at least two independent experiments assayed in quadruplicate + SD. The asterisks represent statistical analysis, which shows that the results observed were significant (p < 0.001).
BAF57 interacts directly with ER
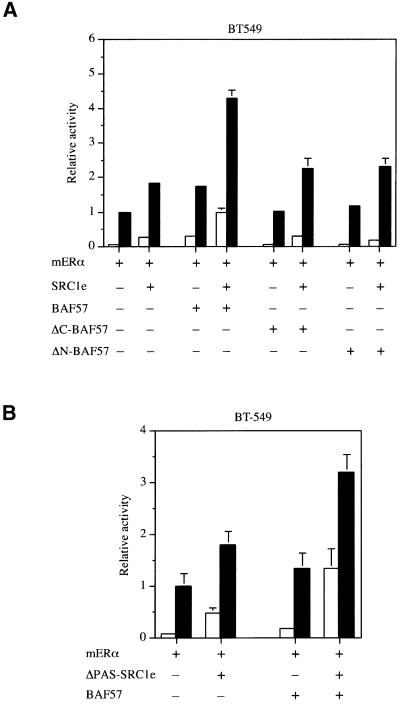
To investigate the importance of the interaction between BAF57 and the bHLH–PAS domain, we tested the ability of deletion mutants to potentiate ERα transcriptional activity. Expression of either N- or C-terminal BAF57 mutants had a negligible effect in BT549 cells (Figure 5A), suggesting that full-length BAF57 is required to rescue the ability of SRC1 to act as a coactivator.
Fig. 5. Effect of BAF57 and SRC1e deletion mutants on transcriptional activation by ER. (A) BAF57 deletion mutants do not restore SRC1e coactivation in BAF57-deficient cells. BT549 cells were transiently transfected with expression vectors for mERα and SRC1e, the 2XERE- pS2-luciferase reporter, 20 ng of full-length BAF57, ΔC-BAF57 or ΔN-BAF57 expression vectors and pRL-CMV as an internal control. Data are presented as described in Figure 3. The results shown represent the average of two independent experiments assayed in quadruplicate + SD. (B) BAF57 enhances ER coactivation by ΔPAS–SRC1e in BAF57-deficient cells. BT549 cells were transiently transfected with expression vectors for mERα and ΔPAS–SRC1e, the 2XERE-pS2-luciferase reporter, 20 ng of BAF57 expression vector and pRL-CMV as an internal control. Data are presented as described in Figure 3. The results shown represent the average of two independent experiments assayed in quadruplicate + SD.
However, rather surprisingly, we found that the SRC1 bHLH–PAS domain, which is required for the in vitro interaction of SRC1 with BAF57, was not required for the ability of ERα to stimulate transcription from reporter genes in COS-1 cells (Belandia and Parker, 2000). Moreover, while ΔPAS-SRC1e, like full-length SRC1e, showed a reduced activity in the BAF57-deficient cell line, its activity was still potentiated by BAF57 (Figure 5B). Although the stimulatory activity exhibited by ΔPAS-SRC1e in the presence of BAF57 was less than that of full-length SRC1e, the result raised the possibility that BAF57 might be recruited to the reporter gene by a mechanism independent of the bHLH–PAS/BAF57 interaction.
Our results are consistent with the observation that a number of NR coactivators have been shown to interact directly both with the p160 coactivators and with NRs (Lee et al., 1999; Chauchereau et al., 2000; Wu et al., 2001). Moreover, the recruitment of BRG1 to ERα was postulated to be indirect and mediated by unknown additional factors, distinct from SRC1 (DiRenzo et al., 2000). We therefore investigated whether there was a direct interaction between ER and BAF57 that might explain the potentiation observed in our transfection experiments with ΔPAS-SRC1e. Such an interaction could provide a mechanism for the recruitment of SWI/SNF complexes to estrogen-responsive genes. To investigate this hypothesis, we incubated 35S-labelled ERα with GST–BAF57 fusion proteins in a pull-down assay. ERα showed ligand-independent binding to GST–BAF57, and this binding was strongly increased (>7-fold) in the presence of E2 (Figure 6B, upper panel). The ligand responsiveness of the interaction indicates that it might be relevant for the ligand-dependent transcriptional activity by ER. The closely related ERβ interacted with GST–BAF57 in a similar ligand-dependent fashion (Figure 6B, lower panel).
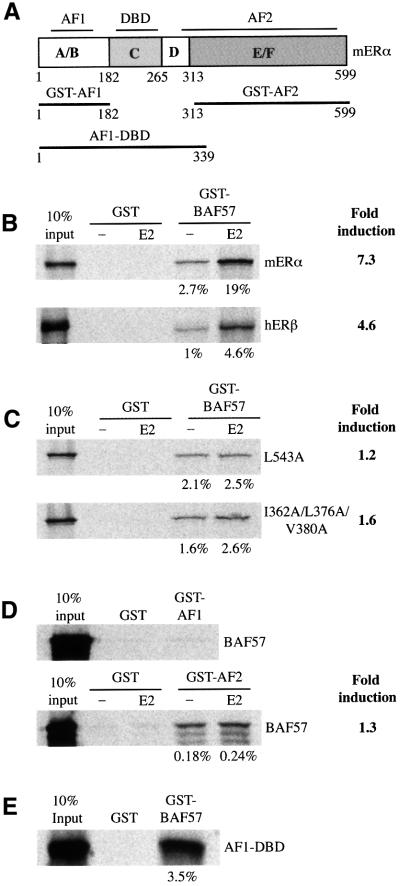
Fig. 6. In vitro interaction of BAF57 with ER. (A) Schematic representation of mERα and the deletion mutants used in the GST pull-down assay. Indicated are the ligand-independent activation function (AF1), the DBD, and the ligand-dependent activation function (AF2). Numbers refer to amino acids in the full-length mERα. (B) Binding of GST fusion proteins of BAF57 to 35S-labelled mERα or hERβ. (C) Binding of GST fusion proteins of BAF57 to 35S-labelled mERα AF2 mutants. (D) Binding of GST fusion proteins of mERα deletion mutants to 35S-labelled full-length BAF57. (E) Binding of GST fusion proteins of full-length BAF57 to 35S-labelled AF1-DBD mERα deletion mutant. When required, the assays were performed in the presence of vehicle (–) or 100 nM 17β-estradiol (E2). Bound proteins were visualized as described in Figure 2A. To the right of each panel, the 35S-labelled proteins used in the assay are indicated and the fold induction in the binding observed in the presence of hormone relative to that detected without added hormone. Below each panel, the percentage of the input pulled down for each assay is shown. In each panel, the input lane represents 10% of the total volume of lysate used in each reaction.
Ligand-dependent recruitment of BAF57 by ERα requires a functional hormone-binding domain
The ligand-dependent recruitment of coactivators to ERα is mediated by the C-terminal hormone-binding domain, named AF2 (Lees et al., 1989; Tora et al., 1989). Having identified a ligand-dependent, direct interaction between BAF57 and ERα, we next investigated whether the recruitment of BAF57 correlated with the transcriptional activity of ERα. To analyse this, we tested the binding of different 35S-labelled ERα AF2 mutants to the GST–BAF57 fusion protein. These mutants were the point mutant L543A (affecting helix 12), and the triple mutant I362A/L376/V38A (mutations in helices 3 and 5). These two mutants retained high affinity binding to estrogen and were able to bind to DNA with affinity similar to the wild-type receptor. The transcriptional activity and in vitro binding to SRC1e of the triple mutant were severely impaired, while the L543A mutant had no detectable interaction in vitro with SRC1e and was transcriptionally inert (Mak et al., 1999). The two mutants retained a ligand-independent association with BAF57 similar to wild-type ERα, but the dramatic increase in the binding of the wild-type receptor upon addition of ligand was lost for both mutants (Figure 6C). The observation that the ligand-dependent interaction between BAF57 and ERα requires an intact, transcriptionally competent AF2 domain suggests that this recruitment is important for the ER activity.
The central DNA-binding region of ERα is required for the in vitro interaction with BAF57
The observation that BAF57 and ER showed both a weak ligand-independent interaction and a strong ligand-dependent interaction that requires a functional AF2 may reflect the use of the two classical activation domains of ERα, AF1 and AF2, for the recruitment of BAF57. To test this hypothesis, we used ERα deletion mutants in GST pull-down assays. A GST fusion protein of the ligand-independent activation function, GST–AF1, failed to interact with BAF57 (Figure 6D, upper panel), suggesting that the recruitment of BAF57 is not one of the mechanisms by which AF1 activates transcription. GST–AF1 was able to interact with TIF2 in a control GST pull-down experiment (data not shown) as described previously (Benecke et al., 2000), demonstrating the integrity of the GST fusion protein used in the assay. A GST fusion of the ligand-dependent activation function, GST–AF2, showed poor in vitro binding to BAF57, without any significant ligand responsiveness (Figure 6D, lower panel). This GST–AF2 mutant contains all the molecular determinants required for strong, estrogen-regulated binding to the classical LXXLL-containing coregulators, including SRC1 (Kalkhoven et al., 1998) and NRIP1 (Cavailles et al., 1995). BAF57 does not possess any LXXLL motifs, and therefore a different mechanism must be used for the interaction with the activated ERα. To analyse the importance of the central region of ERα in the recruitment of BAF57, we carried out another GST pull-down experiment, using an in vitro translated deletion mutant of ERα comprising AF1, the DBD and the hinge region. This ERα mutant bound strongly to a GST–BAF57 fusion protein (Figure 6E). Thus, we conclude that no single domain by itself is sufficient for the interaction with BAF57, and that both the DBD of ERα and a functional AF2 are important for this interaction.
Hormone-dependent association between BAF57 and ERα in cellular extracts
We next investigated whether the interactions between ERα and BAF57 observed in vitro could also be detected between the endogenous proteins in intact cells. After treating ZR75.1 breast carcinoma cells with 17β-estradiol or anti-estrogens, we performed immunoprecipitation and western blotting experiments using whole-cell extracts. Upon immunoprecipitation with antibodies against BAF57 and western blotting using the anti-ERα antibody D12, we observed that BAF57 was able to interact with ERα in cells treated with 17β-estradiol but not the pure anti-estrogen ICI 182,780. Pre-immune antibodies failed to co-immunoprecipitate ERα in similar conditions (Figure 7A). This ligand-dependent interaction of the endogenous proteins strongly supports a functional role for the recruitment of BAF57 in the mechanism of transcriptional activation by ERα.
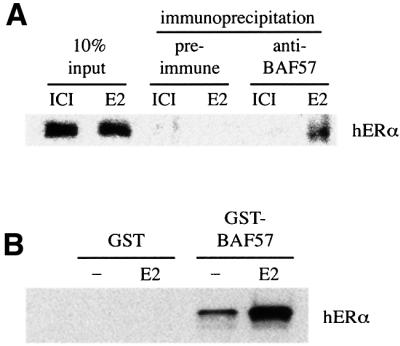
Fig. 7. Hormone-dependent interaction between BAF57 and ERα in cellular extracts. (A) Co-immunoprecipitation of endogenous BAF57 and ERα. ZR75.1 cells were treated with anti-estrogens (ICI) or 17β-estradiol for 30 min. Whole-cell lysates were then immunoprecipitated with antibodies against BAF57. The immunoprecipitated material was subjected to western blotting analysis with anti-ERα monoclonal IgG. (B) Whole-cell extracts from SW13 cells (BRG1/BRM-deficient cell line), previously transfected with hERα, were incubated with GST alone or GST–BAF57 bound to glutathione– agarose in the presence of vehicle (–) or 100 nM 17β-estradiol (E2). The associated hERα was detected by western blotting using anti-ERα monoclonal IgG.
The interaction between ER and BAF57 observed in the GST pull-down assays suggests that those two proteins interact directly, but we could not rule out the possibility that the interaction occurs through BRG1 or BRM proteins present in the rabbit reticulocyte lysate. To investigate this, we examined the interaction in SW13 cells, an adrenal carcinoma cell line devoid of BRG1 and BRM proteins (Muchardt and Yaniv, 1993). After transient transfection of ERα, whole-cell extracts were incubated with bacterially expressed GST–BAF57 fusion proteins, and the amount of ERα bound was analysed by western blotting. ERα was able to interact with BAF57 in a ligand-independent manner, but the binding was dramatically increased in the presence of 17β-estradiol (Figure 7B). Thus we conclude that ER and BAF57 are capable of interacting in the absence of BRG1 or BRM proteins.
BAF57 is recruited to the pS2 estrogen-responsive promoter in a hormone-dependent manner
We have demonstrated that there are at least two protein–protein interactions used by the ER to associate with BAF57 in a ligand-dependent manner. This association seems to be important for the transcriptional activity by the ER because the expression of exogenous BAF57 in BAF57-deficient cells restores the ability of all p160 proteins to serve as ER coactivators. BAF57 is a core subunit of all SWI/SNF complexes, which are well-characterized chromatin remodelling cofactors recruited to specific promoters in a regulated fashion to facilitate RNA polymerase transcription. To examine whether BAF57 and SRC1 are associated with estrogen-regulated promoters, we used chromatin immunoprecipitation (ChIP) assays to probe the chromatin structure around the pS2 gene in hormonally manipulated cells. Chromatin fragments were prepared from mock-treated and E2-stimulated ZR75.1 breast carcinoma-derived cells, after formaldehyde cross-linking. The chromatin preparations were immunoprecipitated using specific antibodies to BAF57 or to SRC1. The amount of ERE-containing pS2 promoter DNA present in immunoprecipitated chromatin fractions was then determined by quantitative real-time PCR. We observed a 5.5-fold enrichment of the ERE region in the chromatin fraction immunoprecipitated with the anti-SRC1 antibody, post-stimulation with estrogen, compared with untreated cells (Figure 8A), thus confirming the expected estrogen-dependent recruitment of this coactivator to the pS2 promoter (Shang et al., 2000). Similarly, the ligand-dependent recruitment of BAF57 to the same region of the pS2 promoter was demonstrated by the 4.5-fold increase in the pS2 promoter DNA present in the immunoprecipitated fraction subsequent to E2 treatment (Figure 8B). Control ChIP assays using pre-immune serum did not show any significant changes with estrogen treatment, strongly suggesting that the fold increases observed with the SRC1- and BAF57-specific antibodies were due to specific immunoprecipitation of these proteins, reflecting the recruitment of both factors to the pS2 promoter in response to the addition of estrogen to the cells.
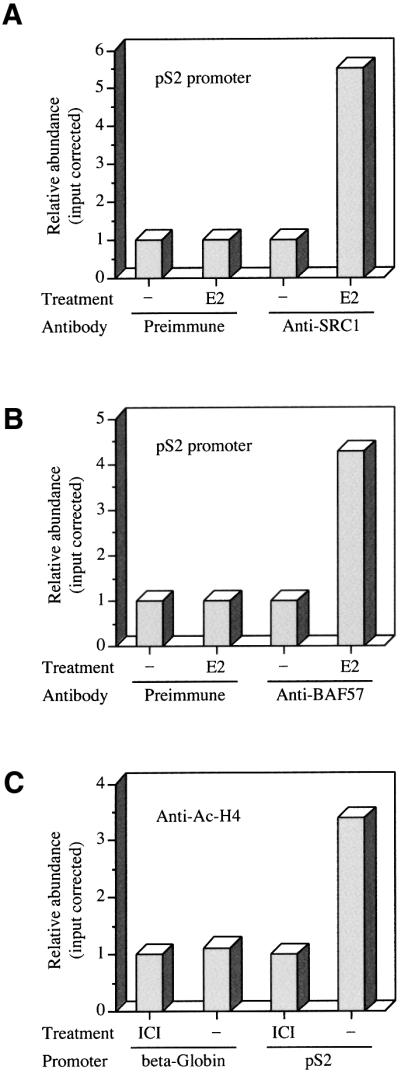
Fig. 8. Hormone-dependent association of BAF57 with the endogenous pS2 promoter. (A) ZR75.1 cells were deprived of estrogen for 48 h and then treated with 100 nM 17β-estradiol (E2) or vehicle (–) for 30 min, and fixed immediately using formaldeyde. Soluble chromatin fragments were obtained by sonication and subjected to immunoprecipitation using anti-SRC1 antibodies. (B) Soluble chromatin fragments from ZR75.1 cells treated as described above were subjected to immunoprecipitation using anti-BAF57 antibodies. (C) ZR75.1 cells maintained in estrogenic conditions were treated with anti-estrogens (ICI) or vehicle (–) for 30 min, and soluble chromatin fragments were subjected to immunoprecipitation using anti-acetylated histone H4 antibodies. ChIP assays were quantified by real-time PCR using primers specific to the ERE-containing region of the pS2 promoter or an enhancer region 5′ of β-globin gene. The ChIP assays were repeated several times, and results of a representative experiment are shown.
A further experiment was performed to confirm that the chromatin associated with the pS2 promoter showed an estrogen-dependent increase in the status of histone H4 acetylation, a covalent modification associated with transcriptional initiation (Kuo et al., 1996). Using an anti-acetylated histone H4 antibody, we detected a 3.5-fold enrichment of the ERE region in chromatin immunoprecipitated from ZR75.1 cells grown under estrogenic conditions compared with cells treated with anti-estrogens (Figure 8C). Parallel assays examining an enhancer region 5′ of β-GLOBIN gene (which is inactive in these cells) showed no change in histone H4 acetylation status upon hormone manipulation (Figure 8C), indicating that the change observed over the pS2 promoter was specific.
Discussion
There is substantial evidence to indicate that a combination of ATP-dependent chromatin remodelling factors together with enzymes that catalyse post-translational modifications of histones are essential for the regulation of gene transcription. Support for a role for SWI/SNF complexes and histone acetyltransferases in transcriptional activation by NRs comes from studies in transiently transfected cells and in vitro transcription experiments using chromatin templates (Muchardt and Yaniv, 1993; Chiba et al., 1994; Dilworth et al., 2000; Lemon et al., 2001). In this paper, we provide evidence for a functional link between SWI/SNF complexes, the p160 family of HATs and the estrogen receptor. We have found that BAF57, a core subunit of all mammalian chromatin remodelling SWI/SNF complexes may serve as a targeting subunit responsible for the recruitment of the complex to the ligand-bound ERα during target gene activation.
There are a number of biochemically distinct ATP-dependent chromatin remodelling complexes in mammals, of which two SWI/SNF complexes are highly similar with at least eight common subunits. SWI/SNF-A (or BAF) contains BAF250, and either BRM/SNFα or BRG1/SNFβ as the ATPase catalytic subunit, while SWI/SNF-B (or PBAF) contains BAF180 and BRG1/SNFβ (Xue et al., 2000). BAF57 is a common subunit for both of these complexes, with orthologues in non-vertebrates, but not in yeast. Genetic studies using mutants of the Drosophila orthologue of BAF57, BAP111, showed that this protein is required for the Drosophila SWI/SNF-like complex to function in vivo (Papoulas et al., 2001) but, to date, it remains to be established how these proteins contribute to the function of SWI/SNF complexes.
We have demonstrated that BAF57 is necessary for the ability of p160 proteins to act as coactivators for ERs in transfected cells. The importance of BAF57 was evident from our observation that SRC1 failed to potentiate the transcriptional activity of ERα in BT-549 cells devoid of the protein, but this activity could be rescued by co-expression of BAF57. Furthermore, the inhibitory effects of overexpressed BAF57 in cells expressing the protein are consistent with its interference with the recruitment of a pre-formed endogenous SWI/SNF coactivator complex. Previous work has established that the chromatin remodelling activity of SWI/SNF complexes is dependent on hBRM or hBRG1, and both proteins are capable of stimulating the transcriptional activity of a number of nuclear receptors in transfected cells. Support for a role of BRG1 in mediating transcriptional activation by the ER comes from ChIP assays, which demonstrate its recruitment to the promoter of the estrogen-responsive gene, pS2, when cells are treated with estrogen. The requirement of NRs for chromatin remodelling is not evident in vitro (Lemon et al., 2001), but interestingly only PBAF was able to stimulate NR-dependent transcription from a chromatin template. Lemon and co-workers suggested that distinct chromatin remodelling complexes may perform specific non-interchangeable functions and that the role of PBAF may not be limited to its chromatin-remodelling activity (Lemon et al., 2001).
The mechanism by which SWI/SNF complexes are recruited to specific promoters in vivo is unclear. One possibility is that the bromodomain found in BRM and BRG1 binds to acetylated lysine residues following their modification by HATs. Alternatively, DiRenzo and co-workers have found a ligand-dependent association between BRG1 and ERα, but they were unable to detect a direct interaction and postulated that additional factors were required (DiRenzo et al., 2000). We propose that BAF57 represents one of the factors that can mediate the recruitment of SWI/SNF complexes to estrogen target genes in the presence of the hormone. In support of this proposal, we have demonstrated that BAF57 is recruited to the promoter of the endogenous pS2 gene when cells are treated with estrogen. In parallel, SRC1 is also recruited to the promoter, and this is accompanied by histone H4 hyper-acetylation. Further data consistent with this view come from in vitro transcription assay studies using chromatin templates that showed that the N-terminal region of SRC1 was also required for maximal progesterone receptor transcription (Liu et al., 2001).
BAF57 is capable of binding to both the p160 family of coactivators, by means of their N-terminal bHLH–PAS domain, and directly to the ER in a ligand-dependent manner. The interaction with ER is distinct from that found for p160 proteins, which contain LXXLL motifs that bind to the AF2 surface on the ligand-binding domain. While AF2 is necessary for the interaction between the full-length receptor and BAF57, the ligand-binding domain is not sufficient for the interaction. Our interaction data indicate an additional requirement for the ER DBD, suggesting that BAF57 and p160 coactivators interact with distinct surfaces on the receptor. This raises the possibility that BAF57 and p160 coactivators may interact simultaneously with the ER, as opposed to sequentially, as has been suggested for other ER-interacting proteins (Shang et al., 2000). Thus, our results suggest a functional link between different classes of chromatin remodelling complexes and are consistent with a model in which SRC1 is not only required for the recruitment of HAT and arginine methyltransferase activities, but may also be involved in the recruitment of ATP-dependent chromatin remodelling factors.
Finally, it is noteworthy that the human BAF57 gene maps to chromosome band 17q21.1, in close proximity to the BRCA1 gene, a locus associated with frequent loss of heterozygosity (LOH) and allelic imbalance in breast and ovarian cancer (see Decristofaro et al., 2001 and references therein). In sporadic breast and ovarian cancers, mutations in the BRCA1 gene are rare events in samples with LOH in the region (Futreal et al., 1994), suggesting that other genes in this region may function as tumour suppressors. It was previously suggested that BAF57, based on its inferred involvement in DNA transcription and repair events, could be that tumour suppressor gene (Decristofaro et al., 2001). Our data, showing how BAF57 is involved in the regulation of the transcriptional activity by ERα, a key regulator of cell proliferation in these tissues, reinforce that idea.
Materials and methods
Two-hybrid screening
The yeast two-hybrid screening using the N-terminal region of SRC1 as bait, and a mouse embryo (9.5–12.5 d.p.c.) cDNA library, has been described previously (Belandia and Parker, 2000).
Plasmids
The complete open reading frame of the full-length murine BAF57 was amplified by PCR using a mouse embryo (9.5–12.5 d.p.c.) cDNA library as a template, and subcloned into pSG5 or pGEX-6P-1 (Amersham Pharmacia Biotech). The cDNA obtained showed two differences at nucleotide level with the previously described BAF57 cDNA (DDBJ/EMBL/GenBank accession No. AF035263) that do not modify the protein sequence encoded.
The following plasmids have been described previously: pMT2-MOR (Lahooti et al., 1995), pJ3MOR1–339, GST–MOR-AF1, GST–MOR- AF2 (Cavailles et al., 1995), pSG5-SRC1e (Kalkhoven et al., 1998), pSG5-TIF2 (Voegel et al., 1996), pCMX.F-RAC3 (Li et al., 1997), pSG5-ΔPAS-SRC1e, GST–SRC1-(1–450) and pGL3-2XERE-PS2 (Belandia and Parker, 2000), pSG5-L543A-mERα and pSG5-I362A/L376A/V380A-mERα (Mak et al., 1999) and hERβ (Cowley et al., 1997). The cDNA regions encoding amino acids 4–197 (clone 3.4), 1–200 (ΔC-BAF57) and 201–411 (ΔN-BAF57) of BAF57 were amplified by PCR and cloned into pSG5 for in vitro translation and mammalian expression, or into pGEX-6P-1 for generation of GST fusion proteins.
Antibodies
The antibodies used were anti-BAF57 (rabbit polyclonal serum raised against amino acids 1–200 of mouse BAF57), anti-SRC1e (rabbit polyclonal serum raised against amino acids 379–1440 of human SRC1e), anti-AcH4 (rabbit polyclonal antibody raised against fully chemically acetylated histone H4 protein from calf thymus) and anti-ERα D12 (mouse monoclonal IgG; Santa Cruz Biotechnology, Inc.).
GST pull-down assays
Recombinant cDNAs in the pSG5 expression vector were transcribed and translated in vitro in the presence of [35S]methionine in reticulocyte lysate (Promega) according to the manufacturer’s protocol. GST fusion proteins were induced, purified, bound to Sepharose beads (Amersham Pharmacia Biotech), and incubated with translated proteins or whole-cell extracts as described previously (Kalkhoven et al., 1998) in NETN buffer (20 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.5% NP-40, 100 mM NaCl). After extensive washing, the samples were separated on 10% SDS–PAGE. Gels were fixed and dried, and the 35S-labelled proteins were visualized by fluorography, or blotted onto nitrocellulose and probed with antibodies.
Cell culture and transient transfections
COS-1, HeLa and BT549 cells were routinely maintained in E4 supplemented with 10% fetal bovine serum. Twenty-four hours before transfection, cells were plated in 96-well microtiter plates in phenol red-free medium, supplemented with 5% dextran charcoal-stripped serum. COS-1 and HeLa cells were transfected using a modified calcium phosphate protocol (Belandia and Parker, 2000), and BT549 cells were transfected using FuGENE 6 transfection reagent (Roche) according to the manufacturer’s instructions. The transfected DNA included a pRL-CMV (Promega) control plasmid (1 ng/well), the luciferase reporter plasmid pGL3-2XERE-PS2 (10 ng/well), pMT2-MOR (2.5 ng/well), pSG5-SRC1e, pSG5-ΔPAS-SRC1e or pCMX.F-RAC3 (10 ng/well), different amounts of pSG5-BAF57 (10, 20 or 40 ng/well), pSG5- ΔC-BAF57 (20 ng/well) or pSG5-ΔN-BAF57 (20 ng/well) as indicated in the legends for Figures 34–5. Empty vectors were used to normalize the amounts of DNA. After incubation for 16 h with the DNAs, the cells were washed and incubated in fresh medium in the presence or absence of 10 nM 17β-estradiol for 24 h. Subsequently, cells were harvested and extracts were assayed for luciferase activity using a dual luciferase reporter assay as described previously (Belandia and Parker, 2000).
Immunoblotting
Whole-cell extracts from COS-1, HeLa and BT549 cells were separated on 10% SDS–PAGE and blotted onto nitrocellulose. The membranes were blocked in TBS-T (20 mM Tris–HCl pH 7.6, 137 mM NaCl, 0.1% Tween 20) containing 3% non-fat milk powder, washed with TBS-T, and incubated for 2 h with anti-BAF57 rabbit polyclonal serum. After washing, the membranes were incubated with biotinylated goat anti-rabbit IgG (Dako), washed with TBS-T, incubated with streptavidin– horseradish peroxidase (Dako) and washed again with TBS-T. Bound immunoglobulins were visualized using the ECL detection system (Amersham Pharmacia Biotech).
Co-immunoprecipitation assay
Following stimulation with 10 nM 17β-estradiol or 100 nM ICI 182,780 (Fasolodex, Astra-Zeneca) for 30 min, ZR75.1 cells were washed twice with ice-cold phosphate-buffered saline (PBS) containing protease inhibitor cocktail (Roche), collected using a rubber policeman and then resuspended in ice-cold PBS. Cells were pelleted at 2000 g and incubated for 10 min in lysis buffer [50 mM Tris–HCl pH 7.5, 100 mM NaCl, 10% (v/v) glycerol, 0.1% (v/v) NP-40, 1 mM dithiothreitol, 1 mM EDTA, protease inhibitor cocktail and phosphatase inhibitor cocktail (Sigma-Aldrich)]. Cells were then sonicated twice for 10 s at one-third full power at 4°C using a Sanyo Gallenkamp sonicator. The lysate was pre-cleared using an anti His-tag antibody and protein A/G–Sepharose at 4°C for 45 min. Following centrifugation at 14 000 g for 10 min, the supernatant was used for immunoprecipitation with pre-immune serum or anti-Baf57 polyclonal antibody at 4°C for 90 min; immune complexes were then captured using protein A/G–Sepharose. Following centrifugation at 6000 g for 10 min, the protein A/G–Sepharose was washed twice in wash buffer [0.1% (v/v) SDS, 1% (v/v) Triton X-100, 2 mm EDTA, 20 mM Tris–HCl pH 8.1, 150 mM NaCl]. Complexes were released from the protein A/G–Sepharose by boiling for 5 min in 2× SDS loading buffer. The immunoprecipitated material was separated on 10% SDS–PAGE and blotted onto nitrocellulose; the membrane was probed using anti-ERα antibody essentially as described above.
ChIP assay
ZR75.1 cells were grown in phenol-free RPMI 1640 supplemented with 10% dextran charcoal-stripped serum for 48 h prior to the addition of 100 nM 17β-estradiol (Sigma) or ethanol vehicle for 30 min. In the ChIP assay using anti-AcH4 antibodies, cells were maintained in estrogenic conditions and treated with anti-estrogen (10 nM ICI 182,780) or vehicle for 30 min. For each treatment, 108 cells were fixed with 1% formaldehyde for 10 min at room temperature, the media was removed and the cells washed in ice-cold PBS containing a protease inhibitor cocktail (Roche). Cells were collected using a rubber policeman and soluble chromatin material was isolated essentially as described previously (Braunstein et al., 1993). Chromatin was sonicated using a Soniprep 150 MSE (Sanyo Gallenkamp) to generate chromatin fragments (typically 200–1000 bp). Chromatin fragments from ∼107 cells were used for each immunoprecipitation. Samples were pre-cleared with protein A/G–Sepharose (pre-adsorbed with sonicated salmon sperm) for 45 min at 4°C. Following precipitation of the beads, the supernatant was incubated with 50 µl immune or pre-immune serum overnight at 4°C. Immune complexes were isolated using 100 µl protein A/G–Sepharose. Non-specific proteins were removed as described previously (Braunstein et al., 1993). Formaldehyde cross-linking was reversed and DNA removed from the immune complex by heating at 65°C overnight. Proteins were digested using proteinase K, and removed by phenol/chloroform extraction. The released DNA was recovered by ethanol precipitation and resuspended in 100 µl TE buffer.
Real-time PCR and data analysis
Real-time PCR was performed and data analysed essentially as described previously (Litt et al., 2001) using an ABI Prism 7700 sequence detector following PE Applied Biosystems’ Taqman SYBR Green Master Mix protocol. The optimal primer concentrations were determined according to the manufacturer’s guidelines, and single amplicon generation checked by agarose gel electrophoresis. Data were analysed during the linear phase of the PCR and the results plotted using Microsoft Excel. Primers to the PS2 promoter and 5′ β-globin enhancer were generated using PE Applied Biosystems software and sequence details are available on request.
Statistical analysis
Statistical significance of the effects of full-length BAF57 in the transient transfection assays was determined using the Student’s _t_-test application from Microsoft Excel software package (Microsoft Corporation).
Acknowledgments
Acknowledgements
We are grateful to H.Gronemeyer, J.Don Chen and B.Katzenellenbogen for gifts of plasmids. We thank A.W.Thorne for the anti-AcH4 antibody, and Ho Yi Mak, Roger White, David Baker and members of the Molecular Endocrinology Laboratory for plasmids, helpful discussion and critical reading of the manuscript. B.B. was supported by the European Community TMR program. This work was funded by the Wellcome Trust and Cancer Research UK.
References
- Bauer U.M., Daujat,S., Nielsen,S.J., Nightingale,K. and Kouzarides,T. (2002) Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO rep., 3, 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belandia B. and Parker,M.G. (2000) Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J. Biol. Chem., 275, 30801–30805. [DOI] [PubMed] [Google Scholar]
- Benecke A., Chambon,P. and Gronemeyer,H. (2000) Synergy between estrogen receptor α activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2. EMBO rep., 1, 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D., Broach,J.R., Tisell,L., Fletterick,R.J., Parker,M.G. and Chatterjee,V.K. (1993) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. [DOI] [PubMed] [Google Scholar]
- Cavailles V., Dauvois,S., L’Horset,F., Lopez,G., Hoare,S., Kushner,P.J. and Parker,M.G. (1995) Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J., 14, 3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D., LaMorte,V.J., Nelson,M.C., Nakajima,T., Schulman,I.G., Juguilon,H., Montminy,M. and Evans,R.M. (1996) Role of CBP/P300 in nuclear receptor signalling. Nature, 383, 99–103. [DOI] [PubMed] [Google Scholar]
- Chauchereau A., Georgiakaki,M., Perrin-Wolff,M., Milgrom,E. and Loosfelt,H. (2000) JAB1 interacts with both the progesterone receptor and SRC-1. J. Biol. Chem., 275, 8540–8548. [DOI] [PubMed] [Google Scholar]
- Chen D., Ma,H., Hong,H., Koh,S.S., Huang,S.M., Schurter,B.T., Aswad,D.W. and Stallcup,M.R. (1999) Regulation of transcription by a protein methyltransferase. Science, 284, 2174–2177. [DOI] [PubMed] [Google Scholar]
- Chen S.L., Dowhan,D.H., Hosking,B.M. and Muscat,G.E. (2000) The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes Dev., 14, 1209–1228. [PMC free article] [PubMed] [Google Scholar]
- Chiba H., Muramatsu,M., Nomoto,A. and Kato,H. (1994) Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res., 22, 1815–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley S.M., Hoare,S., Mosselman,S. and Parker,M.G. (1997) Estrogen receptors α and β form heterodimers on DNA. J. Biol. Chem., 272, 19858–19862. [DOI] [PubMed] [Google Scholar]
- Crews S.T. and Fan,C.M. (1999) Remembrance of things PAS: regulation of development by bHLH–PAS proteins. Curr. Opin. Genet. Dev., 9, 580–587. [DOI] [PubMed] [Google Scholar]
- Decristofaro M.F., Betz,B.L., Rorie,C.J., Reisman,D.N., Wang,W. and Weissman,B.E. (2001) Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J. Cell Physiol., 186, 136–145. [DOI] [PubMed] [Google Scholar]
- Di Croce L., Koop,R., Venditti,P., Westphal,H.M., Nightingale,K.P., Corona,D.F., Becker,P.B. and Beato,M. (1999a) Two-step synergism between the progesterone receptor and the DNA-binding domain of nuclear factor 1 on MMTV minichromosomes. Mol. Cell, 4, 45–54. [DOI] [PubMed] [Google Scholar]
- Di Croce L. et al. (1999b) Steroid and nuclear receptors. EMBO J., 18, 6201–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth F.J. and Chambon,P. (2001) Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene, 20, 3047–3054. [DOI] [PubMed] [Google Scholar]
- Dilworth F.J., Fromental-Ramain,C., Yamamoto,K., Chambon,P., Wu,X., Li,H. and Chen,J.D. (2000) ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR in vitro. Mol. Cell, 6, 1049–1058. [DOI] [PubMed] [Google Scholar]
- DiRenzo J., Shang,Y., Phelan,M., Sif,S., Myers,M., Kingston,R. and Brown,M. (2000) BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol. Cell. Biol., 20, 7541–7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal P.A. et al. (1994) BRCA1 mutations in primary breast and ovarian carcinomas. Science, 266, 120–122. [DOI] [PubMed] [Google Scholar]
- Heery D.M., Kalkhoven,E., Hoare,S. and Parker,M.G. (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature, 387, 733–736. [DOI] [PubMed] [Google Scholar]
- Kalkhoven E., Valentine,J.E., Heery,D.M. and Parker,M.G. (1998) Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J., 17, 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y. et al. (1996) A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell, 85, 403–414. [DOI] [PubMed] [Google Scholar]
- Kuo M.H., Brownell,J.E., Sobel,R.E., Ranalli,T.A., Cook,R.G., Edmondson,D.G., Roth,S.Y. and Allis,C.D. (1996) Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature, 383, 269–272. [DOI] [PubMed] [Google Scholar]
- Lahooti H., White,R., Hoare,S.A., Rahman,D., Pappin,D.J. and Parker,M.G. (1995) Identification of phosphorylation sites in the mouse oestrogen receptor. J. Steroid Biochem. Mol. Biol., 55, 305–313. [DOI] [PubMed] [Google Scholar]
- Lee H.L. and Archer,T.K. (1998) Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J., 17, 1454–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K. et al. (1999) A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J. Biol. Chem., 274, 34283–34293. [DOI] [PubMed] [Google Scholar]
- Lees J.A., Fawell,S.E., Parker,M.G., Wong,J., Tsai,S.Y., Tsai,M.J. and O’Malley,B.W. (1989) Identification of two transactivation domains in the mouse oestrogen receptor. Nucleic Acids Res., 17, 5477–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon B., Inouye,C., King,D.S. and Tjian,R. (2001) Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature, 414, 924–928. [DOI] [PubMed] [Google Scholar]
- Leo C. and Chen,J.D. (2000) The SRC family of nuclear receptor coactivators. Gene, 245, 1–11. [DOI] [PubMed] [Google Scholar]
- Li H., Gomes,P.J. and Chen,J.D. (1997) RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc. Natl Acad. Sci. USA, 94, 8479–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt M.D., Simpson,M., Recillas-Targa,F., Prioleau,M.N., Felsenfeld,G., Parker,M.G. and Chatterjee,V.K. (2001) Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J., 20, 2224–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wong,J., Tsai,S.Y., Tsai,M.J. and O’Malley,B.W. (2001) Sequential recruitment of steroid receptor coactivator-1 (SRC-1) and p300 enhances progesterone receptor-dependent initiation and reinitiation of transcription from chromatin. Proc. Natl Acad. Sci. USA, 98, 12426–12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak H.Y., Hoare,S., Henttu,P.M. and Parker,M.G. (1999) Molecular determinants of the estrogen receptor–coactivator interface. Mol. Cell. Biol., 19, 3895–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf D.J. et al. (1995) The nuclear receptor superfamily: the second decade. Cell, 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna N.J., Lanz,R.B. and O’Malley,B.W. (1999) Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev., 20, 321–344. [DOI] [PubMed] [Google Scholar]
- Muchardt C. and Yaniv,M. (1993) A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J., 12, 4279–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson S. et al. (2001) Mechanisms of estrogen action. Physiol. Rev., 81, 1535–1565. [DOI] [PubMed] [Google Scholar]
- Papoulas O., Daubresse,G., Armstrong,J.A., Jin,J., Scott,M.P. and Tamkun,J.W. (2001) The HMG-domain protein BAP111 is important for the function of the BRM chromatin-remodeling complex in vivo. Proc. Natl Acad. Sci. USA, 98, 5728–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S.Y., Denu,J.M. and Allis,C.D. (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu,X., DiRenzo,J., Lazar,M.A. and Brown,M. (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell, 103, 843–852. [DOI] [PubMed] [Google Scholar]
- Tora L., White,J., Brou,C., Tasset,D., Webster,N., Scheer,E. and Chambon,P. (1989) The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell, 59, 477–487. [DOI] [PubMed] [Google Scholar]
- Torchia J., Rose,D.W., Inostroza,J., Kamei,Y., Westin,S., Glass,C.K. and Rosenfeld,M.G. (1997) The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature, 387, 677–684. [DOI] [PubMed] [Google Scholar]
- Underhill C., Qutob,M.S., Yee,S.P. and Torchia,J. (2000) A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem., 275, 40463–40470. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz P. (2001) ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene, 20, 3076–3085. [DOI] [PubMed] [Google Scholar]
- Voegel J.J., Heine,M.J., Zechel,C., Chambon,P. and Gronemeyer,H. (1996) TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J., 15, 3667–3675. [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. (2001) Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science, 293, 853–857. [DOI] [PubMed] [Google Scholar]
- Wang W., Chi,T., Xue,Y., Zhou,S., Kuo,A. and Crabtree,G.R. (1998) Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl Acad. Sci. USA, 95, 492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Li,H. and Chen,J.D. (2001) The human homologue of the yeast DNA repair and TFIIH regulator MMS19 is an AF-1-specific coactivator of estrogen receptor. J. Biol. Chem., 276, 23962–23968. [DOI] [PubMed] [Google Scholar]
- Xue Y., Canman,J.C., Lee,C.S., Nie,Z., Yang,D., Moreno,G.T., Young,M.K., Salmon,E.D. and Wang,W. (2000) The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl Acad. Sci. USA, 97, 13015–13020. [DOI] [PMC free article] [PubMed] [Google Scholar]