Small Evolutionarily Conserved RNA, Resembling C/D Box Small Nucleolar RNA, Is Transcribed from PWCR1, a Novel Imprinted Gene in the Prader-Willi Deletion Region, Which Is Highly Expressed in Brain (original) (raw)
Abstract
Prader-Willi syndrome is a complex neurodevelopmental disorder caused by the inactivation or deletion of imprinted, paternally expressed genes in chromosome band 15q11.2. We report the identification and characterization of PWCR1, a novel imprinted gene within that region, and its mouse orthologue, Pwcr1, which was mapped to the conserved syntenic region on mouse chromosome 7. Expressed only from the paternal allele, both genes require the imprinting-center regulatory element for expression and are transcribed from the same strand. They are intronless and do not appear to encode a protein product. High human/mouse sequence similarity (87% identity) is limited to a 99-bp region called “HMCR” (for “human-mouse conserved region”). The HMCR sequence has features of a C/D box small nucleolar RNA (snoRNA) and is represented in an abundant small transcript in both species. Located in nucleoli, snoRNAs serve as methylation guidance RNAs in the modification of ribosomal RNA and other small nuclear RNAs. In addition to the nonpolyadenylated small RNAs, larger polyadenylated PWCR1 transcripts are found in most human tissues, whereas expression of any Pwcr1 RNAs is limited to mouse brain. Genomic sequence analysis reveals the presence of multiple copies of PWCR1 and Pwcr1 that are organized within local tandem-repeat clusters. On a multispecies Southern blot, hybridization to an HMCR probe encoding the putative snoRNA is limited to mammals.
Introduction
In somatic cells, some mammalian genes are expressed only from one parental allele. By a process called “gametic imprinting” or “genomic imprinting,” these genes are epigenetically marked during gametogenesis, according to their parental origin (Bartolomei and Tilghman 1997). The nature of the epigenetic mark is not fully understood. Although DNA methylation is required for the maintenance of the imprinted state (Tucker et al. 1996; Jaenisch 1997), the mechanisms regulating genomic imprinting are likely to be more complex and to involve gene-specific as well as chromosomal domain–specific modifications (Barlow 1995; Bartolomei and Tilghman 1997; Nicholls et al. 1998). Recently, a mechanism for control of imprinting by insulator elements that bind the enhancer-blocking protein CTCF, when unmethylated, was discovered (Bell and Felsenfeld 2000; Hark et al. 2000). Regional control of imprinting may explain the clustering of imprinted genes. Human chromosome region 15q11-q13 and the 7C–D1 region in the mouse contain such a conserved imprinted gene cluster. Several genes in this region are expressed only from the paternally derived allele, including SNRPN (Özçelik et al. 1992; Reed and Leff 1994)/Snrpn (Leff et al. 1992), SNURF (Gray et al. 1999), IPW (Wevrick et al. 1994)/Ipw (Wevrick and Francke 1997), NDN/Ndn (Jay et al. 1997; MacDonald and Wevrick 1997; Watrin et al. 1997), ZNF127 (Jong et al. 1999_b_)/Zfp127 (Jong et al. 1999_a_), MAGEL2/Magel2 (Boccaccio et al. 1999), the testis-specific transcript C15orf2 (Farber et al. 2000), and two poorly characterized transcripts, PAR-1 and PAR-5 (Sutcliffe et al. 1994). Thus far, in this region, UBE3A is the only gene known to be expressed exclusively from the maternal allele, and that imprinted expression is found only in brain (Albrecht et al. 1997; Kishino et al. 1997; Matsuura et al. 1997).
Genomic alterations in the 15q11-q13 region that result in the loss of gene products of imprinted genes are associated with two clinically distinct neurodevelopmental disorders. Prader-Willi syndrome (PWS [MIM 176270]) is characterized by severe neonatal hypotonia, failure to thrive during infancy, subsequent hyperphagia leading to obesity, hypogonadism, growth delay, mild dysmorphic features, and mental retardation (Holm et al. 1993). In contrast, individuals with Angelman syndrome (AS) have more severe mental retardation, with extreme speech impairment, ataxia, seizures, and motor hyperactivity; they possess a happy disposition with frequent outbursts of inappropriate laughter (Williams et al. 1995). Approximately 70% of individuals with either PWS or AS have an ∼4-Mb de novo deletion in the 15q11-q13 region. Deletion on the paternal chromosome results in PWS, whereas maternal deletion of the same region results in AS. Approximately 28% of cases of PWS are caused by maternal uniparental disomy for chromosome 15, whereas considerably fewer (∼2%) of all cases of AS are due to paternal uniparental disomy (Knoll et al. 1991; Mascari et al. 1992). Some of the remaining cases of AS are due to inactivating mutations in UBE3A (Kishino et al. 1997; Matsuura et al. 1997). PWS and AS can also be caused by rare “imprinting mutations,” small submicroscopic deletions of variable size (10–1,000 kb) within the 5′ region of SNRPN. Maternally derived microdeletions result in AS. Microdeletions of the paternal copy result in PWS and, in addition, lead to loss of expression of several paternally expressed genes. The observation and delineation of spontaneously occurring microdeletions have been used to define a bipartite _cis_-acting regulatory element, referred to as the “imprinting center” (IC) (Buiting et al. 1995).
Two different deletion mutations were created in mice, in the attempt to generate a mouse model for PWS (Yang et al. 1998). Mice carrying a partial Snrpn deletion, which removes exon 6 and portions of exons 5 and 7, are phenotypically normal. In contrast, mice harboring a 42-kb deletion including exons 1–6 of Snrpn and the putative IC region manifest hypotonia, failure to thrive, and early death, features similar to those seen in human infants with PWS. Moreover, this mouse model also recapitulates loss of expression of paternally expressed nondeleted genes Zfp127, Ndn, Ipw (Yang et al. 1998), and Magel2 (Lee et al. 2000). Mutant mice individually targeted for Snurf (Tsai et al. 1999_b_), Ipw (Jong et al. 1999_a_), Zfp127 (Jong et al. 1999_a_), and Ndn (Gerard et al. 1999; Tsai et al. 1999_a_) are normal, except for the _Ndn_-deficient mice generated by Gerard et al. (1999), which displayed hypotonia and neonatal respiratory distress; in contrast, the _Ndn_-deficient mice generated by Tsai et al. (1999_a_) had no abnormal phenotype. The contradictory results may be partly due to differences in strain backgrounds.
Mice engineered to carry a larger paternal deletion, extending from Snrpn to Ube3A, are hypotonic and severely growth retarded, and ∼80% of them die before weaning (Tsai et al. 1999_b_). In contrast, mice carrying a paternally derived deletion (P30PUb) of the p locus, which extends proximally to include the Ipw gene, do not display neonatal lethality (Johnson et al. 1995). Taken together, these results suggest the existence of a gene that is located between Snrpn and Ipw and that may be important for the phenotype of the mouse models of PWS. Here we report the identification and characterization of a novel human gene, PWCR1, located between SNRPN and IPW. For comparative studies in search of functional domains, we also cloned the mouse orthologue, Pwcr1. Both genes show similar hallmarks. They contain multiple stop codons in all potential reading frames and are intronless. Both PWCR1 and Pwcr1 generate alternative transcripts, the most abundant of which is a short (140-nt) RNA, of highly similar sequence, that may represent a novel small nucleolar RNA (snoRNA). PWCR1 is expressed in a variety of tissues, including brain, whereas Pwcr1 expression is brain specific. Onset of PWCR1/Pwcr1 expression occurs during embryonic development and is maintained throughout adulthood. Both genes are imprinted with exclusive expression from the paternally derived allele. PWCR1 is not expressed in PWS samples with either the classic deletion or an IC microdeletion. Likewise, in the mouse, Pwcr1 expression from the paternal allele depends on the presence of the _cis_-acting IC.
Material and Methods
cDNA Isolation and Analysis
A fetal brain λZAP II cDNA library (Stratagene) was screened with probe hB, by standard plaque-lift hybridization. Filters were hybridized in Church buffer at 65°C. After three rounds of selection, plasmids were excised from the plaque-purified phage in vivo, according to the manufacturer’s instructions_._ DNA sequencing was performed by cycle sequencing with an ABI Prism 377 sequencer (Applied Biosystems).
Isolation and Labeling of Probes
DNA fragments used as probes were generated by PCR amplification of genomic DNA, PAC, or cosmid templates. Sequences of primers used for probes are as follows (nucleotide numbers refer to human [accession number AF241255] or mouse [accession number AF241256] sequences as submitted to GenBank): probe hA, H1TD (5′-AACATGTGCCTGCCCTCCAT-3′ [positions 959–976]) and H2TD (5′-GCAAGGACTAGGTGAATGTCC-3′ [positions 312–292]); probe hB, STS-F (5′-ACCTCAGTTCGACGAGGATG-3′ [positions 1115–1134]) and STS-R (5′-CCTCATTTGCAGGGACAAAT-3′ [positions 1233–1214]); probe mC, M27TD (5′-GGAGCTTCGGCCCATTGTTC-3′ [positions 1184–1203]) and M33TD (5′-CTCCTTCTCTATTTCCTAGC-3′ [positions 1952–1933]); probe mD, M58TD (5′-AACGAGCTTGGATCTATGATG-3′ [positions 1297–1317]) and M59TD (5′-CCGAAGAAGTCAAGAACAATG-3′ [positions 1476–1456]); and probe mE, M26TD (5′-AAGTGCTATGGGCGTCAAGA-3′ [positions 1417–1436]) and M32TD (5′-GACTGAGTCACTGCCCGATA-3′ [positions 1745–1725]).
Prior to being labeled, DNA fragments were purified by agarose gel electrophoresis. For [α-32P]-dCTP labeling of double-stranded DNA probes, the Multiprime DNA labeling system RPN 1601Z (Amersham) was used. Single-stranded DNA probes were made by PCR amplification according to the protocol of Konat et al. (1994), with minor modifications. The forward or reverse primers used in the PCR were from the same primer pair used to generate the DNA fragment template. Both forward and reverse single-stranded probes were generated from the same PCR cocktail, consisting of 1× PCR buffer, 25–30 ng of DNA template, 0.5 μl each of 1 mM each of dATP, dTTP, and dGTP mixture, 0.3 μl of 100 μM dCTP, 50 μCi [α-32P]-dCTP, 2.5 μl of 10 μM forward or reverse primer, and 1 U of Ampli_Taq_ DNA polymerase (Applied Biosystems), in a final volume of 25 μl. Conditions used for PCR were 94°C for 2 min; then 30 cycles of 94°C for 45 s, 58°C for 1 min, and 72°C for 4 min; and then 72°C for 10 min. To generate RNA probes, DNA fragments amplified from mouse genomic DNA by primers M58TD and M59TD were cloned by use of a TOPO TA cloning kit (Invitrogen). RNA probes were synthesized from appropriate clones, by use of an RNA transcription kit (Stratagene).
RNA Methods
Total RNA was purified with RNA STAT (TelTest ‘B' Inc.), according to manufacturer’s directions. For northern blots, 10 μg of total RNA were loaded in each lane. Probes were hybridized either to Hybond N+ membranes with total RNA (figs. 3_d_ and 4_a_ and b) or to commercial filters with poly A+ RNA (figs. 3_a_–c and 8_a_ and b) (CLONTECH), by use of ExpressHyb (CLONTECH) hybridization solution at 65°C. Blots were washed at high stringency, in a final solution of 0.2 × SSC/0.1% SDS, at 65°C for 20 min.
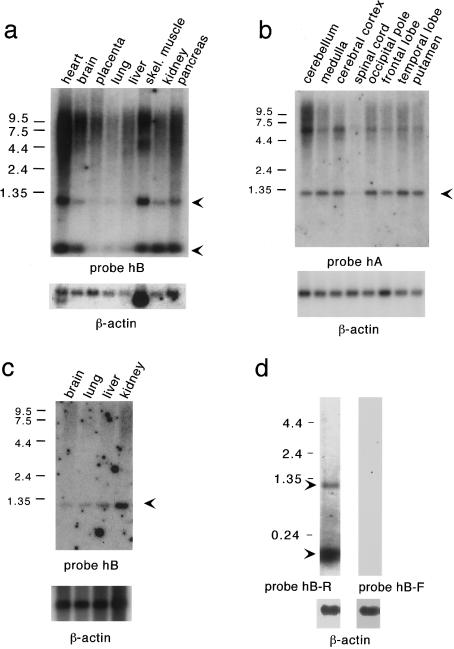
Figure 3.
Northern blot analyses of polyadenylated RNA, with probes indicated. a, Human adult tissues. b, Different regions of human brain. c, Human fetal tissues. Arrowheads indicate major PWCR1 transcripts. d, Direction of transcription of PWCR1. Identical northern blots contain total RNA from a normal control brain. Both the 1.2-kb and the 140-nt transcripts were detected only with probe hB-R. β-actin control probes were used to assess the amount of RNA loaded.
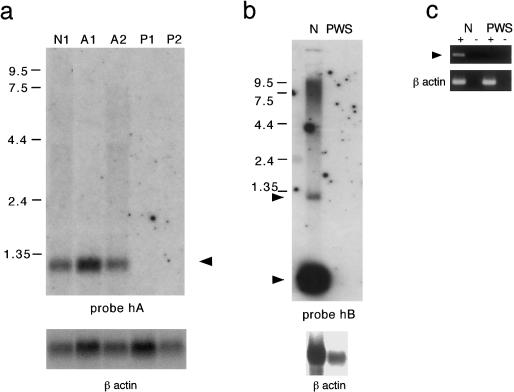
Figure 4.
Expression of PWCR1 transcripts exclusively from the paternal allele. Northern blots were hybridized with either probe hA or probe hB, as indicated. a, Total lymphoblast RNA from control individuals (lane N1) and from individuals with either AS (lanes A1 and A2) or PWS (lanes P1 and P2). b, Total brain RNA from a control (lanes N) and a PWS subject (lanes PWS). A β-actin probe was used as control for RNA loading. Arrowheads indicate PWCR1 transcripts. c, RT-PCR using _PWCR1_-specific primers H58JS and H59JS on brain RNA from a control individual (lanes N), which amplified a 548-bp product. No _PWCR1_-specific product was observed in the PWS brain RNA sample, whereas β-actin–specific primers amplified products in both samples. A plus sign (+) denotes the presence of reverse transcriptase; a minus sign (−) denotes its absence.
For reverse transcriptase–PCR (RT-PCR) assays, RNA was treated with DNaseI (RNAse-free; Boehringer Mannheim) prior to incubation with reverse transcriptase SuperScript II (Gibco-BRL). One-fifth of the reverse-transcriptase reaction was used for PCR. Conditions used for PCR were 94°C for 3 min; then 28 cycles of 94°C for 45 s, 58°C for 45 s, and 72°C for 30 s; and then 72°C for 5 min. Primer sequences used for monoallelic expression analysis were STS-F (5′-ACCTCAGTTCGACGAGGATG-3′), STS-R (5′-CCTCATTTGCAGGGACAAAT-3′), H1TD (5′-AACATGTGCCTGCCCTCCAT-3′), H2TD (5′-GCAAGGACTAGGTGAATGTCC-3′), H58JS (5′-ATGTGGTCTCTTATGGGTGAT-3′ [positions 182–202]), H59JS (5′-ATCCCTCTCAACATCACTGC-3′ [positions 728–709]), M49TD (5′-GCCCATAATCCATGTGGTT-3′ [positions 573–591]), and M50TD (5′-CAGGTGACCTAGGGCAAGT-3′ [positions 780–762]).
Genetic Mapping of Pwcr1
For chromosomal localization of the mouse Pwcr1 gene, we genotyped 94 N2 animals from the interspecific backcross (C57BL/6JEi × SPRET/Ei) F1× SPRET/Ei (BSS) panel (The Jackson Laboratory Mapping Panels, Bar Harbor, ME), for a size polymorphism detected with primers M51TD (5′-GGCACGAGGTTCCTTTCAG-3′) and M52TD (5′-CAAGTGCTTCCTGGGTCC-3′). The typing results were sent to The Jackson Laboratory, for comparison with previous typing data on markers on mouse chromosome 7.
Genomic Southern Blot
Eight micrograms of genomic DNA from various species were digested with either _Eco_RI or _Bam_HI and then were electrophoresed and Southern blotted onto Hybond N membranes. Probe mD was hybridized to filters by use of ExpressHyb (CLONTECH) hybridization solution at 65°C. Blots containing chicken, rabbit, cow, dog, pig, baboon, and human DNA were washed in a final solution of 0.2 × SSC/0.1% SDS at 65°C. The filter containing DNA from Xenopus, opossum, wallaby, mouse, and rat was washed in a final solution of 2 × SSC/0.1% SDS at 58°C.
Cell Lines and Cell Culture
Epstein-Barr virus–transformed lymphoblasts from control individuals (LCL 1497 and LCL 1385), individuals with PWS (LCL 863 [del (15)(q11.2q13.1), cell line GM09133, obtained from the Camden Cell Repository, Camden, NJ] and LCL 1309 [O family microdeletion, obtained from A. Beaudet, Baylor College of Medicine, Houston]), and individuals with AS (LCL 1101 [del (15)(q11q13) mat transformed in our laboratory, with informed consent] and LCL 1201 [del (15)(q11q13) mat, cell line GM11404, obtained from the Camden Cell Repository]) were cultured in RPMI 1640 medium containing 10% fetal bovine serum (GIBCO), glutamine, and antibiotics.
Dendrogram
For _PWCR1_-and _PWCR1_-like genes (PWCR1L1_–_PWCR1L25), as well as for _Pwcr1_- and _Pwcr_-like genes (Pwcr1L-1–L-9), sequences were compared by the multiple-sequence alignment program PileUp from the GCG SeqWeb sequence-analysis software. The gap-creation penalty was set at 5, and the gap-extension penalty was set at 1; maximum-input sequence range and maximum number of gap characters added were set at 5,000 and 2,000, respectively.
Results
Identification of the Human PWCR1 Gene
To identify novel imprinted genes within the PWS deletion region, we searched the databases for expressed-sequence tags (ESTs) that map within the 15q11-q13 PWS deletion region and then tested these ESTs for parent-of-origin–specific expression. Radiation-hybridscoring data of sequence-tagged sites (STSs), mapped to chromosome 15 by use of the Genebridge 4 radiation-hybrid panel, were retrieved from RHdb (radiation-hybrid database at the European Bioinformatics Institute). The retrieved data were then submitted to the Whitehead Institute for Biomedical Research/MIT Center for Genome Research mapping server, to identify ESTs that map close to 15q11-q13. To identify imprinted transcripts, RT-PCR was performed with STS primers from candidate ESTs on RNA extracted from lymphoblastoid cell lines of individuals with PWS or AS and of normal controls. When we tested 60 ESTs by this approach, several monoallelically expressed transcripts were identified. One of them was a 3′ partial transcript of I.M.A.G.E clone 204357 (GenBank accession number H59928), derived from a human fetal liver-and-spleen cDNA library. STS primers specific for this EST amplified a 119-bp product from control and AS cell lines but not from PWS cell lines. The HUGO/GDB Human Gene Nomenclature Committee named the locus “PWCR1_” (for “Prader-Willi chromosome region 1). The corresponding 1.2-kb cDNA clone was sequenced by primer walking. In figure 1_a_,_ monoallelic expression from the paternally derived chromosome 15 was subsequently confirmed by RT-PCR with primers H1TD and H2TD.
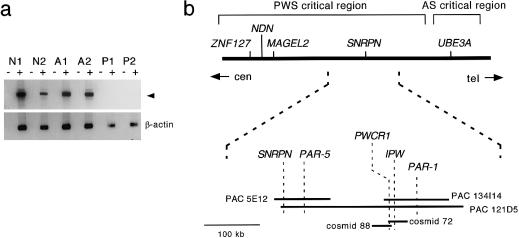
Figure 1.
a, Monoallelic expression of PWCR1, demonstrated by RT-PCR analyses of control (lanes N1 and N2), AS (lanes A1 and A2), and PWS (lanes P1 and P2) lymphoblastoid cell lines. Primers H1TD and H2TD amplified a 690-bp product (arrowhead) from control and AS cell lines but not from PWS cell lines. β-Actin–specific primers were included as positive controls. A plus sign (+) denotes presence of reverse transcriptase; a minus sign (−) denotes its absence. Lane N1, LCL 1497. Lane N2, LCL 1385. Lane A1, LCL 1201. Lane A2, LCL 1101. Lane P1, LCL 863. Lane P2, LCL 1309. b, Schematic map of known imprinted genes in the human 15q11.2 region. PWCR1 is located centromeric to IPW. PWCR1 probes hybridize to PAC pDJ134I14, PAC pDJ121D5, cosmid 88, and cosmid 72.
To determine the exact genomic location of PWCR1, probes representing the 3′ end (probe hB) and the middle region (probe hA) of PWCR1 were hybridized to Southern blots of DNA from PACs and cosmids that had previously been mapped to 15q11-q13 (Sutcliffe et al. 1997). Probes hA and hB hybridized to overlapping cosmids 88 and 72 under stringent conditions, whereas an IPW_-specific probe hybridized only to cosmid 72–specific fragments. PWCR1 probes also detected bands from PAC pDJ134I14 and PAC pDJ121D5 (data not shown). These results indicate that PWCR1 is located at the distal end of the PWS critical region and is excluded from the AS critical region, as shown in figure 1_b_._
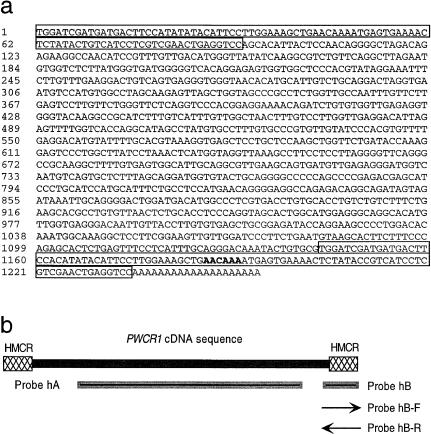
Sequence analysis of PWCR1 cDNA revealed that it contains 50% AT sequences, as shown in figure 2_a_. The PWCR1 cDNA clone is polyadenylated, but it does not contain the consensus polyadenylation signal (AATAAA). The sequence that most closely resembles AATAAA is located at positions 1186–1191 (AACAAA), which are shown in boldface in figure 2a. In all three reading frames, the cDNA sequence contains multiple stop codons and no significant open reading frame (ORF). The longest ORF is 114 bp, but it lacks an acceptable Kozak consensus sequence flanking the potential start ATG. Analysis of the cDNA sequence reveals a near-identical repeat at the 5′ and 3′ ends, with nucleotides 1–93 being 98% identical to nucleotides 1143–1235 (fig. 2_a_). We called these repeats “HMCR” (for “human-mouse conserved region”), because an 87%-identical sequence is present in the mouse orthologue (as shown in fig. 6_b_).
Figure 2.
a, Sequence of PWCR1 cDNA clone. The 5′ and 3′ repeats (called “HMCR” [human-mouse conserved region]) are boxed. The imperfect putative poly-A signal is in boldface. b, Probes used for analysis of PWCR1 expression, by northern blot. Probes hA and hB are double-stranded DNA probes. Probes hB-F and hB-R are PCR-labeled single-stranded DNA probes.
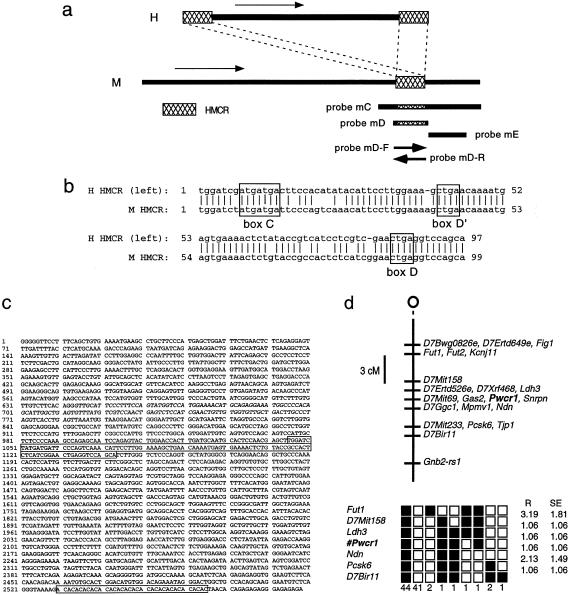
Figure 6.
a, Comparison of human (H) and mouse (M) genes, which reveals that PWCR1 and Pwcr1 cDNAs contain a block of conserved sequences (HMCR). Arrows indicate direction of transcription. Probes mC, mD, and mE are double-stranded DNA probes. Probes mD-F and mD-R are RNA probes. b, Alignment of human (H) and mouse (M) HMCRs. Sequences are 87% identical over 99 nucleotides. Conserved sequences representing C, D′, and D boxes that identify a class of snoRNAs are boxed (Smith and Steitz 1998). c, Sequence of Pwcr1 derived from cDNA clone and HTGS data. The HMCR and the AC-repeat region are boxed. The sequence identical to Ipw exon A1 (nt 644–789) is in italics. d, Mapping of Pwcr1 to mouse chromosome 7, in a conserved syntenic region with human 15q11.2. The Jackson Laboratory Mapping Panels BSS backcross was typed for a Pwcr1 fragment-length polymorphism. Raw typing data for other markers in the region also were obtained from The Jackson Laboratory Mapping Panels Web site. Missing typing was inferred from surrounding data, wherever assignment was unambiguous. Top, BSS backcross map of chromosome 7. The centromere is at the top of the map. Loci mapping to the same position are listed in alphabetical order. Bottom, Haplotypes from the BSS backcross. Loci are listed in order, with the most proximal at the top. Black boxes represent the C57BL6/JEi allele, and white boxes represent the SPRET/Ei allele. Listed at the bottom of each column is the number of animals with each haplotype. R = percent recombination for adjacent loci. SE = standard error for each R.
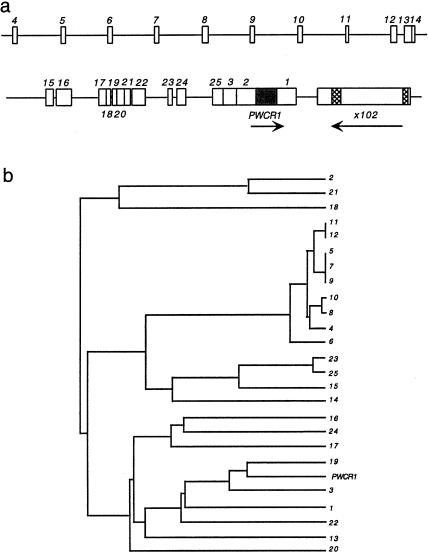
Nucleic-acid and protein-database searches identified one sequence with significant similarity to _PWCR1_—the genomic clone x102 (GenBank accession number AF017338), which has two regions of similarity with PWCR1. Originally, x102 was isolated in a screen for differentially expressed genes in individuals with psychiatric disorders (Yee and Yolken 1997). The x102 genomic clone contains an ORF encoding a hypothetical 138-amino-acid protein with no significant similarity to known proteins in the database. We mapped x102 to a BAC clone (GenBank accession number AC009696) that contains genomic DNA of the chromosome 15q11.2 region (location shown in fig. 5_a_).
Figure 5.
a, Simplified map of the region surrounding PWCR1, showing both the location of _PWCR1_-like truncated copies (PWCR1L1_–_PWCR1L25) and the direction of transcription (arrows). Hatched boxes within x102 represent sequences similar to PWCR1. The arrow below x102 indicates the proposed direction of transcription, according to GenBank annotation (accession number AF017338). b, Dendrogram of relationships between PWCR1 and the various truncated copies, based on sequence similarity (also see table 1).
PWCR1 Transcripts and Expression Pattern
On northern blot analysis, PWCR1 was strongly expressed in multiple tissues. For the locations of the probes used, see figure 2b. With probe hB, we detected a predominant 1.2-kb transcript, a smaller ∼140-nt transcript, and a strong smear of ∼4–10 kb in all human tissues examined, as is shown in figure 3_a_. Signals corresponding to the 1.2-kb transcript and to the ∼4–10-kb smear were most intense for heart and skeletal muscle. The ∼140-nt transcript was also present at higher levels in heart and skeletal muscle, as well as in brain, kidney, and pancreas. Interestingly, the hybridization signal of the ∼140-nt transcript was greatly increased compared with those of the other transcripts on northern blots containing total RNA derived from brain tissues, suggesting that the vast majority of the ∼140-nt transcripts are not polyadenylated, as evidenced in figures 3_d_ and 4_b_.
A comparison of the different transcripts detected by probes hA and hB on northern blots indicated that the ∼140-nt product is transcribed from the HMCR. Probe hA detects the 1.2-kb transcript, as well as the smear, but does not detect the ∼140-nt transcript (fig. 3_b_). Probe hB, on the other hand, detects all transcripts except for the 6-kb transcript (figs. 3_a_ and 4_b_).
Because individuals with PWS have a neurological phenotype, we examined PWCR1 expression levels in different regions of the brain (fig. 3_b_). In addition to the predominant 1.2-kb and large heterologous transcripts, a distinct ∼6-kb transcript was detected in cerebellum, cerebral cortex, medulla, putamen, and in the occipital, frontal, and temporal lobes, with probe hA. PWCR1 is weakly expressed in the spinal cord. In fetal brain, lung, liver, and kidney, PWCR1 transcripts were detected with probe hB (fig. 3_c_), indicating that onset of PWCR1 expression occurs during fetal development. The ∼140-nt transcript was also detected in fetal tissues by probe hB but is not included in figure 3c.
To determine whether only one or both strands of PWCR1 are transcribed, single-stranded probes were hybridized to total RNA from normal brain. Single-stranded probe hB-R detected the 1.2-kb and ∼140-nt transcripts. In contrast, probe hB-F did not detect any transcripts (fig. 3_d_). Similar results were obtained on blots containing LCL RNA (not shown). These data demonstrate that the 1.2-kb and ∼140-nt RNAs are transcribed from the same strand.
Imprinted Expression of PWCR1 from the Paternal Allele
To verify the RT-PCR results that initially identified PWCR1 as an imprinted gene, we performed northern blot analysis by using total lymphoblast RNA from two individuals with PWS (P1 and P2), two individuals with AS (A1 and A2), and a normal control (N1). P1, A1, and A2 contain standard-size deletions of 15q11-q13, whereas P2 has a microdeletion that removes the IC regulatory element. Probe hA detected no expression of PWCR1 in PWS cell lines, whereas control and AS cell lines expressed similar levels of 1.2-kb transcripts (fig. 4_a_). Similar results were obtained with probe hB, which detected both the 1.2-kb and ∼140-nt transcripts in the same control and AS cell lines (data not shown) and in brain RNA derived from a control individual but not in brain RNA from an individual with PWS (fig. 4_b_). These results were confirmed by RT-PCR with primers H58JS and H59JS (fig. 4_c_). Taken together, these results clearly demonstrate that the highly expressed PWCR1 RNAs are transcribed exclusively from the paternally derived allele and that PWCR1 is imprinted in brain and lymphoblasts. Moreover, the absence of PWCR1 transcripts in P2 LCLs indicate that PWCR1 expression is dependent on an intact IC.
_PWCR1_—Location within a Cluster of Truncated Copies
To determine the genomic structure of PWCR1, PCR products derived from cosmid 88 and from PAC pDJ134I14 were sequenced and compared with the PWCR1 cDNA clone. We found a perfect match between the cDNA and genomic sequences, indicating that PWCR1 is an intronless gene. BLASTN analysis of the PWCR1 cDNA, against the high-throughput genome-sequence (HTGS) database, identified a partially sequenced BAC (GenBank accession number AC009696) that had been mapped to 15q11-q13. A contiguous 50-kb sequence contains multiple, truncated copies of PWCR1. Further analysis revealed a complex genomic structure, as shown in figure 5_a_ and table 1. The truncated copies immediately flanking the PWCR1 gene were designated “_PWCR1L1,_” “_PWCR1L2,_” and “_PWCR1L3._” As others were discovered, they were designated “_PWCR1L4_”–“_PWCR1L25,_” in a 5′-to-3′ order. A dendrogram illustrates the relationship of the various copies to each other (fig. 5_b_). The sequence of genomic clone x102 is located ∼1 kb from the 3′ end of PWCR1L1 (fig. 5_a_). To further analyze the 50-kb genomic sequence containing PWCR1, we used NIX software (UK Human Genome Mapping Project Resource Center), a bioinformatics tool that consolidates various DNA analysis programs within one interface. This analysis did not identify significant similarity to known transcription initiation sequences or promoter sequences.
Table 1.
**PWCR1**-Like Genes and Their Location Relative to PWCR1
| Locus | AC009696 Position | Size(bp) | Identity with PWCR1(%) |
|---|---|---|---|
| PWCR1 | 146792–148030 | 1,235 | 100 |
| PWCR1L1 | 148031–149154 | 1,123 | 84 |
| PWCR1L2 | 145649–146792 | 1,143 | 77 |
| PWCR1L3 | 145093–145649 | 556 | 82 |
| PWCR1L4 | 110417–110516 | 99 | 86 |
| PWCR1L5 | 113195–113267 | 72 | 91 |
| PWCR1L6 | 115826–115930 | 104 | 84 |
| PWCR1L7 | 118535–118580 | 45 | 95 |
| PWCR1L8 | 121310–121416 | 106 | 86 |
| PWCR1L9 | 124063–124138 | 75 | 95 |
| PWCR1L10 | 126803–126903 | 100 | 86 |
| PWCR1L11 | 129487–129561 | 74 | 89 |
| PWCR1L12 | 132140–132243 | 103 | 86 |
| PWCR1L13 | 132892–133166 | 274 | 74 |
| PWCR1L14 | 133195–133311 | 116 | 82 |
| PWCR1L15 | 134971–135159 | 188 | 81 |
| PWCR1L16 | 135545–136195 | 650 | 77 |
| PWCR1L17 | 137990–138214 | 224 | 76 |
| PWCR1L18 | 138388–138480 | 92 | 79 |
| PWCR1L19 | 138760–138831 | 71 | 83 |
| PWCR1L20 | 138870–139281 | 411 | 76 |
| PWCR1L21 | 139425–139576 | 151 | 77 |
| PWCR1L22 | 139876–140475 | 595 | 77 |
| PWCR1L23 | 141871–141986 | 115 | 93 |
| PWCR1L24 | 142385–142793 | 408 | 77 |
| PWCR1L25 | 144498–144882 | 384 | 79 |
Identification of the Mouse Orthologue Pwcr1, Revealing a Conserved Region That Encodes a snoRNA
Having characterized human PWCR1 as an intronless gene that is unlikely to be translated into a protein product, we wanted to determine whether these specific features are conserved throughout evolution. We searched for a putative mouse homologue by screening a mouse-brain cDNA library (Stratagene Lambda ZAPII) with human PWCR1 probe hB. Four overlapping clones were isolated that, under stringent conditions, hybridized with the human probe. Sequence analysis of these clones led to the identification of a novel mouse gene containing a 99-bp region of similarity (87% identical) to human PWCR1. This conserved sequence corresponds to the HMCR repeats located at the 5′ and 3′ ends of the PWCR1 cDNA; see figure 6_a_ and b . We named the mouse gene “_Pwcr1,_” because it shows a high degree of similarity to specific regions of the human PWCR1 cDNA. Pwcr1 further resembles human PWCR1 in that it has an AT content of 50% and lacks a canonical polyadenylation signal. The sequence also contains an AC dinucleotide repeat (nt 2529–2565) and one copy of the exon A1 repeat (nt 644–789) first identified in the Ipw gene, where it is present in multiple copies (Wevrick and Francke 1997). The longest ORF of Pwcr1 is 252 bp, which, if translated, would result in an 84-amino-acid protein. The potential ATG start codon, however, is not contained within an acceptable Kozak consensus sequence. When we translated the Pwcr1 ORFs and searched the protein databases, no significant match was found. Comparison of the human and mouse HMCRs did not identify an ORF that can be translated into a conserved peptide sequence more than five or six amino acids long.
These data suggest that Pwcr1, like its human counterpart, is not translated into a protein product but, rather, may function as an RNA. When the HMCR sequence was compared with those of known small nuclear RNAs (snRNAs), no similarity was detected. However, when compared with the box C/D class of snoRNAs, defined by the presence of upstream C (AUGAUGA) and downstream D′ and D (CUGA) boxes (Maxwell and Fournier 1995), striking similarities were found. The HMCR contains one C and two D boxes, in the correct spacing and orientation (fig. 6_b_).
Genomic Structure and Chromosomal Localization of Pwcr1
Most vertebrate snoRNAs that have been identified are located within introns of protein-coding genes and are processed as the introns are removed (Maxwell and Fournier 1995). To learn more about the genomic organization of the Pwcr1 locus, we screened the WI/MIT820 mouse YAC library by PCR with primers from different regions of Pwcr1. Positive clones were identified by primers M49TD and M50TD located 5′ to the HMCR and were subsequently verified by other Pwcr1 primer pairs. PCR products derived from positive YAC clones were sequenced and compared with the cDNA sequence. The genomic and cDNA sequences were identical, indicating that Pwcr1 is intronless.
To genetically map the Pwcr1 locus, we genotyped the interspecies BSS-backcross mapping panel from The Jackson Laboratory Mapping Panels, for a PCR-amplicon size polymorphism. The Pwcr1 segregation pattern was identical to that of Snrpn (fig. 6_d_). Pwcr1 maps to position 29 cM (bands 7C–D1), within a region of known conserved synteny with human 15q11-q13 (Human-Mouse Homology Map).
A Local Cluster of Head-to-Tail Copies of Pwcr1
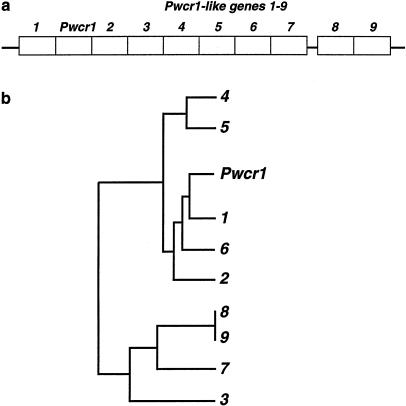
In lower eukaryotes, box C/D snoRNA genes are often organized into clusters (Dunbar et al. 2000). Having already identified truncated copies of PWCR1 in human, we asked whether mouse Pwcr1 was indeed a single-copy gene, as had been suggested by the genomic structure and chromosome-mapping results. We therefore searched the HTGS of the mouse, as it became available, and identified Pwcr1 within sequence AC026683. We restricted our analysis to a contiguous region 23,710 bp long. Pwcr1 is located within this region, adjacent to nine almost-identical sequences that we named “Pwcr1L-1_”–“_Pwcr1L-9,” as in table 2 and figure 7_a_. The dendrogram showing the relatedness of the nine copies is consistent with a model of cluster expansion by local duplication events, although it is based on HTGS data and not on finished sequence (fig. 7_b_). In addition, there are _Pwcr1_-like copies on other contigs. There are a total of 51 _Pwcr1_-related copies within the sequenced genomic clone. However, the total number of _Pwcr1_-related copies in the entire mouse chromosome 7C–D1 region is not yet known.
Table 2.
Pwcr1L Copies and Their Location Relative to Pwcr1
| Name | AC026683 Position | Size(bp) | Identity with Pwcr1(%) |
|---|---|---|---|
| Pwcr1 | 47730–50246 | 2,517 | 100 |
| Pwcr1L-1 | 46011–47692 | 1,682 | 98 |
| Pwcr1L-2 | 50248–52747 | 2,501 | 98 |
| Pwcr1L-3 | 52775–55286 | 2,520 | 97 |
| Pwcr1L-4 | 55288–57799 | 2,518 | 98 |
| Pwcr1L-5 | 57801–60302 | 2,502 | 97 |
| Pwcr1L-6 | 60334–62835 | 2,502 | 98 |
| Pwcr1L-7 | 62861–64787 | 1,930 | 98 |
| Pwcr1L-8 | 65401–67902 | 2,502 | 98 |
| Pwcr1L-9 | 67940–69712 | 1,775 | 97 |
Figure 7.
Local cluster of _Pwcr1_-like repeat units. a, Organization of head-to-tail copies within the GenBank sequence (accession number AC026683). b, Dendrogram of relationships between Pwcr1 and the _Pwcr1_-like genes, based on sequence similarity, which suggests expansion of the cluster by local duplication events.
Expression of Pwcr1
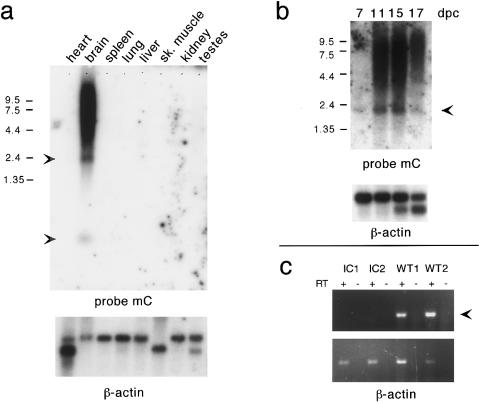
In contrast to the wide range of expression of its human orthologue, Pwcr1 expression in postnatal mice is restricted to the brain. For the location of probes used, see figure 6a. Northern blot analysis with probe mC, shown in figure 8_a_, detected a predominant 2.2-kb transcript, a smaller ∼140-nt transcript, and a ∼4–10-kb smear in brain. No expression was seen in heart, spleen, lung, liver, skeletal muscle, kidney, or testes. Pwcr1 transcripts were detected in whole embryos from day 7 onward (fig. 8_b_). As we had found for human PWCR1, the short ∼140-nt Pwcr1 transcript was much more abundant on northern blots with total RNA than with polyadenylated RNA (data not shown), suggesting that most ∼140-nt transcripts are not polyadenylated. Northern blot analysis with probe mD, which contains the entire HMCR, detected the ∼140-nt transcript, whereas probe mE, which does not contain the HMCR, did not (data not shown). Therefore, the ∼140-nt transcript of Pwcr1 is transcribed from the conserved HMCR, as is the case for PWCR1. Because the HMCR contains the C/D boxes characteristic of snoRNAs, we hypothesize that the abundant nonpolyadenylated 140-kb transcript may represent stable snoRNA molecules.
Figure 8.
Tissue-specific and imprinted expression of Pwcr1. Northern blot analyses of polyadenylated mRNA from adult mouse tissues (a) and total mouse embryos (b). Arrowheads indicate Pwcr1 transcripts. dpc = days postcoitum. c, Pwcr1 expressed exclusively from the paternal allele. RT-PCR was performed with _Pwcr1_-specific primers M49TD and M50TD, on RNA from mutant mice deleted for exons 1–6 of Snrpn and for the putative imprinting center (IC) region (lanes C1 and lanes IC2) and from their wild-type littermates (lanes WT1 and lanes WT2). A 208-bp product was amplified in WT1 and WT2 but not in either IC1 or IC2.
To assess whether Pwcr1 is uni- or bidirectionally transcribed, and whether the putative snoRNA is transcribed from the correct DNA strand, we hybridized single-stranded RNA probes to northern blots. Both the 2.2-kb and the ∼140-nt transcripts were detected with probe mD-R but not with probe mD-F. Low-level hybridization to a high-molecular-weight smear was seen with both single-stranded probes (data not shown). The plus- and minus-strand designation of Pwcr1 matches that of its human counterpart. Therefore, the major transcripts of the human and mouse genes are made from the same strand, and the ∼140-nt transcript is likely to represent a novel human snoRNA. Whether this short RNA is transcribed from its own promoter or is derived, by exonucleolytic processing, from the larger, 2.2-kb transcript is currently unknown.
Monoallelic Expression of Pwcr1
To determine whether Pwcr1 is regulated by the putative cis_-acting IC regulatory element, we studied its expression in a mouse model for imprinting mutations. Mice with a paternally derived chromosome 7 that carries a 42-kb deletion including exons 1–6 of Snrpn and the putative IC are hypotonic, fail to thrive, and die soon after birth (Yang et al. 1998). In addition to lacking expression of Snrpn/Snurf, genes that are disrupted by the deletion, the mice also do not express other genes in the PWS deletion region—_Zfp127, Ndn, Magel2 (Lee et al. 2000), and _Ipw_—that are normally expressed from the paternal allele. We performed RT-PCR with RNA derived from mice carrying the deletion mutation on the paternally derived allele (IC1 and IC2) and from wild-type control mice kindly provided by C. Brannan (Yang et al. 1998). The 208-bp Pwcr1 RT-PCR product seen in the control samples, WT1 and WT2, was absent in the IC1 and IC2 lanes (fig. 8_c_). Therefore, as we had found for human PWCR1, Pwcr1 is expressed exclusively from the paternally derived allele, and its expression is dependent on the IC regulatory element. To our knowledge, PWCR1/Pwcr1 is the first potential snoRNA-encoding gene that is imprinted.
Conserved Region of PWCR1 and of Pwcr1 (HMCR): Presence in Other Species
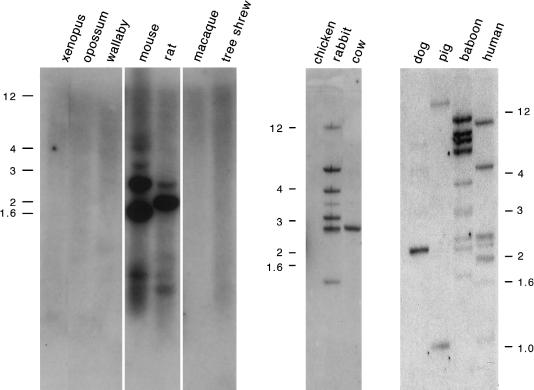
Because snoRNAs have important functions in both pre-rRNA processing and RNA-nucleotide modification, they exist in all organisms, in great abundance and variety. To determine when in evolution the potential C/D box snoRNA-encoding region of PWCR1/Pwcr1 arose, we performed a Zoo blot analysis, as shown in figure 9. A Southern blot with DNA samples from several species was hybridized with mouse probe mD, which contains the HMCR. Bands were observed for most placental mammals studied, with the tree shrew and the macaque being the only exceptions. No hybridizing bands were seen for marsupials, Xenopus, and chicken. The presence of single discrete bands in cow and dog suggests that HMCR-containing sequences exist at single- or low-copy numbers. The restriction-fragment patterns in mouse, rabbit, baboon, and human, however, reveal a complex organization. These signals may be generated by multiple copies in the vicinity of PWCR1, as we detected in the genomic sequence of mouse and human, although the presence of dispersed copies elsewhere in the genome cannot be excluded.
Figure 9.
Southern blot analyses of various animal species, which reveal conservation of _PWCR1_-related sequences in most mammals. Mouse probe mD that contains the HMCR was used (see fig. 6_a_). DNA samples from dog, pig, baboon, and human were digested with _Bam_HI; the other DNA samples were digested with _Eco_RI.
Discussion
Here we have reported the discovery and characterization of PWCR1 and its mouse orthologue, Pwcr1. Located in conserved imprinted regions (human 15q11.2 and mouse 7C–D1), both genes are expressed exclusively from the paternal chromosome. Transcription occurs unidirectionally from the same (plus) strand and is dependent on an intact IC regulatory element on the same expressed allele. Both genes are intronless and do not seem to encode protein products, yet they share a block of highly conserved sequences. The patterns of alternative transcripts of PWCR1 and Pwcr1 are strikingly similar. Both genes express predominant polyadenylated transcripts of either ∼1.2 kb, for PWCR1, or ∼2.2 kb, for Pwcr1, as well as short, ∼140-nt transcripts that are not polyadenylated yet are very abundant on northern blots of total RNA. Intriguingly, these short transcripts are derived from the conserved regions, because only probes containing the 99-bp 87%-identical HMCR sequences detect the ∼140-nt transcripts. The presence and spacing of box C and D sequences in the predicted transcripts of the HMCR suggest that these abundant short transcripts represent snoRNAs (Balakin et al. 1996; Lowe and Eddy 1999).
snoRNAs are stable products of RNA polymerase II. They are located in the nucleolus, in association with ribosomal RNA. Complexed with specific proteins, they form ribonucleoprotein particles (snoRNPs) (Maxwell and Fournier 1995). These particles participate both in the processing of large ribosomal RNA transcripts into the distinct rRNA species and in RNA modification. The C/D box snoRNAs, specifically, associate with the abundant nucleolar protein fibrillarin and are required for ribose 2′O-methylation of rRNA, which has earned them the designation “methylation guide RNAs” (Balakin et al. 1996). A computational screen of the yeast genome revealed 41 different guide snoRNAs responsible for ribose methylation of 51 of the 55 known sites on rRNA (Lowe and Eddy 1999). The consistent sequence features of methylation guide snoRNAs that are shared by the HMCR of PWCR1/Pwcr1 include a box C (AUGAUGA), a box D′ (CUGA) starting 27 or 28 nt downstream, and a box D (CUGA) located 40 or 41 nt farther 3′. These sequence elements are necessary and sufficient for nucleolar localization of the snoRNA. Short complementary sequences near the termini of the HMCR (UGGA at nt 1–4 and UCCA at nt 91–94, in human, and at nt 93–96 in mouse; fig 6_b_) would enable the formation of a terminal stem, which is usually 4–8 bp in length. The “guide sequence” that base-pairs with the rRNA target is located just upstream of box D′ and is usually 10–21 nt in length (Lowe and Eddy 1999). In this region, we find 14 identical nucleotides in the human and mouse HMCR sequences; however, these nucleotides do not match with any known rRNA target. Modification of RNA by methylation via C/D box snoRNAs is not limited to rRNA, however, because U6 snRNA is also methylated by this mechanism (Weinstein and Steitz 1999). Therefore, the RNA target for the PWCR1 small transcript may be another snRNA, yet to be discovered. It is interesting to note that the polypeptide encoded by SNRPN, the first imprinted gene identified within the PWS deletion region, forms part of an snRNP complex (Özçelik et al. 1992).
Another feature that sets PWCR1 and Pwcr1 apart from known mammalian snoRNA genes is their genomic organization. In vertebrates, most snoRNAs are encoded in introns of housekeeping genes, usually one snoRNA per intron. Interesting exceptions are UHG, the single-copy “host” gene for the human U22 snoRNA (Tycowski et al. 1996), and gas5, a “growth arrest gene” (Smith and Steitz 1998). Both genes encode 8–10 different snoRNAs in their introns, but the transcripts assembled from their exons have lost their protein-coding potential. Therefore, when the mouse and human sequences are compared, the introns are more highly conserved than are the exons. One possible reason for the preferred intronic location may be that the requirement for snoRNAs is high, at ∼104 copies per cell, and that the location of their genes in highly transcribed loci may be advantageous. PWCR1/Pwcr1 is the first snoRNA-encoding gene that has been shown tobe monoallelically expressed, and, at first glance, that should be an evolutionary disadvantage. On the paternal chromosome, however, the genes in the PWS region are actively transcribed in most tissues, with PWCR1 giving rise to the abundant ∼140-nt transcript that we observed on northern blot analyses in human and mouse.
Although most snoRNA genes exist as single copies within introns of other genes, we found the HMCR of PWCR1/Pwcr1 embedded in tandemly arrayed larger sequence blocks, quite reminiscent of U2 snRNA genes that are organized within tandem-repeat clusters and undergo concerted evolution (Liao et al. 1997). As for rRNA, U1 and U2 genes, PWCR1 and Pwcr1 elements have been locally duplicated to form clusters of multiple similar copies. In the 15q11.2 region, the repeated elements are all truncated copies of PWCR1, which, depending on their size, may or may not contain the HMCR. Although the _PWCR1_-like copies are more diverged and very different in size, the _Pwcr1_-like elements that we compared seem more closely related and are arranged in a tight head-to-tail fashion.
On Zoo blots with HMCR-containing probes, hybridization signals were observed in most placental mammals but not in wallaby and opossum. The co-occurrence—and subsequent local amplification—of these novel snoRNA-like genes with the appearance of placental mammals may be coincidental or may indicate a functional role of these genes, possibly even in the process of imprinting. As judged from the Southern-hybridization pattern, the copy number and complexity of HMCR-homologous genes are highly variable in mammals. These observations agree with the notion that the region is characterized by genomic instability and may undergo rapid evolutionary change. For example, the 15q11-q13 region contains other genes that have been locally duplicated. The SNRPN upstream IC region contains elements that have undergone multiple duplication events (Farber et al. 1999). Transcribed, low-copy repeats of the HERC2 gene are associated with the breakpoint hotspots of the common deletions found in individuals with either PWS or AS (Ji et al. 1999). In mouse, the Ipw gene is characterized by a multitude of highly similar exons (Wevrick and Francke 1997). Other imprinted regions also contain simple repeats, and it has been proposed that repeats may be involved in attracting and maintaining the methylation imprint of a given region (Neumann et al. 1995).
The chromatin configuration of the PWS region on the maternal and the paternal chromosome 15 is different, as assessed by the presence of parent-of-origin–specific hypersensitive sites and of regions of differential methylation (Schweizer et al. 1999). Therefore, the two parental alleles have different transcription patterns. The chromatin configuration of defined regions of the paternal chromosome allows for increased rates of transcription. The large heterologous PWCR1 transcripts, seen on northern blots of total RNA, may simply be transcriptional noise: these transcripts are monoallelically expressed and appear imprinted simply because they are located within an imprinted region—the “innocent bystander hypothesis” (Varmuza and Mann 1994). The distinct polyadenylated transcripts of 1.2 kb in human and 2.2 kb in mouse, however, may have an additional function, unrelated to the fact that they also harbor a snoRNA-like sequence. Outside the HMCR, however, there is low conservation between the human and mouse sequences, as was seen for another noncoding RNA in the PWS deletion region. The region of conservation between IPW and its mouse orthologue is restricted to exon c and part of the adjacent intron (Wevrick and Francke 1997).
There are many precedents for genes whose products function as noncoding RNAs. For instance, noncoding RNAs found in the 5′ region of SNRPN are proposed to play important roles in switching the paternal epigenotype in the maternal germline. Indeed, deletions and a point mutation of these upstream transcripts are associated with AS (Dittrich et al. 1996). Noncoding RNAs involved in the epigenetic regulation of gene expression are exemplified by Xist in mammals and by roX RNAs in Drosophila. In mammals, equal gene expression from the X chromosome in both sexes is achieved through the inactivation of one of the X chromosomes in females. This process is dependent on Xist, a gene whose product is a large processed RNA that is expressed exclusively from—and that is associated, in cis, with—the inactive X chromosome (reviewed in Panning and Jaenisch 1998). Dosage compensation in Drosophila is achieved by a different mechanism. Instead of inactivation of one of the X chromosomes in females, the rate of transcription from the single male X chromosome is greatly increased. Two noncoding RNAs, roX1 and roX2, are involved in this process (Kelley et al. 1999). In addition, gene regulation by antisense RNAs has been suggested for Igf2r (Wutz et al. 1997) and UBE3A (Rougeulle et al. 1998). Although we did not detect any antisense transcripts at the PWCR1 locus, it remains possible that the 1.2-kb PWCR1 RNA has other functions that affect chromatin structure. PWCR1 RNAs may recruit regulatory protein complexes and/or other RNAs involved in the maintenance of allele-specific expression in the region.
PWCR1 is highly expressed in all regions of the brain, and Pwcr1 is expressed exclusively in the mouse brain. Expression is completely abolished in PWS brain and in the imprinting-mutation mouse model. There is a distinct possibility that lack of the small RNA encoded by these genes causes abnormalities in the modification of an snRNA and, thus, contributes to the hypothalamic dysfunctions and developmental delay in patients with PWS. Further work on the sublocalization of the small PWCR1 RNAs and of the larger transcripts in cell nuclei, as well as identification of target RNAs and associated proteins in snoRNP complexes, will be needed to evaluate these possibilities.
Acknowledgments
We are indebted to Art Beaudet, for PWS-region cosmids and cell lines of the O family; to Camilynn Brannan, for RNA from IC deletion mice; to Mary Barter from The Jackson Laboratory, for the mouse mapping figure; to Todd M. Lowe, for discussion of snoRNAs; and the members of our laboratory, for advice and stimulation. The work was supported by the Howard Hughes Medical Institute (support to T.D.L.S, J.S., and U.F.) and by a fellowship from the Deutsche Forschungsgemeinschaft (to C.R.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for PWCR1 [accession number AF241255] and Pwcr1 [accession number AF241256])
- Human-Mouse Homology Map, http://www.ncbi.nih.gov/Homology
- Jackson Laboratory Mapping Panels, The, http://www.jax.org/resources/documents/cmdata
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for PWS [MIM 176270])
- RHdb (radiation-hybrid database at the European Bioinformatics Institute), http://www.ebi.ac.uk/RHdb
- UK Human Genome Mapping Project Resource Center, http://www.hgmp.mrc.ac.uk
- Whitehead Institute for Biomedical Research/MIT Center for Genome Research, http://www-genome.wi.mit.edu
References
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL (1997) Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet 17:75–78 [DOI] [PubMed] [Google Scholar]
- Balakin AG, Smith L, Fournier MJ (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 86:823–834 [DOI] [PubMed] [Google Scholar]
- Barlow DP (1995) Gametic imprinting in mammals. Science 270:1610–1613 [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Tilghman SM (1997) Genomic imprinting in mammals. Annu Rev Genet 31:493–525 [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482–485 [DOI] [PubMed] [Google Scholar]
- Boccaccio I, Glatt-Deeley H, Watrin F, Roeckel N, Lalande M, Muscatelli F (1999) The human MAGEL2 gene and its mouse homologue are paternally expressed and mapped to the Prader-Willi region. Hum Mol Genet 8:2497–2505 [DOI] [PubMed] [Google Scholar]
- Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls RD, Horsthemke B (1995) Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet 9:395–400 (erratum: Nat Genet 10:249 [1995]) [DOI] [PubMed] [Google Scholar]
- Dittrich B, Buiting K, Korn B, Rickard S, Buxton J, Saitoh S, Nicholls RD, Poustka A, Winterpacht A, Zabel B, Horsthemke B (1996) Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene. Nat Genet 14:163–170 [DOI] [PubMed] [Google Scholar]
- Dunbar DA, Chen AA, Wormsley S, Baserga SJ (2000) The genes for small nucleolar RNAs in Trypanosoma brucei are organized in clusters and are transcribed as a polycistronic RNA. Nucleic Acids Res 28:2855–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber C, Dittrich B, Buiting K, Horsthemke B (1999) The chromosome 15 imprinting centre (IC) region has undergone multiple duplication events and contains an upstream exon of SNRPN that is deleted in all Angelman syndrome patients with an IC microdeletion. Hum Mol Genet 8:337–343 [DOI] [PubMed] [Google Scholar]
- Farber C, Gross S, Neesen J, Buiting K, Horsthemke B (2000) Identification of a testis-specific gene (C15orf2) in the Prader-Willi syndrome region on chromosome 15. Genomics 65:174–183 [DOI] [PubMed] [Google Scholar]
- Gerard M, Hernandez L, Wevrick R, Stewart CL (1999) Disruption of the mouse necdin gene results in early post-natal lethality. Nat Genet 23:199–202 [DOI] [PubMed] [Google Scholar]
- Gray TA, Saitoh S, Nicholls RD (1999) An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci USA 96:5616–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 25:486–489 [DOI] [PubMed] [Google Scholar]
- Holm VA, Cassidy SB, Butler MG, Hanchett JM, Greenswag LR, Whitman BY, Greenberg F (1993) Prader-Willi syndrome: consensus diagnostic criteria. Pediatrics 91:398–402 [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R (1997) DNA methylation and imprinting: why bother? Trends Genet 13:323–329 [DOI] [PubMed] [Google Scholar]
- Jay P, Rougeulle C, Massacrier A, Moncla A, Mattei MG, Malzac P, Roeckel N, Taviaux S, Lefranc JL, Cau P, Berta P, Lalande M (1997) The human necdin gene, NDN, is maternally imprinted and located in the Prader-Willi syndrome chromosomal region. Nat Genet 17:357–361 [DOI] [PubMed] [Google Scholar]
- Ji Y, Walkowicz MJ, Buiting K, Johnson DK, Tarvin RE, Rinchik EM, Horsthemke B, Stubbs L, Nicholls RD (1999) The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum Mol Genet 8:533–542 [DOI] [PubMed] [Google Scholar]
- Johnson DK, Stubbs LJ, Culiat CT, Montgomery CS, Russell LB, Rinchik EM (1995) Molecular analysis of 36 mutations at the mouse pink-eyed dilution (p) locus. Genetics 141:1563–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong MT, Carey AH, Caldwell KA, Lau MH, Handel MA, Driscoll DJ, Stewart CL, Rinchik EM, Nicholls RD (1999_a_) Imprinting of a RING zinc-finger encoding gene in the mouse chromosome region homologous to the Prader-Willi syndrome genetic region. Hum Mol Genet 8:795–803 [DOI] [PubMed] [Google Scholar]
- Jong MT, Gray TA, Ji Y, Glenn CC, Saitoh S, Driscoll DJ, Nicholls RD (1999_b_) A novel imprinted gene, encoding a RING zinc-finger protein, and overlapping antisense transcript in the Prader-Willi syndrome critical region. Hum Mol Genet 8:783–793 [DOI] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI (1999) Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98:513–522 [DOI] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J (1997) UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet 15:70–73 (erratum: Nat Genet 15:411 [1997]) [DOI] [PubMed] [Google Scholar]
- Knoll JH, Glatt KA, Nicholls RD, Malcolm S, Lalande M (1991) Chromosome 15 uniparental disomy is not frequent in Angelman syndrome. Am J Hum Genet 48:16–21 [PMC free article] [PubMed] [Google Scholar]
- Konat G, Laszkiewicz I, Grubinska B, Wiggins R (1994) Generation of labelled DNA probes by PCR. In: Griffin H, Griffin A (eds) PCR technology: current innovations. CRC Press, Ann Arbor, MI, pp 37–42 [Google Scholar]
- Lee S, Kozlov S, Hernandez L, Chamberlain SJ, Brannan CI, Stewart CL, Wevrick R (2000) Expression and imprinting of MAGEL2 suggest a role in Prader-Willi syndrome and the homologous murine imprinting phenotype. Hum Mol Genet 9:1813–1819 [DOI] [PubMed] [Google Scholar]
- Leff SE, Brannan CI, Reed ML, Ozcelik T, Francke U, Copeland NG, Jenkins NA (1992) Maternal imprinting of the mouse Snrpn gene and conserved linkage homology with the human Prader-Willi syndrome region. Nat Genet 2:259–264 [DOI] [PubMed] [Google Scholar]
- Liao D, Pavelitz T, Kidd JR, Kidd KK, Weiner AM (1997) Concerted evolution of the tandemly repeated genes encoding human U2 snRNA (the RNU2 locus) involves rapid intrachromosomal homogenization and rare interchromosomal gene conversion. EMBO J 16:588–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR (1999) A computational screen for methylation guide snoRNAs in yeast. Science 283:1168–1171 [DOI] [PubMed] [Google Scholar]
- MacDonald HR, Wevrick R (1997) The necdin gene is deleted in Prader-Willi syndrome and is imprinted in human and mouse. Hum Mol Genet 6:1873–1878 [DOI] [PubMed] [Google Scholar]
- Mascari MJ, Gottlieb W, Rogan PK, Butler MG, Waller DA, Armour JA, Jeffreys AJ, Ladda RL, Nicholls RD (1992) The frequency of uniparental disomy in Prader-Willi syndrome. Implications for molecular diagnosis. N Engl J Med 326:1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL (1997) De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet 15:74–77 [DOI] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ (1995) The small nucleolar RNAs. Annu Rev Biochem 64:897–934 [DOI] [PubMed] [Google Scholar]
- Neumann B, Kubicka P, Barlow DP (1995) Characteristics of imprinted genes Nat Genet 9:12–13 (erratum: Nat Genet 9:451 [1995]) [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Saitoh S, Horsthemke B (1998) Imprinting in Prader-Willi and Angelman syndromes. Trends Genet 14:194–200 [DOI] [PubMed] [Google Scholar]
- Özçelik T, Leff S, Robinson W, Donlon T, Lalande M, Sanjines E, Schinzel A, Francke U (1992) Small nuclear ribonucleoprotein polypeptide N (SNRPN), an expressed gene in the Prader-Willi syndrome critical region. Nat Genet 2:265–269 [DOI] [PubMed] [Google Scholar]
- Panning B, Jaenisch R (1998) RNA and the epigenetic regulation of X chromosome inactivation. Cell 93:305–308 [DOI] [PubMed] [Google Scholar]
- Reed ML, Leff SE (1994) Maternal imprinting of human SNRPN, a gene deleted in Prader-Willi syndrome. Nat Genet 6:163–167 [DOI] [PubMed] [Google Scholar]
- Rougeulle C, Cardoso C, Fontes M, Colleaux L, Lalande M (1998) An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet 19:15–16 [DOI] [PubMed] [Google Scholar]
- Schweizer J, Zynger D, Francke U (1999) In vivo nuclease hypersensitivity studies reveal multiple sites of parental origin-dependent differential chromatin conformation in the 150 kb SNRPN transcription unit. Hum Mol Genet 8:555–566 [DOI] [PubMed] [Google Scholar]
- Smith CM, Steitz JA (1998) Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol 18:6897–6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JS, Jiang YH, Galijaard RJ, Matsuura T, Fang P, Kubota T, Christian SL, Bressler J, Cattanach B, Ledbetter DH, Beaudet AL (1997) The E6-Ap ubiquitin-protein ligase (UBE3A) gene is localized within a narrowed Angelman syndrome critical region. Genome Res 7:368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JS, Nakao M, Christian S, Orstavik KH, Tommerup N, Ledbetter DH, Beaudet AL (1994) Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet 8:52–58 [DOI] [PubMed] [Google Scholar]
- Tsai TF, Armstrong D, Beaudet AL (1999_a_) Necdin-deficient mice do not show lethality or the obesity and infertility of Prader-Willi syndrome. Nat Genet 22:15–16 [DOI] [PubMed] [Google Scholar]
- Tsai TF, Jiang YH, Bressler J, Armstrong D, Beaudet AL (1999_b_) Paternal deletion from Snrpn to Ube3a in the mouse causes hypotonia, growth retardation and partial lethality and provides evidence for a gene contributing to Prader-Willi syndrome. Hum Mol Genet 8:1357–1364 [DOI] [PubMed] [Google Scholar]
- Tucker KL, Beard C, Dausmann J, Jackson-Grusby L, Laird PW, Lei H, Li E, Jaenisch R (1996) Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev 10:1008–1020 [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Steitz JA (1996) A mammalian gene with introns instead of exons generating stable RNA products. Nature 379:464–466 [DOI] [PubMed] [Google Scholar]
- Varmuza S, Mann M (1994) Genomic imprinting—defusing the ovarian time bomb. Trends Genet 10:118–123 [DOI] [PubMed] [Google Scholar]
- Watrin F, Roeckel N, Lacroix L, Mignon C, Mattei MG, Disteche C, Muscatelli F (1997) The mouse Necdin gene is expressed from the paternal allele only and lies in the 7C region of the mouse chromosome 7, a region of conserved synteny to the human Prader-Willi syndrome region. Eur J Hum Genet 5:324–332 [PubMed] [Google Scholar]
- Weinstein LB, Steitz JA (1999) Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol 11:378–384 [DOI] [PubMed] [Google Scholar]
- Wevrick R, Francke U (1997) An imprinted mouse transcript homologous to the human imprinted in Prader-Willi syndrome (IPW) gene. Hum Mol Genet 6:325–332 [DOI] [PubMed] [Google Scholar]
- Wevrick R, Kerns JA, Francke U (1994) Identification of a novel paternally expressed gene in the Prader-Willi syndrome region. Hum Mol Genet 3:1877–1882 [DOI] [PubMed] [Google Scholar]
- Williams CA, Angelman H, Clayton-Smith J, Driscoll DJ, Hendrickson JE, Knoll JH, Magenis RE, Schinzel A, Wagstaff J, Whidden EM (1995) Angelman syndrome: consensus for diagnostic criteria. Angelman Syndrome Foundation. Am J Med Genet 56:237–238 [DOI] [PubMed] [Google Scholar]
- Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP (1997) Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389:745–749 [DOI] [PubMed] [Google Scholar]
- Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, Francke U, Jenkins NA, Copeland NG, Brannan CI (1998) A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet 19:25–31 [DOI] [PubMed] [Google Scholar]
- Yee F, Yolken RH (1997) Identification of differentially expressed RNA transcripts in neuropsychiatric disorders. Biol Psychiatry 41:759–761 [DOI] [PubMed] [Google Scholar]