Identification of Four Gene Variants Associated with Myocardial Infarction (original) (raw)
Abstract
Family history is a major risk factor for myocardial infarction (MI). However, known gene variants associated with MI cannot fully explain the genetic component of MI risk. We hypothesized that a gene-centric association study that was not limited to candidate genes could identify novel genetic associations with MI. We studied 11,053 single-nucleotide polymorphisms (SNPs) in 6,891 genes, focusing on SNPs that could influence gene function to increase the likelihood of identifying disease-causing gene variants. To minimize false-positive associations generated by multiple testing, two studies were used to identify a limited number of nominally associated SNPs; a third study tested the hypotheses that these SNPs are associated with MI. In the initial study (of 340 cases and 346 controls), 637 SNPs were associated with MI (P<.05); these were evaluated in a second study (of 445 cases and 606 controls), and 31 of the 637 SNPs were associated with MI (P<.05) and had the same risk allele as in the first study. For each of these 31 SNPs, we tested the hypothesis that it is associated with MI, using a third study (of 560 cases and 891 controls). We found that four of these gene variants were associated with MI (P<.05; false-discovery rate <10%) and had the same risk allele as in the first two studies. These gene variants encode the cytoskeletal protein palladin (KIAA0992 [odds ratio (OR) 1.40]), a tyrosine kinase (ROS1 [OR 1.75]), and two G protein–coupled receptors (TAS2R50 [OR 1.58] and OR13G1 [OR 1.40]); all ORs are for carriers of two versus zero risk alleles. These findings could lead to a better understanding of MI pathophysiology and improved patient risk assessment.
Introduction
Myocardial infarction (MI) is a prevalent and often fatal manifestation of coronary heart disease. MI occurs when thrombosis, induced by a ruptured or eroded atherosclerotic plaque, occludes a coronary artery, which leads to necrosis of the myocardium. Because of the high prevalence of risk factors, each year, ∼865,000 Americans are diagnosed with MI and ∼180,000 will die from the disease (American Heart Association 2002).
MI is a complex disease with a strong genetic component. Gene variants affecting traditional risk factors for MI, such as hypertension, hypercholesterolemia, and diabetes, have been described (reviewed by Lusis et al. [2004]). Family history of disease is a risk factor independent of traditional risk factors and is driven largely by genetic variation (Shea et al. 1984; Marenberg et al. 1994). The complexity of MI is evidenced by the many cell types that are involved in the formation of atherosclerotic plaques (Faxon et al. 2004; Lusis et al. 2004) and by the multiple processes that can affect risk of MI, such as inflammation, plaque calcification, extracellular matrix turnover, apoptosis, and thrombosis (Naghavi et al. 2003). Given this complexity, it is not obvious which genes will harbor the genetic variation responsible for the genetic component of MI.
Previous efforts to identify gene variants associated with MI have included both linkage studies and case-control association studies. Genomewide linkage studies of coronary heart disease or MI have identified linkage peaks in several chromosomal regions (reviewed by Lusis et al. [2004]). However, detailed analysis of these regions will be required to determine if they contain gene variants involved in disease. Most case-control association studies have focused on variants in genes related to known risk factors for coronary heart disease. Gene variants involved in dyslipidemia (e.g., apolipoprotein E [_APOE_], low-density lipoprotein receptor [_LDLR_], apolipoprotein(a) [_LPA_], and hepatic lipase [_LIPC_]) and hypertension (e.g., angiotensinogen [_AGT_], angiotensin converting enzyme [_ACE_], angiotensin II receptor, type 1 [_AGTR1_], and β2-adrenergic receptor [_ADRB2_]) have been found to be associated with disease. Gene variants have also shown association with MI in case-control association studies of candidate genes selected on the basis of disease pathophysiology. These include paroxonase 1 (PON1), toll-like receptor 4 (TLR4), and matrix metalloproteinase 3 (MMP3) (for recent reviews of candidate genes implicated in coronary heart disease, see the works of Gibbons et al. [2004] and Lusis et al. [2004]). The complex pathophysiology of MI suggests the involvement of multiple genes; therefore, an extensive search of human gene variants could lead to novel associations with MI. Ozaki et al. (2002) conducted a broad survey of 65,671 gene-based SNPs in a case-control association study of Japanese subjects and found several SNPs in the lymphotoxin-α (LTA) gene to be associated with MI. However, only 1,491 of the SNPs in that survey were nonsynonymous SNPs, and it has been suggested (Botstein and Risch 2003) that broad association studies would benefit from focusing on SNPs that are likely to change protein function or expression.
It seems unlikely that those gene variants shown so far to have convincing, reproducible association with MI can fully explain the genetic component of risk for MI. We therefore hypothesized that we could identify novel gene variants associated with MI in an extensive gene-centric association study that was not limited to candidate genes. We reasoned that testing SNPs that could influence gene function—for example, those affecting the amino acid sequence of the encoded protein or SNPs located in transcription-factor binding sites—would increase the likelihood of identifying disease-causing gene variants (Risch and Merikangas 1996; Botstein and Risch 2003).
Material and Methods
Strategy
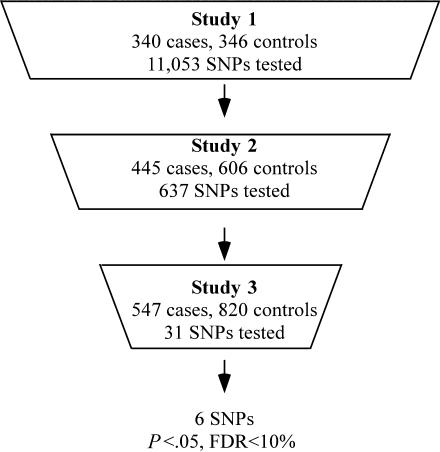
We conducted two sequential case-control studies (study 1 and study 2) to identify SNPs putatively associated with MI and then a third study (study 3) to test the hypotheses that these SNPs are associated with MI (fig. 1). This approach was used because of the difficulties often encountered in reproducing reported SNP associations with common complex diseases (Hirschhorn and Daly 2005). In study 1, DNA samples from cases and controls were pooled separately to increase the number of SNPs that could be tested (Sham et al. 2002). The allele frequency of each SNP was determined in pools of DNA (Germer et al. 2000). SNPs associated with MI in study 1 (P<.05) were evaluated in study 2 by use of a similar pooling strategy. SNPs associated with MI in study 2 (P<.05) and with the same risk allele as in study 1 were then tested for association with MI by genotyping individual DNA samples of study 3 subjects. Since testing multiple SNPs in study 3 could have led to false-positive associations due to multiple testing, we estimated the false-discovery rate (FDR) in study 3 and used an FDR of <10% as a criterion for further analysis of SNPs associated with disease. The three studies included only whites, to minimize the likelihood of population stratification.
Figure 1.
Diagram of the process used to reduce the number of hypotheses tested in subsequent studies. Study size and the number of SNPs tested in each study are indicated. Only SNPs with unadjusted P values <.05 for association with MI advanced to the next study. Additionally, only SNPs with the same risk allele in study 1 and study 2 advanced to study 3. Six SNPs in study 3 had P values <.05 for association with MI and an FDR <10%.
SNP Selection
We determined the allele frequency of 11,053 SNPs (in 6,891 genes) in study 1. These SNPs are located in genes and have the potential to affect gene function or expression. The majority (7,496) of these SNPs are nonsynonymous SNPs, expected to modify the amino acid sequence encoded by the gene. Other SNPs are located in transcription-factor binding sites or in UTRs of mRNA, which we reasoned could affect mRNA expression or stability. The SNPs tested were selected from public databases, from the Celera database (see the Applied Biosystems Web site), and from SNPs identified by sequence analysis of ∼24,000 genes in 39 individuals (Clark et al. 2003). We excluded SNPs that had <1% allele frequency in a white population, as well as SNPs that were described in a single public database and lacked white allele-frequency information.
Allele Frequency and Genotype Determination
Allele frequencies in pooled DNA were determined by kinetic PCR (Germer et al. 2000). In brief, DNA concentrations were standardized to 10 ng/μl with use of PicoGreen (Molecular Probes) fluorescent dye (Singer et al. 1997). DNA pools, typically from 50 cases or controls, were made by mixing equal volumes of standardized DNA from each individual member of the pool. With 3 ng of pooled DNA, one of two allele-specific primers, and a common reverse primer, each allele was amplified separately by PCR with use of an Applied Biosystems PRISM 7900HT Sequence Detection System. The allele frequency was calculated from the two allele-specific PCR amplification curves (Germer et al. 2000) on the basis of at least eight independent pools of DNA, each determined in duplicate. Individual genotyping was similarly performed using 0.3 ng of DNA. All genotyping was performed at the Celera Diagnostics high-throughput genotyping facility. Genotyping accuracy in this facility is typically >99%, as reported elsewhere (Li et al. 2004). The genotyping accuracy for the four gene variants reported here was confirmed to be >99.5%, by repeat genotyping of samples from study 3 with use of an oligonucleotide ligation assay, which is an independent genotyping technology.
Subjects
Cases had a history of MI; controls had no history of MI. Since genetic makeup is a more important MI risk factor in younger than in older individuals (Marenberg et al. 1994), we preferentially recruited cases who experienced an MI at a young age. Participants in study 1 (males) and study 3 (males and females) were enrolled between July 1989 and October 2003 by the University of California–San Francisco (UCSF) Genomic Resource in Arteriosclerosis. Participants included patients who underwent diagnostic or interventional cardiac catheterization, patients of the UCSF Lipid Clinic, and healthy older individuals. Participants in study 2 (males and females) were patients of the Cleveland Clinic Foundation Heart Center who had undergone diagnostic or interventional cardiac catheterization between July 2001 and March 2003. All subjects gave informed consent and completed an institutional review board–approved questionnaire. All subjects chose “white” as their ethnicity in response to a multiple-choice questionnaire; subjects who chose “Hispanic” or other nonwhite ethnicities were excluded from the study.
Study 1 subjects were aged <75 years. Cases had a history of MI, as defined by The International Classification of Diseases, 9th Revision (ICD9) codes for MI (codes 410 or 411); by a clinical chart review; or by a self-reported history. An ongoing effort by UCSF physicians to obtain clinical verification of the MI status for patients who had self-reported an MI has resulted in >98% verification. Controls had no history of MI or unstable angina. Most controls (95%) had undergone cardiac catheterization, and 63% of the controls had stable angina.
Study 2 cases were males aged <66 years and females aged <75 years who had a history of MI that had been verified by electrocardiogram, cardiac enzymes, or perfusion imaging. Controls had no history of MI and had <50% coronary luminal narrowing, on the basis of clinical angiography, and 81% had stable angina.
Study 3 cases had a history of MI verified by ICD9 codes for MI (codes 410 or 411) or a clinical chart review. Controls had no history of MI, symptomatic vascular disease, or diabetes and did not have a known first-degree relative with a history of symptomatic coronary disease prior to age 65 years; 10% of the controls had undergone cardiac catheterization. Since we had no way of knowing whether young controls in the study would confound the analysis by experiencing an MI shortly after their recruitment, we recruited older individuals for the study 3 control group (see table 1).
Table 1.
Distribution of Traditional MI Risk Factors[Note]
| Study 1 | Study 2 | Study 3 | ||||
|---|---|---|---|---|---|---|
| Characteristics | Cases(_n_=340) | Controls(_n_=346) | Cases(_n_=445) | Controls(_n_=606) | Cases(_n_=560) | Controls(_n_=891) |
| Percentage male | 100 | 100 | 62 | 62 | 45 | 37a |
| Age (years) | 63±10 | 61±10a | 60±7 | 59±10 | 62±10 | 66±13a |
| Age (in years) at MI | 54±10 | NA | 53±10b | NA | 52±9 | NA |
| Percentage with smoking history | 66 | 60 | 74 | 54c | 65 | 43c |
| Percentage with diabetes | 22 | 14a | 39 | 10c | 20 | 0d |
| Percentage with dyslipidemiae | 86 | 76c | 97 | 57c | 83 | 53c |
| Percentage with hypertensionf | 59 | 52 | 96 | 78c | 62 | 33c |
| BMI (kg/m2) | 28±5 | 27±5 | 31±6 | 30±7c | 28±6 | 26±5c |
Statistical Analysis
The Wilcoxon rank sum test was used to assess associations between continuous phenotypes and disease status, and the Kruskal-Wallis test was used to assess associations of continuous phenotypes with genotypes. Differences of proportions for categorical phenotypes and association between MI status and allele frequencies were assessed by two-tailed Fisher’s exact tests. Associations between diabetes status and MI risk-allele frequencies were assessed by one-tailed Fisher’s exact tests. The exact tests for pooled allele frequencies were performed by rounding the estimated allele counts to the nearest integer. Assessment of genotypic association, including comparisons of risk homozygotes to heterozygotes as well as adjustment of odds ratios (ORs) for other covariables, was performed using logistic regression. P values from logistic regression were calculated using a Wald test. Two-sided P values are presented for comparisons of risk homozygotes to heterozygotes. One-sided P values are presented for study 3 results when the a priori hypothesis was that the risk allele for the association matches the risk allele observed in the previous two studies. Values in the FDR column reported in table 2, denoted here by Q i for the _i_th value in increasing order, were calculated using the MULTTEST procedure (Benjamini and Hochberg 1995) as follows: given m hypotheses _H_1,_H_2,…,H m and corresponding P values _P_1,_P_2,…,P m, the P values are ordered such that P m_⩾_P(_m_-1)⩾…⩾_P_1. Then let Q _m_=P m,Q(_m_-1)=min(Q m,P(_m_-1)×[_m_÷(_m_-1)]),…,Q(_m_-j)=min(Q(_m_-j+1),P(_m_-j)×[_m_÷(_m_-j)]),…,_Q_1=min(_Q_2,_m_×_P_1). Population stratification was assessed by randomly drawing 1,000 SNPs from among the 11,053 SNPs evaluated in study 1 and then by using the method of Reich and Goldstein (2001) to estimate an inflation factor λ for the χ2 test statistic. We repeated this random draw 100 times, and we report the mean λ from the 100 draws. We report an approximate 95% CI for λ that extends 1.96 times the SD of λ on either side of the mean.
Table 2.
FDR for SNPs Tested in Study 3
| P | ||||
|---|---|---|---|---|
| SNP | Gene | Study 2a | Study 3b | FDR |
| rs1376251 | TAS2R50 | .013 | .0018 | .03 |
| rs12510359 | KIAA0992/palladin | .006 | .0028 | .03 |
| rs4804611 | ZNF627 | .013 | .0034 | .03 |
| rs529038 | ROS1 | .010 | .0067 | NAc |
| rs619203 | ROS1 | .022 | .012 | .07 |
| rs1151640 | OR13G1 | .009 | .013 | .07 |
| rs2290526 | TOX | .023 | .048 | .23 |
| rs3776096 | PCDHB6 | .002 | .09 | .35 |
| Chromosome 10:103890719 G/Ad | PPRC1 | .049 | .10 | .35 |
| rs1799883 | FABP2 | .014 | .13 | .40 |
| rs1861956 | FLJ40217 | .035 | .14 | .40 |
| rs10455 | CYBRD1 | .040 | .18 | .41 |
| rs707602 | TPM1 | .003 | .18 | .41 |
| Chromosome 15:39585434 C/Td | ITPKA | .039 | .19 | .41 |
| rs3820594 | SYT11 | .036 | .23 | .47 |
| rs3129272 | HLA-DPB2 | .039 | .31 | .61 |
| rs6691840 | GRIK3 | .007 | .37 | .66 |
| rs1054735 | FLJ45300 | .015 | .42 | .66 |
| rs11955611 | FLJ46010 | .036 | .42 | .66 |
| rs236212 | Nonee | .046 | .44 | .66 |
| rs4875 | MLF1 | .024 | .46 | .66 |
| rs2747701 | KIAA1411 | .038 | .51 | .70 |
| rs2240089 | COBL | .005 | .55 | .73 |
| rs6788438 | LXN | .006 | .56 | NAc |
| rs6512265 | LRRC25 | .003 | .62 | .78 |
| rs7905784 | MCM10 | .009 | .77 | .93 |
| Chromosome 17:2174405 G/Td | FLJ10534 | .036 | .89 | .95 |
| rs6546 | C1QTNF3 | .008 | .92 | .95 |
| rs1611775 | KCNA7 | .015 | .93 | .95 |
| rs11117358 | BANP | .029 | .94 | .95 |
| rs1124649 | FLJ20254 | .021 | .95 | .95 |
Results
Study Populations
The prevalence of traditional risk factors was generally higher in the MI case group than in the control group in all three studies (table 1). In study 1, cases were significantly older than controls and the prevalence of dyslipidemia and diabetes was significantly higher in the case group than in the control group. BMI and the prevalence of hypertension and smoking trended higher. In study 1, we investigated potential population stratification, which could influence the matching of the case and control samples, and found no significant evidence of stratification—the χ2 test statistic inflation factor was not significantly different from 1.0 (λ=1.06; 95% CI .96–1.16). In study 2, BMI and the prevalence of smoking, diabetes, dyslipidemia, and hypertension were significantly higher in the case group than in the control group. In study 3, BMI and the prevalence of smoking, dyslipidemia, and hypertension were significantly higher in the case group than in the control group. The mean age was higher in the study 3 control group than in the study 3 case group because of the recruitment of older controls. In all three studies, the mean age at MI was <55 years, which reflects the preferential enrollment of cases who experienced an MI at a young age.
Gene Variants Associated with MI
In study 1, the first hypotheses-generating study, we tested which of 11,053 SNPs were associated with MI by comparing the allele frequency of each SNP in cases with the frequency in controls. We identified 637 of these SNPs (in 593 genes) as candidates for further investigation in study 2, using an unadjusted P value of <.05 for association with MI as the selection criterion (fig. 1; see the authors' Web site for a supplemental PDF document). We then used study 2, the second hypotheses-generating study, to identify the set of SNPs to be tested in study 3, the hypotheses-testing study. The criteria used to select SNPs for study 3 were that the SNPs had the same risk allele in both study 1 and study 2 and also had an unadjusted _P_ value of <.05 in both studies. We found that 31 of the 637 SNPs met these criteria. These 31 SNPs are located in 30 different genes (table 2). Two of the SNPs tested in study 2 had been reported elsewhere to be associated with coronary heart disease—_rs1041981_ in _LTA_ (Ozaki et al. 2002) and _rs3732563_ in _AGTR1_ (Jones et al. 2003). However, the _LTA_ and _AGTR1_ SNPs had unadjusted _P_ values >.7 in study 2 and therefore were not tested in study 3.
For each of the 31 SNPs we identified in study 2, we tested, in study 3, the hypothesis that the SNP is associated with MI. Since the putative risk allele of each of these 31 SNPs had been identified in the hypotheses-generating portion of this investigation (study 1 and study 2), we tested, in study 3, the specific hypothesis that the same risk allele is associated with risk of MI. For FDR calculations, we considered only 29 of the 31 SNPs tested in study 3, for the following reasons. The two SNPs in ROS1 on chromosome 6 were not independent of one another. They were concordant in all but four individuals in study 3; that is, each individual was a minor homozygote for both SNPs, a major homozygote for both SNPs, or a heterozygote for both SNPs, so we included only one (rs619203) in FDR calculations and in subsequent analyses. Similarly, two SNPs on chromosome 3 (rs4875 and rs6788438) are only 67 kb apart and were discordant in only five individuals in study 3; we included only one of these SNPs in FDR calculations.
The SNPs with an FDR <10% were in palladin, a cytoskeletal protein (KIAA0992 [MIM 608092]), a tyrosine kinase (ROS1 [MIM 165020], 2 SNPs), two G protein-coupled receptors (TAS2R50 and OR13G1), and a zinc finger protein (ZNF627) (table 3). Four of the six SNPs cause amino acid residue substitutions. The SNP in palladin is located in an intron, and the SNP in ZNF627 is located in the 3′ UTR of the gene. Each SNP had the same risk allele as well as a similar magnitude of risk in all three studies (table 4).
Table 3.
Characteristics of SNPs Associated with MI
| RefSNP Number | Gene Name | Chromosome | BaseChangea | SNP Type |
|---|---|---|---|---|
| rs12510359 | KIAA0992/palladin | 4 | G/A | Intron |
| rs529038 | ROS1 | 6 | A/G | Asn2213Asp |
| rs619203 | ROS1 | 6 | G/C | Cys2229Ser |
| rs1376251 | TAS2R50 | 12 | C/T | Cys203Tyr |
| rs1151640 | OR13G1 | 1 | A/G | Ile132Val |
| rs4804611 | ZNF627 | 19 | A/G | 3′ UTR |
Table 4.
Allelic Association with MI in Three Studies
| Study 1 | Study 2 | Study 3 | ||||
|---|---|---|---|---|---|---|
| Gene Name (SNP) | ORa (95% CI) | Pb | OR (95% CI) | Pb | OR (90% CI)c | Pd |
| KIAA0992/palladin (rs12510359) | 1.28 (1.02–1.59) | .031 | 1.29 (1.07–1.55) | .006 | 1.25 (1.10–1.43) | .0028 |
| ROS1 (rs619203) | 1.28 (1.01–1.64) | .046 | 1.34 (1.08–1.66) | .010 | 1.23 (1.06–1.42) | .012 |
| TAS2R50 (rs1376251) | 1.27 (1.01–1.59) | .049 | 1.27 (1.05–1.54) | .013 | 1.28 (1.11–1.46) | .0018 |
| OR13G1 (rs1151640) | 1.26 (1.01–1.55) | .034 | 1.27 (1.06–1.50) | .009 | 1.19 (1.05–1.36) | .013 |
| ZNF627 (rs4804611) | 1.33 (1.05–1.68) | .019 | 1.29 (1.06–1.57) | .013 | 1.25 (1.09–1.44) | .0034 |
MI Risk Associated with Genotypes
We estimated the risk of MI associated with each genotype of the five gene variants described in table 4. Since the data did not support the use of a dominant, additive, or recessive model for all of these SNPs, we show only genotypic results for all SNPs, to provide a uniform presentation. When a particular genetic model could have been justified, a model-based analysis would have resulted in lower P values. Carriers of one or two risk alleles of the ZNF627 SNP were at a significantly increased risk of MI versus those who did not carry a risk allele. Likewise, carriers of one or two risk alleles of the OR13G1 SNP were at a significantly increased risk of MI versus those who did not carry a risk allele (table 5). The difference in risk between the two risk genotypes was not significant (_P_=.30 for ZNF627; _P_=.66 for OR13G1); thus, the heterozygotes as well as the risk homozygotes were at similar and significantly increased risk of MI. In the case of the TAS2R50, palladin, and ROS1 variants, the risk of MI for carriers of two risk alleles was greater than the risk for carriers of one risk allele (for comparison of MI risk of the risk homozygotes vs. the heterozygote reference group, _P_=.002 for palladin; _P_=.03 for TAS2R50; _P_=.08 for ROS1).
Table 5.
Genotypic Association with MI in Study 3
| Findings with Adjustment | ||||||||
|---|---|---|---|---|---|---|---|---|
| No. (%) of Subjects | None | Risk Factorsa | RiskFactorsand SNPsb | |||||
| Gene andGenotype | Case | Control | ORc | Pd | OR | Pd | OR | Pd |
| KIAA092/palladin: | ||||||||
| AA | 73 (13.1) | 132 (14.9) | ||||||
| GA | 233 (41.7) | 430 (48.4) | .98 | .47 | .97 | .56 | .94 | .62 |
| GG | 253 (45.3) | 326 (36.7) | 1.40 | .024 | 1.39 | .04 | 1.35 | .07 |
| ROS1: | ||||||||
| GG | 298 (54.1) | 522 (58.9) | ||||||
| GC | 213 (38.7) | 324 (36.6) | 1.15 | .11 | 1.21 | .078 | 1.21 | .09 |
| CC | 40 (7.3) | 40 (4.5) | 1.75 | .010 | 1.54 | .051 | 1.49 | .07 |
| TAS2R50: | ||||||||
| TT | 54 (9.8) | 116 (13.1) | ||||||
| CT | 226 (40.9) | 396 (44.8) | 1.23 | .14 | 1.06 | .39 | 1.09 | .34 |
| CC | 273 (49.4) | 372 (42.1) | 1.58 | .007 | 1.37 | .06 | 1.40 | .06 |
| OR13G1: | ||||||||
| AA | 154 (27. 8) | 293 (34.0) | ||||||
| AG | 286 (51.7) | 416 (48.2) | 1.31 | .017 | 1.26 | .056 | 1.37 | .002 |
| GG | 113 (20.4) | 154 (17.8) | 1.40 | .019 | 1.39 | .036 | 1.40 | .04 |
| ZNF627: | ||||||||
| GG | 49 (8.8) | 118 (13.4) | ||||||
| AG | 224 (40.1) | 359 (40.7) | 1.50 | .018 | 1.63 | .014 | 1.57 | .02 |
| AA | 285 (51.1) | 405 (45.9) | 1.69 | .002 | 1.95 | .001 | 1.99 | .001 |
We investigated whether the risk associated with each of these five gene variants was confounded by either traditional risk factors or the other four gene variants. Using multivariate analysis, we estimated the risk associated with each genotype, after adjustment for five traditional risk factors: dyslipidemia, sex, smoking history, hypertension, and BMI (table 5). Adjustment of the genotypic risk for traditional risk factors, either alone or in addition to the other four gene variants, resulted in modest changes in the risk estimates for these SNPs (all resultant adjusted P values were <.1). Since we had intentionally selected controls who were somewhat older than the cases in study 3, we asked whether age was associated with any of the five SNPs in the control group. We found an association of young age with the risk allele of the SNP in _ZNF627_ (_P_=.0005). The other four SNPs were not associated with age (all _P_ values were >.24). Similarly, since we selected controls for study 3 who did not have diabetes, we asked whether the MI risk allele of any of the five SNPs was associated with diabetes in the case group of study 3. We found that there was a trend toward association with diabetes for the MI risk allele of the SNP in OR13G1 (_P_=.16). The association of this SNP with MI remained significant (P<.05) after exclusion of all subjects with diabetes from the analysis. The MI risk alleles of the other four SNPs were not associated with diabetes (all _P_ values were >.45).
Discussion
We identified four novel gene variants associated with MI. They encode a cytoskeletal protein (palladin [_KIAA0992_]), a tyrosine kinase (ROS1), and two G protein–coupled receptors (TAS2R50 and OR13G1). These genes have not been previously reported to be associated with MI and are not typical candidate genes for cardiovascular disease—which indicates the potential value of unbiased, gene-centric genetic association studies in the search for novel insights into cardiovascular disease pathogenesis.
Association studies that test thousands of SNPs without multiple-testing correction can lead to false-positive associations, whereas a conservative correction can lead to false-negative associations. We addressed this multiple-testing dilemma by interrogating a smaller number of SNPs in each of three successive studies and calculating the FDR for the SNPs tested in the third study. We used an FDR of 10% as a criterion for further analysis. The six SNPs with the lowest P values met this FDR criterion of 10% in study 3. The FDRs reported in table 2 were calculated by the method of Benjamini and Hochberg (1995); however, a simple, intuitive way to estimate the FDR of the six most significant SNPs is to estimate the number of expected false-positive SNPs and divide it by the observed number of SNPs. Multiplying the number of independent SNPs tested (29) by the highest P value (.013) in the group of the six most significant SNPs gives the expected number of false-positive SNPs (29×.013=0.377). The observed number of independent SNPs was five (excluding one of the two SNPs in ROS1); thus, the FDR estimate for the group estimated by this simplified method would be 0.075 (0.38÷5=0.075).
Mindful of the fact that conventional risk factors were enriched in the case groups as compared with the control groups, we investigated confounding by traditional risk factors. With use of multivariate analysis to adjust for dyslipidemia, sex, smoking history, hypertension, and BMI, none of these risk factors was found to be a strong confounder for the associations observed in study 3. Similarly, the risk estimates for each of these SNPs were only incrementally changed when adjusted for the risk associated with the other four gene variants. When association with age was evaluated in the study 3 control group, the putative MI risk allele of the SNP in ZNF627 was associated with young age; thus, the apparent association of this SNP with MI could be an artifact caused by the preferential recruitment of older controls, who were less likely to have the risk allele. However, one possible explanation for the association of the risk allele with young age would be that carriers of this allele have a shorter life expectancy, which could be due to increased risk of MI or some other disease. None of the other SNPs was associated with age. We also found that the MI risk allele of OR13G1 trended toward association with diabetes. However, the OR13G1 SNP remained associated with MI when subjects with diabetes were excluded from the case group, making it unlikely that the association of OR13G1 with MI is an artifact of selecting controls without diabetes for study 3. These results suggest that the palladin, ROS1, TAS2R50, and OR13G1 variants are associated with MI and that these associations are not confounded by dyslipidemia, hypertension, BMI, sex, or smoking history.
The presumed function of these genes gives some hints as to how they may be associated with MI; however, the precise biological mechanism that underlies these associations is unknown. For instance, palladin is a cytoskeletal protein that interacts with α-actinin and localizes to stress fibers, focal adhesions, and cell-cell junctions (Parast and Otey 2000; Ronty et al. 2004). A change in palladin expression or function could affect endothelial cell junctions, mononuclear cell trafficking into arterial walls, or platelet adhesion. ROS1, a receptor tyrosine kinase and a proto-oncogene (Birchmeier et al. 1987), is involved in cell proliferation and differentiation (Zong et al. 1997) and is expressed at low levels in most tissues (Boon et al. 2002); changes in cell proliferation could affect arterial plaque formation and stability. The underlying biological mechanisms for the association of the G protein–coupled receptors we describe (TAS2R50 and OR13G1) are not clear. However, TAS2R50 is a bitter taste–receptor homolog (Conte et al. 2003). Duffy et al. (2004) suggested that this family of receptors may contribute to the risk of cardiovascular disease by influencing dietary choices. Similarly, the association with MI of OR13G1, an olfactory receptor homolog, might be mediated through altered dietary preferences.
The association of some of these five SNPs with MI might be explained by linkage disequilibrium with other SNPs. In the case of palladin, ROS1, and OR13G1, the haplotype-block structure reported by the International HapMap Project (International HapMap Consortium 2003) indicates that these SNPs are located in haplotype blocks that do not encompass other genes. Thus, the available linkage data are consistent with the hypothesis that the variants that contribute to risk of MI are located within these three genes. In contrast, the SNP in TAS2R50 is located in a haplotype block of ∼500 kb that contains 13 taste-receptor homologs as well as three genes encoding proline-rich proteins. This raises the possibility that the risk of MI associated with the TAS2R50 SNP could be due to a SNP in one of these other genes.
It has been suggested that population stratification could confound the interpretation of case-control studies (Marchini et al. 2004). To reduce potential confounding of our results by population stratification, we studied only self-described white individuals, because several studies have reported only modest population stratification in white populations (Hao et al. 2004; Marchini et al. 2004; Helgason et al. 2005). Additionally, we conducted three different studies and enrolled the subjects at two independent sites. Thus, if the associations we observed were caused by population stratification, the stratification would have to have been of similar magnitude and direction in all three studies. We also estimated the potential magnitude of population stratification, using an inflation factor (λ) for the χ2 test statistic to estimate population stratification in study 1, and found no significant evidence of stratification. Thus, it seems unlikely that the associations we observed between MI and the genetic variants reported here can be explained by population stratification.
Several limitations were imposed by the samples used in this investigation. Subjects with a history of MI were recruited after the qualifying event. This introduced a survival bias; consequently, gene variants most strongly associated with fatal MI might not have been detected in this case-control study. In addition, study 1 subjects were all males; hence, it is unlikely that genetic markers that are primarily associated with MI in females would have been discovered in this study. The prevalence of traditional risk factors differed between cases and controls in all three studies (table 1), suggesting that differences in the prevalence of genetic risk factors could also be detectable in these studies. However, because of the retrospective nature of these studies, the traditional risk status prior to MI was typically not available, which may have resulted in overreporting traditional risk factors in the case group. Thus, a complete understanding of the relationship of traditional risk factors to the observed associations should be sought in a population-based cohort study. Nevertheless, the associations we identified could be a step toward an improved understanding of disease regardless of whether adjustment for traditional risk factors indicates that these novel associations contribute to pathways involving traditional risk factors or to other aspects of disease pathophysiology.
MI is a prevalent disease with high morbidity and mortality. However, changes in lifestyle as well as pharmacological intervention can markedly reduce an individual’s risk. Thus, better estimates of an individual’s risk could lead to more effective prevention and treatment. In this study, we have observed the association of four gene variants with MI. If confirmed in prospective, population-based studies, these markers—individually or in combination—may be useful in identifying individuals at a risk for MI higher than that predicted by traditional risk factors alone. Moreover, the SNPs we identified are located in genes that have not been previously implicated in coronary heart disease. Thus, investigation of the function of these genes in the context of coronary heart disease could lead to a better understanding of the pathophysiology of this complex disease and to new therapeutic strategies.
Acknowledgments
This study was funded by the University of California Discovery Grant Program, the Mildred V. Strouss Charitable Trust, the Joseph Drown Foundation, and gifts from Donald Yellon and from Judge and Mrs. Lawrence Mana. The authors thank Thomas White, Ann Begovich, Michele Cargill, Andrew Grupe, Maria Langdown, Linda McAllister, David Ross, Steve Schrodi, and Bradford Young for helpful comments on study design and on this manuscript.
Web Resources
The URLs for data presented herein are as follows:
- Applied Biosystems, http://myscience.appliedbiosystems.com/ (for the Celera database)
- Authors' Web site, http://www.celeradiagnostics.com/pdf/MI4_variants_supplemental.pdf
- International HapMap Project, http://www.hapmap.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for KIAA0992 and ROS1)
References
- American Heart Association (2002) Heart disease and stroke statistics: 2005 update. American Heart Association, Dallas [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:1289–1300 [Google Scholar]
- Birchmeier C, Sharma S, Wigler M (1987) Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc Natl Acad Sci USA 84:9270–9274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K, Osorio EC, Greenhut SF, Schaefer CF, Shoemaker J, Polyak K, Morin PJ, Buetow KH, Strausberg RL, De Souza SJ, Riggins GJ (2002) An anatomy of normal and malignant gene expression. Proc Natl Acad Sci USA 99:11287–11292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Risch N (2003) Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nat Genet Suppl 33:228–237 [DOI] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, Ferriera S, Wang G, Zheng X, White TJ, Sninsky JJ, Adams MD, Cargill M (2003) Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science 302:1960–1963 [DOI] [PubMed] [Google Scholar]
- Conte C, Ebeling M, Marcuz A, Nef P, Andres-Barquin PJ (2003) Evolutionary relationships of the Tas2r receptor gene families in mouse and human. Physiol Genomics 14:73–82 [DOI] [PubMed] [Google Scholar]
- Duffy VB, Lucchina LA, Bartoshuk LM (2004) Genetic variation in taste: potential biomarker for cardiovascular disease risk? In: Prescott J, Tepper BJ (eds) Genetic variations in taste sensitivity: measurement, significance and implications. Marcel Dekker, New York, pp 197–229 [Google Scholar]
- Faxon DP, Fuster V, Libby P, Beckman JA, Hiatt WR, Thompson RW, Topper JN, Annex BH, Rundback JH, Fabunmi RP, Robertson RM, Loscalzo J (2004) Atherosclerotic vascular disease conference: Writing Group III: pathophysiology. Circulation 109:2617–2625 [DOI] [PubMed] [Google Scholar]
- Germer S, Holland MJ, Higuchi R (2000) High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res 10:258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons GH, Liew CC, Goodarzi MO, Rotter JI, Hsueh WA, Siragy HM, Pratt R, Dzau VJ (2004) Genetic markers: progress and potential for cardiovascular disease. Circulation Suppl 109:47–58 [DOI] [PubMed] [Google Scholar]
- Hao K, Li C, Rosenow C, Wong WH (2004) Detect and adjust for population stratification in population-based association study using genomic control markers: an application of Affymetrix Genechip Human Mapping 10K array. Eur J Hum Genet 12:1001–1006 [DOI] [PubMed] [Google Scholar]
- Helgason A, Yngvadottir B, Hrafnkelsson B, Gulcher J, Stefansson K (2005) An Icelandic example of the impact of population structure on association studies. Nat Genet 37:90–95 [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ (2005) Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 6:95–108 [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796 [DOI] [PubMed] [Google Scholar]
- Jones A, Dhamrait SS, Payne JR, Hawe E, Li P, Toor IS, Luong L, Wootton PT, Miller GJ, Humphries SE, Montgomery HE (2003) Genetic variants of angiotensin II receptors and cardiovascular risk in hypertension. Hypertension 42:500–506 [DOI] [PubMed] [Google Scholar]
- Li Y, Tacey K, Doil L, van Luchene R, Garcia V, Rowland C, Schrodi S, Leong D, Lau K, Catanese J, Sninsky J, Nowotny P, Holmans P, Hardy J, Powell J, Lovestone S, Thal L, Owen M, Williams J, Goate A, Grupe A (2004) Association of ABCA1 with late-onset Alzheimer’s disease is not observed in a case-control study. Neurosci Lett 366:268–271 [DOI] [PubMed] [Google Scholar]
- Lusis AJ, Mar R, Pajukanta P (2004) Genetics of atherosclerosis. Annu Rev Genomics Hum Genet 5:189–218 [DOI] [PubMed] [Google Scholar]
- Marchini J, Cardon LR, Phillips MS, Donnelly P (2004) The effects of human population structure on large genetic association studies. Nat Genet 36:512–517 [DOI] [PubMed] [Google Scholar]
- Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U (1994) Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med 330:1041–1046 [DOI] [PubMed] [Google Scholar]
- Naghavi M, Libby P, Falk E, Casscells SW, Litovsky S, Rumberger J, Badimon JJ, et al (2003) From vulnerable plaque to vulnerable patient: a call for new definitions and risk assessment strategies: Part I. Circulation 108:1664–1672 [DOI] [PubMed] [Google Scholar]
- Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Sato H, Hori M, Nakamura Y, Tanaka T (2002) Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nat Genet 32:650–654 [DOI] [PubMed] [Google Scholar]
- Parast MM, Otey CA (2000) Characterization of palladin, a novel protein localized to stress fibers and cell adhesions. J Cell Biol 150:643–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Goldstein DB (2001) Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol 20:4–16 [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Ronty M, Taivainen A, Moza M, Otey CA, Carpen O (2004) Molecular analysis of the interaction between palladin and α-actinin. FEBS Lett 566:30–34 [DOI] [PubMed] [Google Scholar]
- Sham P, Bader JS, Craig I, O’Donovan M, Owen M (2002) DNA Pooling: a tool for large-scale association studies. Nat Rev Genet 3:862–871 [DOI] [PubMed] [Google Scholar]
- Shea S, Ottman R, Gabrieli C, Stein Z, Nichols A (1984) Family history as an independent risk factor for coronary artery disease. J Am Coll Cardiol 4:793–801 [DOI] [PubMed] [Google Scholar]
- Singer VL, Jones LJ, Yue ST, Haugland RP (1997) Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantitation. Anal Biochem 249:228–238 [DOI] [PubMed] [Google Scholar]
- Zong CS, Chan JL, Yang SK, Wang LH (1997) Mutations of Ros differentially effecting signal transduction pathways leading to cell growth versus transformation. J Biol Chem 272:1500–1506 [DOI] [PubMed] [Google Scholar]