Functional Analysis of the Neurofibromatosis Type 2 Protein by Means of Disease-Causing Point Mutations (original) (raw)
Abstract
Despite intense study of the neurofibromatosis type 2 (NF2) tumor-suppressor protein merlin, the biological properties and tumor-suppressor functions of merlin are still largely unknown. In this study, we examined the molecular activities of NF2-causing mutant merlin proteins in transfected mammalian cells, to elucidate the merlin properties that are critical for tumor-suppressor function. Most important, we found that 80% of the merlin mutants studied significantly altered cell adhesion, causing cells to detach from the substratum. This finding implies a function for merlin in regulating cell-matrix attachment, and changes in cell adhesion caused by mutant protein expression may be an initial step in the pathogenesis of NF2. In addition, five different mutations in merlin caused a significant increase in detergent solubility of merlin compared to wild type, indicating a decreased ability to interact with the cytoskeleton. Although not correlated to the cell-adhesion phenotype, four missense mutations decreased the binding of merlin to the ERM-interacting protein EBP-50, implicating this interaction in merlin inhibition of cell growth. Last, we found that some NF2 point mutations in merlin most closely resembled gain-of-function alleles in their cellular phenotype, which suggests that mutant NF2 alleles may not always act in a loss-of-function manner, as had been assumed, but may include a spectrum of allelic types with different phenotypic effects on the function of the protein. In aggregate, these cellular phenotypes provide a useful assay for identifying the functional domains and molecular partners necessary for merlin tumor-suppressor activity.
Introduction
Neurofibromatosis type 2 (NF2) (MIM 101000) is an autosomal disorder characterized by the formation of nervous system tumors—specifically, bilateral schwannomas of the eighth cranial nerve and, to a lesser extent, schwannomas of other cranial and peripheral nerves and spinal nerve roots, meningiomas, ependymonas, and gliomas, as well as posterior lens-capsule opacities and retinal abnormalities (Martuza and Eldridge 1988; Kaiser-Kupfer et al. 1989; Evans et al. 1992). Mutational analysis of the NF2 gene in inherited and sporadic schwannomas, meningiomas, and ependymonas has shown that both copies of NF2 are often mutated and/or deleted in >50% of tumors, which suggests that the NF2 protein has a tumor-suppressor activity (Rouleau et al. 1993; Trofatter et al. 1993; Bourn et al. 1994; Jacoby et al. 1994; MacCollin et al. 1994). In support of the NF2 protein’s proposed role as negative regulator of cell growth, overexpression of the wild-type NF2 gene inhibits cell growth both in vitro and in vivo (Lutchman and Rouleau 1995; Sherman et al. 1997) and reverses the Ras-induced phenotype of anchorage-independent growth in v-Has-Ras transformed fibroblasts (Tikoo et al. 1994).
The NF2 protein, nicknamed “merlin,” shares 45% sequence identity with the family proteins ezrin, radixin, and moesin (ERM) (MIM 309845), which link the actin cytoskeleton to plasma membrane proteins at specialized, dynamic regions (Lamb et al. 1997; Martin et al. 1997). Like the ERM proteins, merlin localizes to the plasma membrane at regions rich in filamentous actin, both in cultured cells and endogenous tissues (den Bakker et al. 1995_a_; Gonzalez-Agosti et al. 1996; Stemmer-Rachamimov et al. 1997). Depletion of merlin protein by antisense oligonucleotide treatment (Huynh and Pulst 1996), as well as overexpression of merlin in cultured cells, causes cellular changes associated with alterations in cell-matrix interactions, such as cell morphology, cell motility, and cell-substrate attachment (den Bakker et al. 1995_b_; Koga et al. 1998; Gutmann et al. 1999). Furthermore, studies in cultured NIH3T3 cells demonstrate that endogenous levels of merlin expression and phosphorylation are responsive to changes in cell adhesion, cell confluency, and growth factor stimuli (Shaw et al. 1998), which suggests that merlin may function as part of a signal transduction pathway regulating cell-cell and cell-matrix interactions. Several studies investigating merlin function have focused on identifying cellular interacting proteins. Merlin can bind actin filaments in vitro (Xu and Gutmann 1998), and immunoprecipitation studies have identified the actin-binding protein βII-spectrin, the Na+-H+ exchanger regulatory cofactor EBP50, and the matrix receptor β1-integrin as interactors of merlin (Murthy et al. 1998; Obremski et al. 1998; Scoles et al. 1998). However, the importance of these interactions to merlin tumor-suppressor activity and NF2 pathogenesis has not been clarified.
To elucidate the molecular mechanisms of NF2 tumor-suppressor function, we analyzed the effect of NF2 missense and deletion mutations on merlin cellular and molecular properties in cultured mammalian cells, hypothesizing that these changes would produce stable proteins that are functionally defective. In this report, we demonstrate that the majority of single amino acid (aa) mutations in merlin produce proteins that significantly inhibit cell adhesion and show increased detergent solubility compared to wild-type merlin, which suggests that merlin tumor-suppressor activity involves cell-adhesion signaling. In addition, we provide evidence that missense mutations can act as dominant alleles, which suggests the possibility that a subset of NF2 mutant alleles in patients may act in a dominant manner to promote tumorigenesis.
Material and Methods
Protein Expression Constructs
To create the untagged mammalian expression constructs, all cDNAs were cloned into the mammalian expression vector pcDNA3 (Invitrogen). The green fluorescent protein (GFP) tag was fused N-terminal to all cDNAs by means of the expression vector pEGFP-C1 (Clontech). All cDNAs were cloned into the pGEX-5X-2 bacterial expression vector (Pharmacia Biotech), to fuse glutathione-S-transferase (GST) to the amino terminus of the protein of interest. The full-length merlin cDNA clone for isoform 1 was isolated from a human testes library in the vector pBK-CMV (Stratagene) and was designated M7. All missense merlin constructs were generated from the plasmid M7 by means of the oligonucleotide-mediated mutagenesis protocol of Kunkel et al. (1987), with primers engineered to contain the exact nucleotide change reported in the literature as the disease-associated mutation. The C-terminal coding sequence for isoform 2 was PCR amplified from human lymphocyte cDNA and was subcloned into M7, to generate the full-length isoform 2 cDNA. For ΔCterm, a unique internal _Xho_1 site and the _Xho_1 multiple cloning site were used to “pop out” the carboxyl terminus of M7, to produce a construct encoding aa 1–341 of merlin. ΔNterm (aa 342–595) was generated by PCR amplification of the C-terminal half of merlin. Mer559 was engineered by PCR amplification by means of internal merlin primers. ΔBB was engineered essentially as described elsewhere (LaJeunesse et al. 1998) for the Drosophila “Blue Box” deletion. The entire EBP50 coding sequence was isolated by _Hin_dIII/Not1 digestion, from I.M.A.G.E. clone 33147 (Lennon et al. 1996). The full-length human moesin cDNA was isolated by PCR from I.M.A.G.E. clone 645267. The sequence of all constructs was confirmed by restriction digestion and by direct DNA sequencing with the use of the Sequenase 2.0 DNA Sequencing Kit (United States Biochemical) or the automated fluorescent sequencing machine ABI 377 (PE Biosystems).
Cell Culture and Transient Expression of Merlin Proteins in Culture Cells
COS-7, Hela, and 293 cells (ATCC) were cultured in Dulbecco’s modified Eagle’s minimum essential medium, supplemented with 10% fetal bovine serum and antibiotics (Gibco), at 37°C in a 5% CO2 humidified incubator. Exponentially growing COS-7, Hela, and 293 cells were seeded 24 h before transfection. At 50% cell confluency, DNA transfer was performed by liposome-mediated gene transfer, with the use of either Effectene reagent or Superfect reagent (Qiagen). Cells were incubated for 3 h (Superfect) or overnight (Effectene) in the presence of the DNA-liposome reagent and were washed with PBS; fresh medium was then added. Cells were assayed for protein production 24–72 h after transfection. Overnight exposure to Effectene did not show differences in cell death, morphology, or protein expression compared to a 3-h exposure to lipofect reagent Superfect (Qiagen).
Antibodies
The merlin polyclonal antibodies N50 and C53 were generated by injecting female New Zealand white rabbits with synthetic peptides conjugated to keyhole limpet hemocyanin: N50 (YAEHRGRARDEAEMEYLK-C; residues 192–209, including a carboxyl-terminal cysteine residue) and C53 (C-LHNENSDRGGSSKHNTIK; residues 561–578, including an amino-terminal cysteine residue). N50 antibody was affinity-purified on peptide columns by means of the HiTrap NHS–activated reagent (Pharmacia Biotech). All other antibodies were commercially available: rabbit polyclonal antimerlin antibody A-19 (Santa Cruz Biotechnology), mouse monoclonal anti-GFP antibody (Clontech, Boehringer-Mannheim), rhodamine- and Texas Red–coupled goat antirabbit antibodies, Oregon Green 488–coupled goat antimouse antibody, Texas Red–conjugated X phalloidin, and anti-β-tubulin mouse monoclonal antibody (Molecular Probes).
Immunofluorescence and Microscopy
Transfected cells grown on untreated or poly-D lysine-coated coverslips (Sigma) were fixed in 4% paraformaldehyde and permeabilized with 0.05% saponin (Sigma). Fixed cells were incubated with 0.4–2.0 μg/ml primary antibody. When staining for actin, we also added a 1:40 dilution of Texas Red–conjugated anti-X phalloidin. Cells were rinsed in PBS containing 1% Triton X-100; incubated with 1:500 dilution of the appropriate secondary fluorescent antibody; conjugated to either rhodamine, Texas Red, or Oregon Green 488; and then mounted in Prolong medium and DAPI (Molecular Probes). Cells were visualized with a Zeiss epifluorescence microscope or with a Molecular Dynamics confocal laser-scanning microscope 2010. For analysis of GFP expression and viability in cells, GFP-transfected cells were incubated with propidium iodide (Molecular Probes), washed with PBS, resuspended in culture media without phenol red (Gibco), and visualized with the epifluorescence microscope.
Preparation of Cell Extracts
Suspended COS-7 cells were collected 48–72 h after transfection, washed with PBS, and resuspended in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate) containing proteinase inhibitors (1 mM Pefabloc [Boehringer-Mannheim], 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 10 μg/ml pepstatin). These were used as the detached fractions. Attached cells were incubated with lysis buffer for 30 min at 4°C and were used as the attached fractions. Protein concentrations were determined by means of the BioRad DC Protein Assay Kit.
Detergent Extraction
COS-7 transfected cells were extracted for 40–50 s with 30-μl extraction buffer (50 mM MES, pH 6.4, 3 mM EGTA, 5 mM MgCl2, 0.5% Triton X-100). The detergent-soluble material was precipitated with 85% acetone overnight at −20°C, and the pellet was recovered after centrifugation at 14,000 g for 15 min at 4°C. Detergent-insoluble material was scraped in PBS containing proteinase inhibitors and was centrifuged at 14,000 g for 15 min at 4°C. All pellets were resuspended in the same volume of Laemmli sample buffer.
Immunoblotting
Cell lysates were boiled in 1X Laemmli sample buffer (2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.002% bromophenol blue, and 62.5 mM Tris HCL, pH 6.8), loaded onto a 10% or 7.5% SDS-PAGE gel, and electrotransferred onto polyvinylidene fluoride (PVDF) membrane. Filters were blocked in PBS containing 5% skim milk, incubated with 0.2 μg/ml primary antibody in PBS containing 0.1% Tween 20 (PBS-T), washed extensively with PBS-T, and incubated with 1:3000 dilution of the appropriate secondary antibody. After washing with PBS-T, detection was accomplished with the use of horseradish peroxidase–coupled secondary antibodies (Amersham) and enhanced chemiluminescence substrate (NEN) or by alkaline phosphotase–conjugated secondary antibodies and chemifluorescence ECF substrate (Amersham) or biotin-conjugated secondary antibodies (Sigma), streptavidin and 32P-3DNA dendrimer (Genisphere).
Quantification of Western Blots
Radioactive and fluorescent blots were scanned with the Molecular Dynamics STORM imager and quantitated by means of the ImageQuant software program (Molecular Dynamics). Luminescent blots were exposed to film, digitized with the Silverscan III flatbed scanner (La Cie), and quantitated with the National Institutes of Health Image Program, by means of a calibrated step tablet (Kodak).
Expression and Purification of GST Fusion Proteins
Logarithmically growing cultures of E. Coli BL21, transformed with the pGEX-5X-2 recombinant plasmids, were incubated overnight at room temperature, with 0.05 mM isopropyl-b-D-thiogalactoside. The cells were pelleted and were resuspended in 1/20 volume ice-cold PBS, containing 1% Triton X-100 and proteinase inhibitors, and were sonicated on ice for 30 s. Cellular debris was removed by centrifugation at 10,000 g for 15 min at 4°C. The supernatant was applied to a glutathione sepharose 4B column (Pharmacia Biotech) and the GST fusion proteins were eluted with 5 mM reduced glutathione in 50 mM Tris-HCL, pH 9.5. Purified proteins were analyzed by separation on a 10% SDS-PAGE gel stained with Commassie Blue and were quantified with the BioRad Protein Assay Reagent. Purified proteins were stored at −80°C.
In Vitro Protein Binding Assays
The merlin pcDNA3 plasmids were used as template for a T7-coupled transcription-translation system (Promega TnT kit) in the presence of 35S-methionine, and radiolabeled protein was made. Samples of the TnT reactions were boiled in 1X Laemmli’s sample buffer and were separated on a 10% SDS-PAGE gel. The dried gel was exposed to film to determine the size, specificity, and quantity of the radiolabeled protein. Known amounts of radiolabeled protein (10–25 μl of a 100-μl TnT rxn) were incubated with equal amounts of GST or GST–merlin fusion proteins (5–25 μg) and were immobilized on glutathione-sepharose beads in TEN buffer (10 mM Tris-HCL, pH7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1 mg/ml BSA, and proteinase inhibitors) for 2–4 h at 4°C. The supernatant was removed, and the beads were washed four times with TEN buffer. Protein bound to the beads was eluted by boiling in 2X Laemmli’s sample buffer for 5 min. Samples were separated on a 10% SDS-PAGE gel. The dried gel was analyzed by autoradiography and phosphoimagery, by means of the STORM imager and the ImageQuant software package.
In Vitro Microtubule Binding Assay
One hundred twenty-five micrograms of purified bovine tubulin (Molecular Probes) were assembled into microtubules in vitro with 50 μl of 35S-labeled merlin protein in a total of 1 ml PGEM buffer (80 mM PIPES, 1 mM MgCl2, 1 mM EGTA, 1 mM GTP [Sigma], 30% glycerol, pH 6.8), with 20 M paclitaxel (Molecular Probes), for 30 min at 37°C. Prior to incubation, 50 μl were removed and labeled as the “total” fraction. After assembly, the polymerized microtubules were collected by centrifugation at 18,000 rpm in a Sorvall SS-34 rotor, for 30 min at 30°C. The supernatant was removed to a fresh tube and was labeled “supernatant,” and the microtubule pellet was resuspended in 1 ml PGEM buffer and was labeled “pellet.” Fifty microliters of each fraction were separated on a 12% SDS-PAGE gel. The dried gel was analyzed with the use of the STORM imager and the ImageQuant software package.
Results
Construction of Wild-Type and Mutant Merlin Proteins
To identify biological differences between mutant and wild-type merlin, we constructed GFP-tagged and -untagged expression plasmids for the two wild-type merlin isoforms; for the N-terminal domain, which is analogous to a common NF2-associated nonsense mutation Arg341X; for the unique C-terminal domain; and for five NF2-associated missense mutants (fig. 1). In addition, we designed mammalian expression plasmids for a “dominant-negative” and for an “activated” form of merlin, ΔBB and Mer559, respectively, which have been characterized elsewhere (LaJeunesse et al. 1998) in the Drosophila homologue of merlin. The mammalian expression constructs were confirmed to produce stable proteins of the predicted size by in vitro transcription/translation (fig. 2_A_). As shown in figures 3 and 4, western blot analysis revealed that the tagged and untagged wild-type merlin products migrate as a distinct doublet ∼70 kD for the untagged protein (fig. 3_A_) and ∼100 kD for the GFP-tagged protein (fig. 3_A_). This pattern of migration was demonstrated to be due to phosphorylation of merlin, with the lower migrating band corresponding to the dephosphorylated form (Shaw et al. 1998). We observed both migrating species for both the tagged and the untagged constructs containing the missense mutations and did not observe any difference in the phosphorylated/dephosphorylated ratio between the wild-type and missense proteins, which suggests that the mutant proteins produce phosphorylated and dephosphorylated products in vivo similar to the wild-type protein (fig. 3_A_). For all quantitative assays done in this study, we included both migrating species of merlin in all calculations.
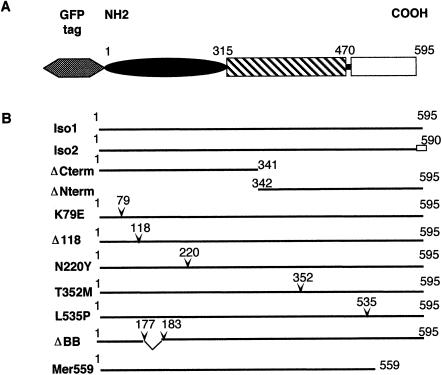
Figure 1.
Merlin structural domains and the different cDNA constructs used in the study. A, Predicted merlin structure. On the basis of the primary sequence and homology to the ERM proteins, merlin is predicted to consist of a globular amino terminus, an alpha-helical central region, followed by a charged carboxyl-terminal domain (Rouleau et al. 1993; Turunen et al. 1998). B, Details of the various cDNA constructs. Numbers correspond to the aa position of isoform 1. All constructs had two versions that either were the natural N-terminus or contained the GFP moiety fused to the N-terminus of merlin (gray hexagon). The unblackened box corresponds to the specific C-terminus of isoform 2 (Arakawa et al. 1994). The position of aa substitutions and deletions are indicated by inverted carets (∨) (MacCollin et al. 1993; Bourn et al. 1994; Sainz et al. 1994; Evans et al. 1995). Letters in the construct names correspond to the single-letter designation for the aa. The left letter identifies the wild-type residue, and the right letter identifies the mutant substitution. The constructs ΔBB and Mer559 were engineered to contain the analogous mutations, a dominant-negative and activated allele, respectively, described elsewhere for Drosophila merlin (LaJeunesse et al. 1998).
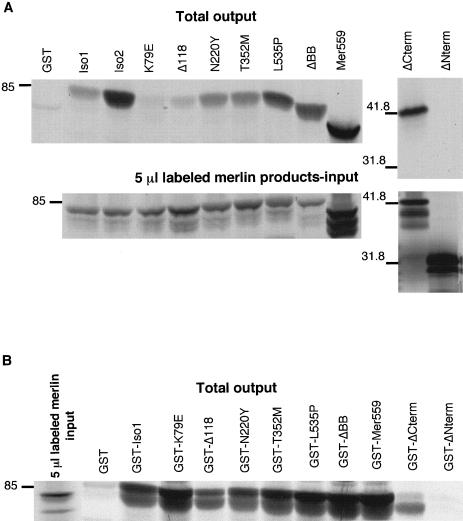
Figure 2.
Analysis of NF2 mutations on merlin protein interactions in vitro. The ability of wild-type and mutant merlin proteins to form intermolecular complexes was quantitated by means of a GST-fusion binding assay. A, 35S-labeled wild-type and mutant merlin proteins produced in vitro. A 5-μl aliquot of each reaction (input) was separated by SDS-PAGE analysis, and the amount of labeled full-length product was quantitated on a phosphoimager. Each radiolabeled protein (10–20 μl) was incubated with immobilized GST-merlin. The total amount of bound radiolabeled protein (output) was run on an SDS-PAGE gel and quantitated on a phosphoimager. The amount of radiolabeled protein bound to GST-merlin was calculated as a percentage of the total radiolabeled input for each reaction. These percentage values were used as data points for table 1. B, The reciprocal in vitro binding assay. In vitro translated 35S-labeled wild-type merlin (25 μl) was incubated with equal amounts of immobilized GST-merlin containing NF2 mutations. The percentage of total radiolabeled merlin bound to the GST fusion proteins was determined by phosphoimage analysis, as described above, and these percentage values were used as data points for table 1. Mutant K79E and ΔCterm gave inconsistent results between the two reciprocal assays. The position of molecular mass standards is indicated, in kilodaltons, on the left.
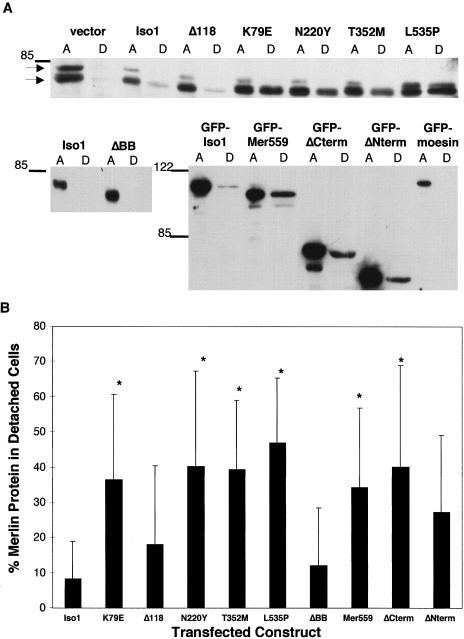
Figure 3.
Analysis of mutant merlin expression on cell attachment. A, Immunoblot of attached and detached COS-7 cells transfected with wild-type and mutant cDNA constructs. Forty-eight hours after transfection the amount of transfected merlin protein expressed in detached (lanes D) and attached (lanes A) cells was determined by western analysis, with a 1:1000 dilution of polyclonal antimerlin antibody A-19 for the untagged constructs and a 1:1000 dilution of monoclonal anti-GFP antibody for the tagged constructs. The two bands indicated by arrows correspond to the phosphorylated (top) and dephosphorylated (bottom) species of endogenous merlin. Molecular mass standards are indicated, in kilodaltons, on the left. B, Quantification, by means of a fluorescent imager and the ImageQuant software program, of amount of transfected protein in detached and attached fractions. Both species of merlin were included in the calculations. The percentage of total transfected protein present in the detached cells was determined for each construct (D/[A+D ]). The average value for each construct was calculated from at least four separate experiments, and the SEM is depicted for each line. Statistically significant differences from isoform 1, determined by the student _t_-test (P<.02), are indicated with an asterisk (*).
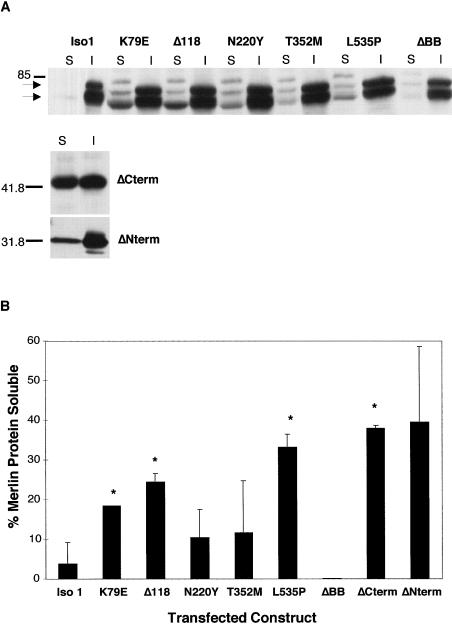
Figure 4.
Analysis of NF2 mutations on protein solubility. A, Immunoblot of soluble (S) and insoluble (I) fractions of COS-7 cells transfected with untagged cDNA constructs. Transfected cell lysates were recovered 48 h after transfection, and equal amounts of soluble and insoluble fractions were analyzed, by western analysis, with 1:1000 dilution of polyclonal anti-merlin antibody A-19. Molecular mass standards are indicated, in kilodaltons, on the left. B, Quantification, by means of a fluorescent imager and the ImageQuant software program, of amount of transfected protein in the soluble and insoluble fractions. The percentage of total transfected protein that was present in the Triton X-100 soluble fraction was calculated for all constructs (S/[S+I]). Average value for each construct was calculated from multiple experiments and the SEM is indicated for each line. Statistically significant differences from isoform 1, determined by the student _t_-test (_P_=.03), are indicated with an asterisk (*).
Subcellular Distribution of Wild-Type and Mutant Merlin
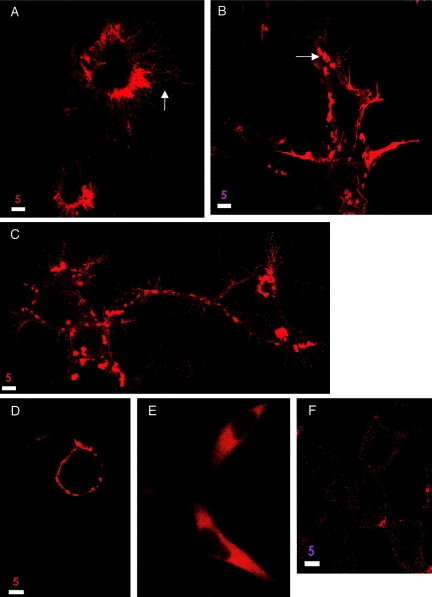
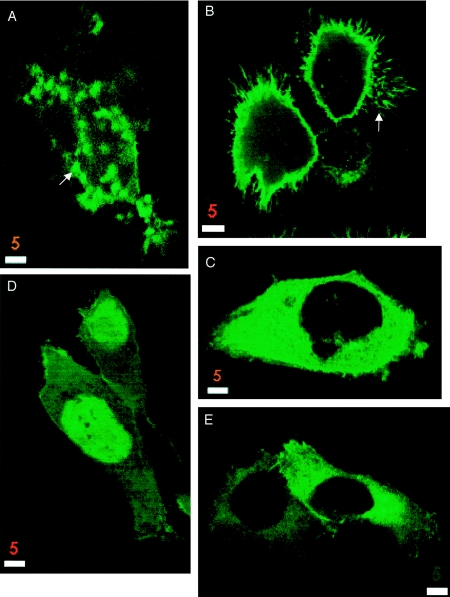
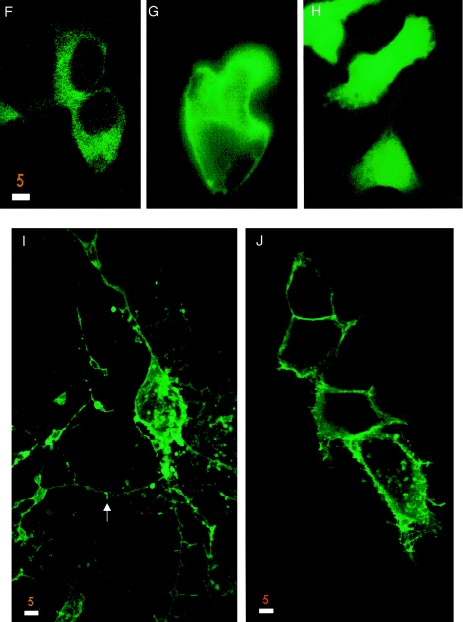
To investigate whether mutations in merlin affect proper subcellular distribution of the protein, we transiently transfected HeLa, 293, and COS-7 cells with the merlin expression constructs and observed the patterns of merlin localization in both fixed and live cells. Endogenous merlin in these cells was detected clearly by western blot (fig. 3A, vector lane) but was difficult to visualize by immunohistochemistry (fig. 5_F_), which suggests a low endogenous level of merlin expression. Polyclonal anti-Nterm, A-19, and anti-Cterm, C53, antibodies were used to stain for untagged merlin expression in fixed cells, and GFP autofluorescence was used to study protein localization in live cells. The results were similar for the tagged and untagged proteins and for the various cell types, and were consistent for multiple independent transfection experiments. The two wild-type isoforms localized to the plasma membrane and were particularly abundant in the submembranous regions of filopodia, microspikes, and ruffling membrane (figs. 5_A_ and6_B_). In addition to the membrane localization, in a large fraction of cells merlin also localized to punctate cytoplasmic granules (figs. 5_B_ and6_A_). The localization of the N-terminal and C-terminal truncations of merlin differed markedly from the full-length protein. The ERM homology domain ΔCterm localized mainly to the nucleus, although some protein did localize to the plasma membrane and was never seen as cytoplasmic granules (6_D_). The C-terminal half of merlin ΔNterm was localized diffusely in the cytoplasm and showed no localization to the dynamic regions of the membrane (6_C_). Of the five missense mutants, only two displayed an abnormal cellular distribution pattern. The K79E missense mutant was strongly localized to the perinuclear cytoplasm, with only minor staining at the plasma membrane (6_E_). The single aa deletion Δ118 protein showed diffuse cytoplasmic staining (figs. 5_E_ and 6_F_). Of the remaining mutant proteins, the missense L535P mutant (6_J_) and the dominant-negative mutant ΔBB (fig. 5_D_) showed submembranous membrane localization but were rarely seen as cytoplasmic granules, which matched the localization pattern of the ΔBB mutant protein reported in Drosophila (LaJeunesse et al. 1998). Mutants N220Y (data not shown), T352M (fig. 5_C_), and Mer559 (6_I_) localized strongly to the submembranous regions in membrane extensions, in addition to cytoplasmic granules in some cells.
Figure 5.
293 cells expressing untagged wild-type and mutant forms of merlin. Untagged wild-type merlin shows the same localization pattern as GFP-tagged merlin (A and B). The mutant T352M localizes to microspikes and filopodia and to cytoplasmic granules, similar to wild-type merlin (C). Overexpression of wild-type merlin in cells leads to an increase in membrane extensions (A) compared to cells transfected with vector alone (F), whereas the dominant-negative mutant ΔBB often stained round cells with no membrane extensions (D). The mutant Δ118 does not affect cell morphology (E). Cells were stained with a 1:500 dilution of antimerlin antibody A-19 and Texas Red–conjugated secondary antibody and were visualized by confocal microscopy. Bar equals 5 μm.
Figure 6.
293 cells expressing GFP-tagged wild-type and mutant forms of merlin. Wild-type merlin (A and B) and the mutants Mer559 (I) and L535P (J) localize to submembranous regions, especially microspikes and filopodia (B, arrow), and wild-type merlin is sometimes seen as granules throughout the cytoplasm (A, arrow). In contrast, the carboxyl-terminal half of isoform 1 localizes to the cytoplasm (C), and the amino-terminal half of merlin is seen mainly in the nucleus (D). The NF2 mutations K79E (E) and Δ118 (F) alter the localization of the protein, with the protein localized mainly in the perinuclear cytoplasm with little or no membrane localization. Overexpression of wild-type merlin in cells (A and B) leads to an increase in membrane extensions and cell spreading compared to expression of GFP alone (H) or cells expressing GFP-tagged full-length moesin (G). Overexpression of NF2 mutant proteins affects cell morphology in varying degrees, with the activated mutant Mer559 leading to round cell bodies with extremely long membrane extensions (arrow, I), whereas the deletion mutant Δ118 does not affect cell morphology (F). Cells were visualized by confocal microscopy. Bar equals 5 μm.
Studies of the subcellular distribution of Drosophila merlin found the protein initially localized to the membrane but then internalized to form punctate cytoplasmic structures (LaJeunesse et al. 1998). We had observed merlin-rich punctate cytoplasmic structures in a subset of the transfected cells and wanted to determine if, over time, human merlin also moved from the membrane into cytoplasmic granules. Thus, we monitored the subcellular localization of the GFP-tagged merlin proteins in live cells during a period of 48 h, to determine if merlin localization altered with time. No time-dependent changes in protein localization were observed (data not shown). Since three of the five missense mutants are indistinguishable from wild-type in cellular localization, proper targeting of merlin to the plasma membrane may not be sufficient for tumor-suppressor activity, although it may be necessary.
Cell-Adhesion Properties of Merlin Proteins
Although morphology showed variability from cell to cell, we observed a general trend of altered morphology in merlin-transfected cells compared to cells transfected with vector alone (figs. 5_F_ and 6_H_). Cells transfected with the wild-type isoforms and the mutants N220Y and T352M showed an increase in membrane spikes, long filopodia, and surface blebs, and some cells had extremely large surface areas (figs. 5_A_, 5_C_, and 6_B_). In contrast, cells overexpressing full-length GFP-moesin did not display these morphological changes (6_G_). Cells expressing the activated mutant Mer559 often appeared as round cell bodies with multiple extremely long, thin cellular processes (6_I_), whereas cells expressing the dominant-negative mutant ΔBB were often completely round with no cellular extensions (fig. 5_D_). Cells expressing the mutants L535P and ΔCterm showed an increase in microspikes but seldom had extremely long cellular processes (figs. 6_D_ and 6_I_). Merlin constructs that mainly localized to the cytoplasm led to no noticeable morphological changes; cells expressing the missense protein Δ118 (figs. 5_E_ and 6_F_) or ΔNterm (6_C_) were similar in cell shape to vector-transfected cells (figs. 5_F_ and 6_H_).
These qualitative morphological changes suggested to us that merlin may be involved in cell-matrix interactions. To determine if NF2 mutant proteins could alter cell-matrix interactions, we assayed the adhesion properties of cells expressing the wild-type and mutant merlin proteins. COS-7 cells were transiently transfected with merlin expression constructs, and 48 h posttransfection detached and attached cell fractions were collected. Equal amounts of attached and detached cell lysates were analyzed by immunoblotting, to determine the percentage of total merlin protein found in the detached fraction. Cells expressing the mutant merlin proteins K79E, N220Y, T352M, L535P, ΔNterm, and ΔCterm showed an increased tendency to detach from the plate surface compared to cells expressing either wild-type merlin isoforms, which were seldom found in the detached fraction (fig. 3_A_). Interestingly, the mutant proteins ΔBB and Mer559, which correspond to the Drosophila dominant-negative mutation and to the activating mutation, respectively, showed opposite effects on the adhesion property of the cell. The activating mutation behaved similarly to the NF2-associated mutant proteins by decreasing cell adhesion, whereas the dominant-negative mutation had no effect on adhesion (fig. 3_A_). To test for reproducibility of this assay, the experiment was performed four separate times with both the tagged and untagged proteins. Although the absolute percentage of detached cells was variable between the experiments, the general trends were consistent, and differences between the wild-type and the mutant proteins were statistically significant with the student _t_-test (fig. 3_B_). Interestingly, only the lower migrating merlin band was found in the detached fraction for wild-type protein, which suggests that only dephosphorylated wild-type merlin is present in the detached cells (fig. 3_A_). In contrast, the upper migrating band was present in the detached fraction of some of the mutant proteins, such as L535P (fig. 3_A_), which implies that the phosphorylated form is no longer restricted from the detached cells. However, these results were not consistent among the four experiments.
To test for the possibility that the increase in detachment for the mutant-expressing cells was due to increased cell death, detached cells were examined for viability by staining with propidium iodide. More than 80% of the detached cells were viable by this assay, and there was no distinguishable difference in viability between the cells expressing mutant merlin and the wild-type (data not shown).
Cytoskeletal Interactions of Wild-Type and Mutant Merlin
It is possible that the effect on cell adhesion and morphology by merlin proteins is mediated through interactions with the actin cytoskeleton. The ERM proteins bind F-actin through a highly conserved region in the C-terminal domain (Turunen et al. 1994), and overexpression of this domain in transfected cells disrupts actin-based cytoskeleton organization (Edwards et al. 1994). Although merlin lacks this conserved actin-binding domain, studies have demonstrated that merlin colocalizes with cortical actin and that chemical disruption of the actin cytoskeleton leads to a redistribution of merlin in the cell (den Bakker et al. 1995_b_; Deguen et al. 1998). To investigate whether mutant merlin protein could disrupt actin cytoskeleton organization through aberrant merlin-cytoskeletal interactions, we studied the distribution of actin in 293 and CHO cells transfected with GFP-tagged wild-type and mutant merlin proteins by immunostaining with Texas Red–conjugated phalloidin. Wild-type and mutant merlin, which showed normal subcellular distribution, colocalized with actin at the motile regions of the plasma membrane-filopodia, ruffling edges, and microspikes, but showed no colocalization with actin stress fibers (data not shown). The merlin mutations that decreased cell adhesion had no effect on organization or distribution of the actin cytoskeleton (data not shown).
To further our studies of possible effects on merlin-cytoskeletal interactions by NF2 mutations, we performed cell fractionation assays on merlin-transfected cells. It is well established that nonionic detergent extraction procedures preserve the cytoskeleton and isolate tightly linked cytoskeletal proteins in the insoluble fraction (Algrain et al. 1993). After mild nonionic detergent extraction, cells expressing the different merlin constructs were stained with the antimerlin antibodies A-19 and C53, to determine the solubility of the wild-type and mutant proteins. The wild-type isoforms and mutants N220Y, T352M, ΔBB, and Mer559 displayed no noticeable change in overall protein subcellular localization or intensity of staining after detergent extraction, which suggests that these proteins are linked to cytoskeletal elements. In contrast, the Δ118 and ΔNterm mutant proteins display an altered staining pattern after mild detergent extraction, changing from a diffuse cytoplasmic to a strong punctate perinuclear localization, after detergent extraction (data not shown). The K79E mutant protein shows a change in cellular localization after extraction similar to but less pronounced than the Δ118 mutant protein. Furthermore, the ΔCterm and the L535P mutant proteins display an overall decrease in intensity of staining, which implies that these mutations increased the solubility of the protein (data not shown). To confirm these observations, quantitative immunoblot analysis of equal amounts of soluble and insoluble fractions from the transfected cells were performed. The immunoblotting confirmed that the missense mutations K79E, Δ118, and L535P and the deletion of the N- or C-terminal domains increased the solubility of the protein, which suggests that these mutations decrease the stability of merlin-cytoskeletal interactions (fig. 4_A_). Since detergent extraction experiments can be difficult to reproduce, the experiment was performed several times, and the differences in solubility from wild-type were statistically significant for the mutants K79E, Δ118, L535P, and ΔCterm (fig. 4_B_). It is interesting to note that the main species of merlin recovered in the soluble fraction is the dephosphorylated form.
Last, we performed in vitro microtubule binding assays with radiolabeled wild-type and mutant merlin proteins. Xu and Gutmann (1998) reported a microtubule-binding region in the N-terminal domain of merlin, which was normally masked in the wild-type protein but was exposed in NF2 mutant proteins containing C-terminal truncations, exon deletions, and an L64P missense mutation. They hypothesized that merlin protein that exists in an “open” conformation would be defective in growth suppression. We wanted to investigate whether the majority of missense mutations in merlin would alter the folding of the full-length molecule such that normally “masked” binding domains would be revealed in the mutant molecule. Purified bovine tubulin was polymerized in the presence of in vitro transcribed and translated radiolabeled merlin protein. Polymerized microtubules and tightly linked proteins were pelleted by high-speed centrifugation, and proteins left in the supernatant were unassociated with microtubules. Equal amounts of each fraction were loaded on an SDS-PAGE gel, and the amount of radiolabeled merlin found in each fraction was determined by phosphoimage analysis. No significant differences in microtubule binding were seen between the wild-type isoforms 1 and 2 (14% and 15% bound, respectively) and the missense mutants K79E, N220Y, T352M, and L535P (18%, 18%, 14%, and 10% bound, respectively). In fact, contrary to other reports, the N-terminal domain (aa 1–341) did not display significant microtubule binding (8% bound), whereas the C-terminal domain (aa 342–595) showed no microtubule binding activity (0.9% bound). Furthermore, immunofluorescence staining of cells transiently transfected with GFP-merlin fusion constructs with antitubulin antibody showed no colocalization of merlin and microtubules (data not shown).
Merlin-Merlin Interactions
It has been demonstrated by the two-hybrid system and in vitro binding assays that merlin could form homodimers, and it was hypothesized that self-association could be a form of functional regulation (Gutmann et al. 1998; Scoles et al. 1998). To determine if NF2 mutations could disrupt merlin self-interactions, we used an in vitro affinity precipitation method to quantitate the binding of wild-type merlin to itself and missense forms of merlin. In a set of experiments, radiolabeled wild-type merlin was generated in vitro and equal amounts of radiolabeled product were incubated with equal amounts of immobilized, bacterially expressed GST-merlin proteins, with or without missense mutations. The GST moiety showed little or no binding of radiolabeled merlin (fig. 2_B_). The strength of the merlin interactions was quantitated by measuring the percentage of total radiolabeled protein that remained bound to the GST fusion proteins after extensive washing. The reciprocal experiment of measuring the amount of in vitro generated radiolabeled mutant merlin bound to immobilized GST–wild-type merlin was also performed (fig. 2_A_). The different radiolabeled merlin constructs all showed little or no binding to the GST moiety (fig. 2_A_ shows the result for Mer559). Because the amount of in vitro generated radiolabeled product differed between the merlin constructs, the total amount of radiolabeled protein input for each incubation was determined by quantification on a phosphoimager (fig. 2). The amount of bound radiolabeled protein was calculated as a percentage of the total radiolabeled input for each separate incubation, thereby normalizing the results to allow direct quantitative comparison of the strength of binding among the different merlin constructs. The experiments were performed three times, and the results were consistent for all constructs except K79E and ΔCterm, rendering the results for these two proteins inconclusive (fig. 2_A_ and 2_B_). In addition, the GST-isoform 1 displayed greatly increased binding of radiolabeled isoform 2 compared to isoform 1 (fig. 2_A_); however, the reciprocal binding experiment has not yet been performed to confirm this result. Five of the remaining seven mutant merlin proteins showed aberrant homodimerization. One missense construct, L535P, displayed a threefold increase in binding to wild-type merlin in vitro (table 1), which suggests that this aa substitution increased merlin dimerization. In contrast, the Δ118 mutant protein gave a fivefold decrease in binding to wild-type merlin, and ΔNterm showed a complete inability to bind full-length merlin, which implicates the amino terminus as an essential domain for homodimerization (table 1). The dominant-negative ΔBB mutation in merlin and the activated mutant Mer559 both demonstrate an increased ability to dimerize with wild-type merlin (table 1), which suggests that homodimers may be an active form of the protein. The differences in binding ability in vitro are not likely to be due to gross differences in protein translation and stability among the GST fusion constructs or in vitro translated products, since by SDS-PAGE analysis, all merlin constructs displayed the expected-size protein products and showed similar protein stability over time (data not shown). Therefore, missense-containing mutations in merlin can have opposite effects on merlin homodimerization. Interestingly, we observed that full-length merlin binds more strongly to another full-length molecule than to either the amino- or carboxyl-terminal domain alone, which is opposite to that seen for the ERM proteins (Magendantz et al. 1995; Bhartur and Goldenring 1998), thereby demonstrating that merlin may be functionally regulated in a different manner from the ERM proteins. In addition, we observed that native merlin did not bind the closely related, full-length moesin protein (table 1).
Table 1.
Summary of Merlin Dimerization In Vitro
| Protein Construct | Dimerization with Wild-Type Merlin Relative to Isoform 1a |
|---|---|
| GST (control) | |
| Isoform 1 | 1 |
| K79E | 2.18±1.9 |
| Δ118 | .21±.09b |
| N220Y | .94±.5 |
| T352M | .76±.03 |
| L535P | 2.36±.58b |
| ΔBB | 2.93±1.29b |
| Mer559 | 6.17±2.36b |
| ΔCterm | .58±.71 |
| ΔNterm | .001±.001b |
| Moesin | .07±.02b |
Merlin-EBP50 Interactions
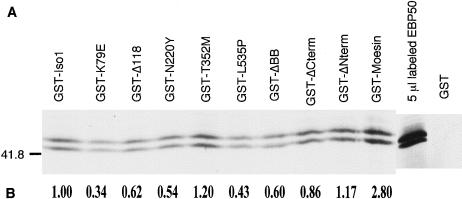
From a two-hybrid screen for merlin interactors and an affinity precipitation experiment for ERM binding partners, two groups independently identified a phosphoprotein, EBP50, which associated strongly with ERM and merlin through the highly conserved amino-terminal domain (Reczek et al. 1997; Murthy et al. 1998). Murthy et al. also reported that merlin containing the missense mutation N220Y showed reduced binding to EBP50, which suggests that this protein interaction may be important for merlin function. To determine if other NF2-associated mutations also affected merlin-EBP50 interactions, we used an affinity precipitation assay to quantitate the ability of mutant forms of merlin to bind EBP50. In vitro translated, radiolabeled EBP50 was incubated with wild-type and mutant merlin GST-fusion proteins immobilized on GSH beads. The amount of EBP50 bound to the GST-merlin proteins was quantitated by phosphoimage analysis of SDS-PAGE gels. In vitro translated EBP50 produced a distinct doublet ∼50 kD (fig. 7_A_), which matches a report (Reczek et al. 1997) that stated that endogenous placental EBP50 produces three species ∼50–53 kD because of differential phosphorylation. Therefore, in vitro generated EBP50 seems to undergo posttranslational modification similar to the endogenous protein. Binding of both EBP50 bands to merlin was quantified, and no differences in merlin binding between the two EBP50 species were detectable (fig. 7_A_). We confirmed the ability of EBP50 to bind wild-type merlin and moesin and found that four of five NF2-associated missense mutations of merlin decreased EBP50 binding by 35%–65% (fig. 7_B_). In addition, we confirmed that the ERM homology domain ΔCterm was sufficient for binding EBP50, as has been reported elsewhere (Murthy et al. 1998). We also found that a missense mutation in the extreme carboxyl terminus at position 535 of merlin reduced binding by 50% compared to wild-type, a region which was not thought to be necessary for EBP50 interaction (fig. 7_B_). To confirm our results, the binding experiment was repeated. Although the overall amount of binding of EBP50-merlin was too low to allow sensitive quantification, the results were consistent in that only wild-type merlin, moesin, and the mutants T352M, ΔCterm, and ΔNterm gave detectable binding to EBP50, whereas the mutants K79E, Δ118, N220Y, L535P, and ΔBB displayed no detectable binding of EBP50 (data not shown). It is worth noting that the effect of NF2 mutations on merlin-EBP50 binding does not correlate with their effect on merlin-merlin interactions (table 2).
Figure 7.
NF2 mutations decrease binding of merlin to EBP50 in vitro. A, Binding of mutant merlin proteins to EBP50, as was determined by means of a GST fusion binding assay. In vitro 35S-labeled full-length EBP50 (10 μl) was incubated with equal amounts of immobilized GST-merlin fusion proteins containing NF2 mutations. GST-merlin and GST-moesin fusion proteins were used as positive controls for EBP50 binding. B, Amount of EBP50 bound to the mutant proteins, calculated as a fraction of EBP50-isoform 1 binding for one experiment, and the calculated value is shown below each lane.
Table 2.
Summary of Molecular and Cellular Differences between Mutant and Wild-Type Merlin Proteins[Note]
| Mutant Protein | Cell Localization | Protein Solubilitya | Cell Adhesion | Dimerization | EBP50 Binding | Phenotype |
|---|---|---|---|---|---|---|
| K79E | Cytoplasmic | Increasedb | Decreasedb | Inconclusive | Decreased | Sporadic schwannoma |
| Δ118 | Cytoplasmic | Increasedb | NS | Decreasedb | Decreased | Severe NF2 |
| N220Y | Membrane | NS | Decreasedb | NS | Decreased | Severe and mild NF2 |
| T352M | Membrane | NS | Decreasedb | NS | NS | Severe NF2 |
| L535P | Membrane | Increasedb | Decreasedb | Increasedb | Decreased | Mild NF2 |
| ΔCterm | Nuclear | Increasedb | Decreasedb | Inconclusive | NS | Severe NF2 |
| ΔNterm | Cytoplasmic | Increased | Decreasedb | Decreasedb | NS | ND |
| ΔBB | Membrane | NS | NS | Increasedb | Decreased | Dominant-negative |
| Mer559 | Membrane | NS | Decreasedb | Increasedb | ND | Activated |
Discussion
In this study, we used five NF2-associated missense mutations and one truncating mutation to identify cellular and molecular activities of the protein that are likely to be necessary for merlin tumor-suppressor action. Of all the cellular assays we performed, the most striking result was the effect on cell adhesion due to mutant protein expression. The expression of five of six NF2 mutant proteins resulted in a significant decrease in cell adhesion, which was observed as an increase in transfected cells detaching from the substratum. In contrast, endogenous and transfected wild-type merlin was seldom seen in detached cells. In agreement with our results, Koga et al. (1998) reported that human fibroblasts microinjected with mutant merlin constructs, containing either NF2-causing truncating mutations or exon deletions, would round up and detach from the substratum. Furthermore, it has been shown that STS26T cells transfected with antisense merlin oligonucleotides to give complete loss of merlin protein also detached from the substratum, in addition to displaying an increase in cell proliferation (Huynh and Pulst 1996). Together, these experimental results suggest that tumorigenesis may result from disruption of cell-matrix attachment because of a complete loss of merlin protein or because of the expression of a mutant merlin protein defective for the cell-adhesion function of wild-type merlin protein. It is well known that cell adhesion in adherent cells has an important function for normal cell growth; in fact, anchorage-independent growth is a criterion for cell transformation (Hynes 1992; Nur et al. 1992). Cultured NF2 Schwann cells, containing either truncating or missense NF2 alleles, lack contact inhibition of growth and display anchorage-independent growth and increased cell proliferation (Rosenbaum et al. 1998), whereas overexpression of full-length merlin in Ras-transformed fibroblast suppresses anchorage-independent growth and restores contact inhibition of cell growth (Tikoo et al. 1994). Therefore, we hypothesize that merlin acts as a positive regulator of cell-matrix attachment and that disruption of this activity is an initial step in tumor progression.
We investigated several possible molecular mechanisms by which merlin could regulate cell attachment. The various mutant merlin proteins were analyzed for their interaction with the cytoskeleton, since endogenous merlin has been shown to colocalize with several cytoskeletal elements, including F-actin, keratohyalin granules, and intermediate filaments (den Bakker et al. 1995_a_; Gonzalez-Agosti et al. 1996), and such interactions are likely to be involved in cell-matrix signaling pathways. Consistent with a protein associated with the cytoskeleton, we and others have demonstrated that wild-type merlin is mainly detergent insoluble (Deguen et al. 1998). We observed no change in merlin subcellular localization or intensity of staining in cells extracted with nonionic detergent, and western blot analysis of detergent soluble and insoluble fractions showed that >90% of merlin is retained in the insoluble fraction. In contrast, 55% of the NF2 mutant proteins displayed a significant increase in detergent extractability, indicating a relaxed association with the cytoskeleton. Furthermore, the mutant proteins K79E, Δ118, ΔCterm, and ΔNterm, which fail to localize properly to the plasma membrane and thus do not colocalize with cortical actin, all demonstrated a significant increase in protein solubility, further supporting the contention that these mutant proteins do not form stable interactions with the cytoskeleton. However, merlin protein solubility did not correlate with the protein’s effect on cell adhesion, since two NF2 mutant proteins that decreased cell adhesion—N220Y and T352M—displayed no increase in detergent solubility and/or difference in subcellular localization compared to wild-type (table 2). Thus, merlin regulation of cell adhesion is not likely to be mediated simply through interaction with the actin cytoskeleton.
One possible mechanism that may account for the differences we observed between wild-type and mutant merlin proteins is differential phosphorylation. Evidence from several in vitro and in vivo studies of the closely related ERM proteins suggests a model where inactive ERM proteins in the cytoplasm are activated by phosphorylation of a conserved threonine in the carboxyl terminus, causing translocation of the activated form to the plasma membrane (Nakamura et al. 1995; Hirao et al. 1996; Serrador et al. 1997; Matsui et al. 1998; Hayashi 1999). Merlin also contains this conserved threonine at position 576, and, likewise, in vivo merlin exhibits both phosphorylated and dephosphorylated forms due to phosphorylation of serine and/or threonine residues (Takeshima et al. 1994; Shaw et al. 1998). The ratio of the phosphorylated and dephosphorylated forms of merlin in cultured cells are affected by cell adhesion. Loss of adhesion results in a complete dephosphorylation of merlin, whereas reattachment of cells to a substrate leads to a rapid increase in the phosphorylated species (Shaw et al. 1998). In this study we found, by western analysis, that the five missense mutants produced both the phosphorylated and dephosphorylated species and that detached cells only express the dephosphorylated form of wild-type merlin. However, we sometimes observed, by western analysis, the presence of the phosphorylated form of the missense mutants K79E, T352M, and L535P in the detached fraction, a condition we never saw for the two wild-type isoforms. Since the expression of these mutant proteins in cells causes loss of adhesion, and since the phosphorylated form of these mutants is abnormally retained in the detached cells, these results suggest that the phosphorylation of these mutant proteins may no longer be responsive to environmental cues, such as changes in cell spreading and attachment, which may lead to improper regulation of tumor-suppressor function. It will be interesting to determine if the addition of phosphatase inhibitors to cell cultures modulates merlin’s effect on cell adhesion. In a recent study, the presence of phosphatase inhibitors in homogenized cells was demonstrated to increase the percentage of merlin recovered in the insoluble fraction, which suggests that merlin is translocated from the insoluble to the soluble fraction by dephosphorylation (Maeda et al. 1999). Under this model, we would expect to see a decrease in the ratio of phosphorylated to dephosphorylated merlin for mutants that showed increased solubility. We did not observe any significant difference in the ratio of total phosphorylated/dephosphorylated forms between the wild-type and the missense mutants.
Intriguingly, four of the five disease-causing missense mutants, as well as the dominant-negative Drosophila mutant ΔBB, displayed decreased binding to EBP50, a protein that binds all the MERM proteins (Murthy et al. 1998; Reczek and Bretscher 1998) (table 2). EBP50 is the human homologue of rabbit NHE-RF, a cofactor that has been shown to mediate protein kinase A inhibition of the renal Na+-H+ exchanger NHE3 in cultured cells (Yun et al. 1997). The physiological importance of merlin-EBP50 interaction remains highly speculative, but it is interesting to note that it has been well documented that stimulation of Na+-H+ exchangers is associated with increased cell proliferation and differentiation in response to growth factors and mitogenic agents (Ritter et al. 1997). We found no direct correlation between the strength of EBP50-merlin interactions and any of the cellular phenotypes we assayed; however, our finding that 63% of the mutant proteins studied reduced merlin-EBP50 binding merits further investigation into the possibility that this interaction is a necessary but not sufficient component of merlin tumor-suppressor function.
Another merlin-protein interaction that may be a contributing factor in merlin regulation of cell growth is merlin homodimerization. This hypothesis was suggested by Sherman et al. (1997), who demonstrated that alterations in the C-terminus that abolished merlin growth suppression also resulted in no merlin self-association. However, to date, no single aa NF2 mutations have been shown to alter merlin self-association. In this report, we demonstrated that two NF2-associated missense mutations—Δ118 and L535P—significantly altered the ability of the full-length mutant molecule to dimerize with wild-type merlin (table 2). Surprisingly, the two missense mutations had opposite effects on merlin interaction. Although the Δ118 mutation almost abolished merlin dimerization, the missense L535P greatly increased dimerization, implicating regulation of merlin self-association as an important component of merlin function. In support of this, the ΔBB mutation in merlin and the merlin mutant Mer559, genetically defined as a dominant-negative allele and a “gain-of-function” allele in Drosophila, respectively (LaJeunesse et al. 1998), both demonstrate an increased ability to dimerize with wild-type merlin, which suggests that homodimers may be an active form of the protein. In this model, functionally defective merlin that can form homodimers with remaining wild-type molecules and mutant merlin that is unable to dimerize could both lead to a decrease in merlin activity. Overall, all of the merlin-protein interactions we assayed—cytoskeletal, EBP50, and homodimerization—were disrupted by NF2 mutations, but none of the changes was correlated with another or with the cell-adhesion phenotype. Thus, it is likely that merlin acts within a complex of molecules and that disruption of any of several interactions by NF2 mutations leads to abnormal cell growth.
In light of the observation that mutations in merlin can greatly alter cell adhesion, a potentially fruitful area of investigation may be in merlin direct interaction with adhesion transmembrane proteins. Recently, merlin was shown to partially colocalize and coimmunoprecipitate with the β1-integrin transmembrane receptor in differentiating Schwann/neuron cell cultures (Obremski et al. 1998). β1-integrin binding to the basal lamina and the actin cytoskeleton have been implicated as part of a signal transduction complex necessary for Schwann cell myelination (Fernandez-Valle et al. 1994). Like merlin, overexpression of integrins can suppress anchorage-independent growth and prevent abnormal cell growth (Giancotti and Ruoslahti 1990). Thus, NF2 mutant proteins may have aberrant interactions with β1-integrins, thereby altering the transduction of signals from the extracellular matrix necessary for Schwann cell differentiation and growth. This model could easily be tested by determining the ability of NF2 mutant merlin to coimmunoprecipitate with β1-integrin from cells and investigating for aberrant localization and responses of β1-integrin in NF2 Schwann cells.
Last, our demonstration of cellular phenotypes associated with exogenous expression of NF2 mutant alleles shows that these missense mutations may act in a dominant manner. We found that the decrease in cellular adhesion by mutant NF2 proteins could be a dominant effect, since COS-7 cells contain two wild-type NF2 alleles and have low levels of endogenous merlin expression detectable by western blotting. However, the mutant alleles may be acting as loss-of-function mutations in the adhesion phenotype. If overexpression of wild-type merlin causes greatly increased cell attachment, whereas mutant merlin has no effect on cell attachment, the results would be similar to what we observed in this study. In agreement with a dominant mode of action for mutant NF2 alleles, Koga et al. (1998) reported that loss of cell attachment still occurred when NF2 deletion mutants and wild-type merlin were coexpressed in fibroblasts. Experimental evidence in a mouse NF2 model supports a dominant mode of action for some NF2 mutant alleles. Recently, a report by Giovannini et al. (1999) showed that NF2 +/+ transgenic mice expressing an NF2-associated mutant allele, lacking exons 2 and 3, under control of a Schwann cell-specific promoter, had a high prevalence of Schwann-cell–derived tumors and schwannosis. Furthermore, the tumorigenic activity of the NF2 mutant allele was modulated by the number of NF2 wild-type alleles in vivo, giving rise to the possibility that the mutant allele may act as in a dominant-negative manner (Giovannini et al. 1999). Providing some results to directly address this possibility, we observed that in the cell-adhesion assay, the expression of the Drosophila dominant-negative mutant ΔBB had no effect on cell attachment, whereas expression of the Drosophila gain-of-function mutant Mer559 in cells led to a significant increase in cell detachment. Our findings suggest that some NF2-associated missense mutations, which account for 10% of naturally occurring mutations and a subset of truncating mutations, may act as dominant gain-of-function alleles. It is possible that our findings and the findings in the mouse and Drosophila models are artifacts of overexpression of inherently unstable proteins or of the model system. In support of the stability of the missense proteins in vivo, the half-life of two missense proteins—L64P and L535P—in stable rat schwannoma cell lines was highly similar to the half-life of isoform 1 (Gutmann et al. 1998), and a meningioma expressing only a missense NF2 transcript—C300Y—still produced full-length protein detectable by western analysis (Takeshima et al. 1998). For most of the missense alleles used in this study—Δ118, N220Y, T352M, and L535P—the status of the second NF2 allele in the tumors was not reported, and the mutation K79E was found in a sporadic schwannoma with deletion of the second NF2 allele (Sainz et al. 1994). Thus, no direct genetic evidence for dominance of missense alleles in human tumors is available. However, western analysis of 22 vestibular schwannoma for merlin expression found that three tumors (14%) expressed both full-length merlin and a truncated protein containing the N-terminal domain of merlin (Harwalkar et al. 1998), which supports the possibility of a dominant mode of action for a small fraction of NF2 truncating alleles. This mode of action could partially explain the conflicting finding that complete deletions of the gene are found in mildly affected patients (Evans et al. 1998; Zucman-Rossi et al. 1998), whereas it would be predicted that if merlin acts as a traditional tumor-suppressor gene, complete loss of the entire NF2 gene would be associated with a more-severe disease course. Since several recurring mutations have been described in patients with varying degrees of disease severity, it is also likely that additional genetic and environmental factors influence the course of the disease. Further detailed characterization of NF2 alleles and merlin expression in naturally occurring tumors is necessary to determine the mechanism by which specific NF2 mutant alleles disrupt merlin tumor-suppressor function.
Acknowledgments
We are grateful to Dr. Nila Patil, for her valuable advice and technical assistance, and to Chris Carlson, for his generous gift of human lymphocyte cDNA. This work was supported by National Institutes of Health training grant (GM07790) to R.P.S.
Electronic-Database Information
The URL for data in this article is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for NF2 [MIM 101000] and ERM [MIM 309845]) [Google Scholar]
References
- Algrain M, Turunen O, Vaheri A, Louvard D, Arpin M (1993) Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane-cytoskeletal linker. J Cell Biol 120:129–139 [DOI] [PMC free article] [PubMed]
- Arakawa H, Hayashi N, Nagase H, Ogawa M, Nakamura Y (1994) Alternative splicing of the NF2 gene and its mutation analysis of breast and colorectal cancers. Hum Mol Genet 3:565–568 [DOI] [PubMed]
- Bhartur SG, Goldenring JR (1998) Mapping of ezrin dimerization using yeast two-hybrid screening. Biochem Biophys Res Commun 243:874–877 [DOI] [PubMed]
- Bourn D, Carter SA, Mason S, Gareth D, Evans R, Strachan T (1994) Germline mutations in the neurofibromatosis type 2 tumour suppressor gene. Hum Mol Genet 3:813–816 [DOI] [PubMed]
- Deguen B, Mérel P, Goutebroze L, Giovannini M, Reggio H, Arpin M, Thomas G (1998) Impaired interaction of naturally occurring mutant NF2 protein with actin-based cytoskeleton and membrane. Hum Mol Genet 7:217–226 [DOI] [PubMed]
- den Bakker MA, Riegman PH, Hekman RA, Boersma W, Janssen PJ, van der Kwast TH, Zwarthoff EC (1995_a_) The product of the NF2 tumour suppressor gene localizes near the plasma membrane and is highly expressed in muscle cells. Oncogene 10:757–763 [PubMed]
- den Bakker MA, Tascilar M, Riegman PH, Hekman AC, Boersma W, Janssen PJ, de Jong TA, et al (1995_b_) Neurofibromatosis type 2 protein co-localizes with elements of the cytoskeleton. Am J Pathol 147:1339–1349 [PMC free article] [PubMed]
- Edwards KA, Montague RA, Shepard S, Edgar BA, Erikson RL, Kiehart DP (1994) Identification of Drosophila cytoskeletal proteins by induction of abnormal cell shape in fission yeast. Proc Natl Acad Sci USA 91:4589–4593 [DOI] [PMC free article] [PubMed]
- Evans DG, Bourn D, Wallace A, Ramsden RT, Mitchell JD, Strachan T (1995) Diagnostic issues in a family with late onset type 2 neurofibromatosis. J Med Genet 32:470–474 [DOI] [PMC free article] [PubMed]
- Evans DG, Huson SM, Donnai D, Neary W, Blair V, Newton V, Harris R (1992) A clinical study of type 2 neurofibromatosis. Q J Med 84:603–618 [PubMed]
- Evans DG, Trueman L, Wallace A, Collins S, Strachan T (1998) Genotype/phenotype correlations in type 2 neurofibromatosis (NF2): evidence for more severe disease associated with truncating mutations. J Med Genet 35:450–455 [DOI] [PMC free article] [PubMed]
- Fernandez-Valle C, Gwynn L, Wood PM, Carbonetto S, Bunge MB (1994) Anti-beta 1 integrin antibody inhibits Schwann cell myelination. J Neurobiol 25:1207–1226 [DOI] [PubMed]
- Giancotti FG, Ruoslahti E (1990) Elevated levels of the alpha 5 beta 1 fibronectin receptor suppress the transformed phenotype of Chinese hamster ovary cells. Cell 60:849–859 [DOI] [PubMed]
- Giovannini M, Robanus-Maandag E, Niwa-Kawakita M, van der Valk M, Woodruff JM, Goutebroze L, Mérel P, et al (1999) Schwann cell hyperplasia and tumors in transgenic mice expressing a naturally occurring mutant NF2 protein. Genes Dev 13:978–986 [DOI] [PMC free article] [PubMed]
- Gonzalez-Agosti C, Xu L, Pinney D, Beauchamp R, Hobbs W, Gusella J, Ramesh V (1996) The merlin tumor suppressor localizes preferentially in membrane ruffles. Oncogene 13:1239–1247 [PubMed]
- Gutmann DH, Geist RT, Xu H, Kim JS, Saporito-Irwin S (1998) Defects in neurofibromatosis 2 protein function can arise at multiple levels. Hum Mol Genet 7:335–345 [DOI] [PubMed]
- Gutmann DH, Sherman L, Seftor L, Haipek C, Hoang Lu K, Hendrix M (1999) Increased expression of the NF2 tumor suppressor gene product, merlin, impairs cell motility, adhesion and spreading. Hum Mol Genet 8:267–275 [DOI] [PubMed]
- Harwalkar JA, Lee JH, Hughes G, Kinney SE, Golubic M (1998) Immunoblotting analysis of schwannomin/merlin in human schwannomas. Am J Otol 19:654–659 [PubMed]
- Hayashi K, Yonemura S, Matsui T, Tsukita S (1999) Immunofluorescence detection of ezrin/radixin/moesin (ERM) proteins with their carboxyl-terminal threonine phosphorylated in cultured cells and tissues. J Cell Sci 112:1149–1158 [DOI] [PubMed]
- Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, et al (1996) Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J Cell Biol 135:37–51 [DOI] [PMC free article] [PubMed]
- Huynh DP, Pulst SM (1996) Neurofibromatosis 2 antisense oligodeoxynucleotides induce reversible inhibition of schwannomin synthesis and cell adhesion in STS26T and T98G cells. Oncogene 13:73–84 [PubMed]
- Hynes RO (1992) Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69:11–25 [DOI] [PubMed]
- Jacoby LB, MacCollin M, Louis DN, Mohney T, Rubio MP, Pulaski K, Trofatter JA, et al (1994) Exon scanning for mutation of the NF2 gene in schwannomas. Hum Mol Genet 3:413–419 [DOI] [PubMed]
- Kaiser-Kupfer MI, Freidlin V, Datiles MB, Edwards PA, Sherman JL, Parry D, McCain LM, et al (1989) The association of posterior capsular lens opacities with bilateral acoustic neuromas in patients with neurofibromatosis type 2. Arch Ophthalmol 107:541–544 [DOI] [PubMed]
- Koga H, Araki N, Takeshima H, Nishi T, Hirota T, Kimura Y, Nakao M, et al (1998) Impairment of cell adhesion by expression of the mutant neurofibromatosis type 2 (NF2) genes which lack exons in the ERM-homology domain. Oncogene 17:801–810 [DOI] [PubMed]
- Kunkel TA, Roberts JD, Zakour RA (1987) Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol 154:367–382 [DOI] [PubMed]
- LaJeunesse DR, McCartney BM, Fehon RG (1998) Structural analysis of Drosophila merlin reveals functional domains important for growth control and subcellular localization. J Cell Biol 141:1589–1599 [DOI] [PMC free article] [PubMed]
- Lamb RF, Ozanne BW, Roy C, McGarry L, Stipp C, Mangeat P, Jay DG (1997) Essential functions of ezrin in maintenance of cell shape and lamellipodial extension in normal and transformed fibroblasts. Curr Biol 7:682–688 [DOI] [PubMed]
- Lennon G, Auffray C, Polymeropoulos M, Soares MB (1996) The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics 33:151–152 [DOI] [PubMed]
- Lutchman M, Rouleau GA (1995) The neurofibromatosis type 2 gene product, schwannomin, suppresses growth of NIH 3T3 cells. Cancer Res 55:2270–2274 [PubMed]
- MacCollin M, Mohney T, Trofatter J, Wertelecki W, Ramesh V, Gusella J (1993) DNA diagnosis of neurofibromatosis 2; altered coding sequence of the merlin tumor suppressor in an extended pedigree. JAMA 270:2316–2320 (erratum: JAMA 272:1104 [1994]). [DOI] [PubMed]
- MacCollin M, Ramesh V, Jacoby LB, Louis DN, Rubio MP, Pulaski K, Trofatter JA, et al (1994) Mutational analysis of patients with neurofibromatosis 2. Am J Hum Genet 55:314–320 [PMC free article] [PubMed]
- Maeda M, Matsui T, Imamura M, Tsukita S (1999) Expression level, subcellular distribution and rho-GDI binding affinity of merlin in comparison with ezrin/radixin/moesin proteins. Oncogene 18:4788–4797 [DOI] [PubMed]
- Magendantz M, Henry MD, Lander A, Solomon F (1995) Interdomain interactions of radixin in vitro. J Biol Chem 270:25324–25327 [DOI] [PubMed]
- Martin M, Roy C, Montcourrier P, Sahuquet A, Mangeat P (1997) Three determinants in ezrin are responsible for cell extension activity. Mol Biol Cell 8:1543–1557 [DOI] [PMC free article] [PubMed]
- Martuza RL, Eldridge R (1988) Neurofibromatosis 2 (bilateral acoustic neurofibromatosis). N Engl J Med 318:684–688 [DOI] [PubMed]
- Matsui T, Maeda M, Doi Y, Yonemura S, Amano M, Kaibuchi K, Tsukita S (1998) Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J Cell Biol 140:647–657 [DOI] [PMC free article] [PubMed]
- Murthy A, Gonzalez-Agosti C, Cordero E, Pinney D, Candia C, Solomon F, Gusella J, et al (1998) NHE-RF, a regulatory cofactor for Na(+)-H+ exchange, is a common interactor for merlin and ERM (MERM) proteins. J Biol Chem 273:1273–1276 [DOI] [PubMed]
- Nakamura F, Amieva MR, Furthmayr H (1995) Phosphorylation of threonine 558 in the carboxyl-terminal actin-binding domain of moesin by thrombin activation of human platelets. J Biol Chem 270:31377–31385 [DOI] [PubMed]
- Nur EKMS, Sizeland A, D'Abaco G, Maruta H (1992) Asparagine 26, glutamic acid 31, valine 45, and tyrosine 64 of Ras proteins are required for their oncogenicity. J Biol Chem 267:1415–1418 [PubMed]
- Obremski VJ, Hall AM, Fernandez-Valle C (1998) Merlin, the neurofibromatosis type 2 gene product, and beta1 integrin associate in isolated and differentiating Schwann cells. J Neurobiol 37:487–501 [PubMed]
- Reczek D, Berryman M, Bretscher A (1997) Identification of EBP50: a PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J Cell Biol 139:169–179 [DOI] [PMC free article] [PubMed]
- Reczek D, Bretscher A (1998) The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J Biol Chem 273:18452–18458 [DOI] [PubMed]
- Ritter M, Wöll E, Haller T, Dartsch PC, Zwierzina H, Lang F (1997) Activation of Na+/H(+)-exchanger by transforming Ha-ras requires stimulated cellular calcium influx and is associated with rearrangement of the actin cytoskeleton. Eur J Cell Biol 72:222–228 [PubMed]
- Rosenbaum C, Kluwe L, Mautner VF, Friedrich RE, Müller HW, Hanemann CO (1998) Isolation and characterization of Schwann cells from neurofibromatosis type 2 patients. Neurobiol Dis 5:55–64 [DOI] [PubMed]
- Rouleau GA, Merel P, Lutchman M, Sanson M, Zucman J, Marineau C, Hoang-Xuan K, et al (1993) Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature 363:515–521 [DOI] [PubMed]
- Sainz J, Huynh DP, Figueroa K, Ragge NK, Baser ME, Pulst SM (1994) Mutations of the neurofibromatosis type 2 gene and lack of the gene product in vestibular schwannomas. Hum Mol Genet 3:885–891 [DOI] [PubMed]
- Scoles DR, Huynh DP, Morcos PA, Coulsell ER, Robinson NG, Tamanoi F, Pulst SM (1998) Neurofibromatosis 2 tumour suppressor schwannomin interacts with betaII-spectrin. Nat Genet 18:354–359 [DOI] [PubMed]
- Serrador JM, Alonso-Lebrero JL, del Pozo MA, Furthmayr H, Schwartz-Albiez R, Calvo J, Lozano F, et al (1997) Moesin interacts with the cytoplasmic region of intercellular adhesion molecule-3 and is redistributed to the uropod of T lymphocytes during cell polarization. J Cell Biol 138:1409–1423 [DOI] [PMC free article] [PubMed]
- Shaw RJ, McClatchey AI, Jacks T (1998) Regulation of the neurofibromatosis type 2 tumor suppressor protein, merlin, by adhesion and growth arrest stimuli. J Biol Chem 273:7757–7764 [DOI] [PubMed]
- Sherman L, Xu HM, Geist RT, Saporito-Irwin S, Howells N, Ponta H, Herrlich P, et al (1997) Interdomain binding mediates tumor growth suppression by the NF2 gene product. Oncogene 15:2505–2509 [DOI] [PubMed]
- Stemmer-Rachamimov AO, Gonzalez-Agosti C, Xu L, Burwick JA, Beauchamp R, Pinney D, Louis DN, et al (1997) Expression of NF2-encoded merlin and related ERM family proteins in the human central nervous system. J Neuropathol Exp Neurol 56:735–742 [PubMed]
- Takeshima H, Izawa I, Lee PS, Safdar N, Levin VA, Saya H (1994) Detection of cellular proteins that interact with the NF2 tumor suppressor gene product. Oncogene 9:2135–2144 [PubMed]
- Takeshima H, Nishi T, Yamamoto K, Kino T, Nakamura H, Saya H, Kochi M, et al (1998) Loss of merlin-p85 protein complex in NF2-related tumors. Int J Oncol 12:1073–1078 [DOI] [PubMed]
- Tikoo A, Varga M, Ramesh V, Gusella J, Maruta H (1994) An anti-Ras function of neurofibromatosis type 2 gene product (NF2/Merlin). J Biol Chem 269:23387–23390 [PubMed]
- Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, et al (1993) A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell 75:826 [DOI] [PubMed]
- Turunen O, Sainio M, Jääskeläinen J, Carpén O, Vaheri A (1998) Structure-function relationships in the ezrin family and the effect of tumor-associated point mutations in neurofibromatosis 2 protein. Biochim Biophys Acta 1387:1–16 [DOI] [PubMed]
- Turunen O, Wahlström T, Vaheri A (1994) Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J Cell Biol 126:1445–1453 [DOI] [PMC free article] [PubMed]
- Xu HM, Gutmann DH (1998) Merlin differentially associates with the microtubule and actin cytoskeleton. J Neurosci Res 51:403–415 [DOI] [PubMed]
- Yun CH, Oh S, Zizak M, Steplock D, Tsao S, Tse CM, Weinman EJ, et al (1997) cAMP-mediated inhibition of the epithelial brush border Na+/H+ exchanger, NHE3, requires an associated regulatory protein. Proc Natl Acad Sci USA 94:3010–3015 (erratum: Proc Natl Acad Sci USA 94:10006 [1997]) [DOI] [PMC free article] [PubMed]
- Zucman-Rossi J, Legoix P, Der Sarkissian H, Cheret G, Sor F, Bernardi A, Cazes L, et al (1998) NF2 gene in neurofibromatosis type 2 patients. Hum Mol Genet 7:2095–2101 [DOI] [PubMed]