Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations (original) (raw)
Abstract
fw2.2 is a major quantitative trait locus that accounts for as much as 30% of the difference in fruit size between wild and cultivated tomatoes. Evidence thus far indicates that fw2.2 alleles modulate fruit size through changes in gene regulation rather than in the FW2.2 protein itself. To investigate the nature of these regulatory changes and the manner in which they may affect fruit size, a pair of nearly isogenic lines has been subjected to detailed developmental, transcriptional, mitotic, and in situ hybridization studies. The results indicate that the large- and small-fruited alleles of fw2.2 differ in peak transcript levels by ≈1 week. Moreover, this difference in timing of expression is associated with concomitant changes in mitotic activity in the early stage of fruit development. The changes in timing of gene expression (heterochronic allelic variation), combined with overall differences in total transcript levels, are sufficient to account for a large portion phenotypic differences in fruit weight associated with the two alleles.
Cell division and expansion are generally considered two distinct phases during tomato fruit development (1). The rate of both cell growth and cell proliferation, which might be coordinated and instructed by cellular mechanisms, is critically important in determining final fruit size (2). Despite the fact that cell expansion may account for the greatest increase in volume, cell division is also an essential factor of fruit organogenesis because it determines the final cell number within the fruit. Therefore, fruit size is, in part, the result of a defined number of cell divisions that occur during development (3). High levels of cell division often take place in the first few weeks after anthesis, but mitotic cells can still be found in later phases of development, even in ripening fruit (4).
Recently, increasing knowledge in molecular genetics has allowed the characterization of a number of molecular events that influence cell division or cell expansion. Many genes have been shown to control final organ size in Drosophila via cell division and cell expansion, such as dMYC (5), CyclinD/Cdk4 (6), RAS (7), TSC1 and TSC2 (8), etc. In plants, recent research has yielded much evidence of the molecular and genetic control of cell division and expansion as well as organ size; for example, AINTEGUMENTA (ANT) (9, 10), ANGUSTIFOLIA and ROTUNDIFOLIA3 (11), ABP1 (12), CLV and WUS (13), NtKIS1a (14), and CYC1At (15). Despite this recent progress, the precise molecular mechanisms governing organ size by cell division or expansion in plants remain far less clear than in Drosophila. Moreover, most genes characterized to control organ size in plants are associated with the development of leaves, roots, and shoot apical meristems. Less attention has been paid to developmental and molecular mechanisms that regulate fruit weight and size.
Recently, molecular marker studies have found 28 quantitative trait loci (QTLs) affecting tomato fruit weight and size in crosses between the domesticated tomato (Lycopersicon esculentum Mill.) and related wild tomato species (16, 17). One of the QTLs with largest effect, fw2.2, has been cloned and characterized. fw2.2 accounts for an ≈30% difference in fruit weight between the domesticated tomato and its wild relatives and has been postulated to function as a regulator of cell division (18, 19). Moreover, comparative sequence analysis of fw2.2 from the large- and small-fruited alleles indicates that fw2.2's effect on fruit weight and size is probably due to changes in gene regulation, rather than differences in the sequence and structure of the protein (18, 20). Yet the molecular and developmental mechanisms underlying modulation of fruit size by fw2.2 remain unknown.
To shed light on the manner in which different fw2.2 alleles modulate fruit size, a pair of nearly isogenic lines, differing for alleles at the fw2.2 locus, were subjected to developmental studies of fruit growth, cell division (mitotic index), cell expansion (cell size), and fw2.2 transcript levels.
Materials and Methods
Plant Material.
TA1143 and TA1144 are nearly isogenic lines (NILs) that contain large- and small-fruited alleles at fw2.2, respectively (16, 21). Ten pairs of TA1143 and TA1144 plants were grown in continuous cycles in the greenhouse in Ithaca, NY, as a source of flower/fruit tissue for developmental studies. As a supplement, 30 pairs of the same NILs were grown in a field in the summer seasons of 2000 and 2001. For developmental studies, flowers were tagged at anthesis and harvested at specific time points during fruit development.
Phenotypic Analyses.
Carpels/fruit were harvested throughout the entire period of fruit development starting at anthesis and continuing to 42 days postanthesis (DPA), at which point fruit ripening commences. Fruits were collected and weighed every other day during the first 18 days, followed by sampling at 6-day intervals until 42 DPA. More than 10 fruits from individual plants were weighed for each developmental time stage. After weighing, fruits were transversally cut in half and imaged with vistascan software (UMAX Technologies, Dallas). Images were used to measure length, width (diameter), pericarp thickness (fruit wall), and placental areas of each fruit.
Cell Number and Cell Size Measurements.
Ten flowers or fruits derived from 10 individual TA1143 and TA1144 plants were harvested at the following stages: 0, 4, 8, 12, 18, 24, and 36 DPA. Subsequently, they were fixed with 4% (wt/vol) polyformaldehyde in PBS buffer (pH 7.0) overnight at 4°C, dehydrated in increasing ethanol concentrations and Histoclear (National Diagnostics), and embedded in paraplast (Sigma). Seven-micrometer sections were cut and flattened on slides. Before staining in 0.1% Fluorescent Brightener 28 (Sigma) for 5 min, the sections were dewaxed and rehydrolated, then observed under a fluorescence microscope (Olympus, Tokyo). Digital images were taken from 10 samples at each stage by using a Polaroid DMC camera (Polaroid). The cell number was counted from images of the mesocarp at each stage (excluding the epidermis and hypodermis, vascular bundles, and internal epidermis) by using scion image software (Scion, Frederick, MD). Cell size was calculated from the cell number counted per unit area (1 mm2) measurements. The same procedure was used to determine the placental cell number (excluding developing seeds).
Mitotic Index Determinations.
The flowers or fruit of TA1143 and TA1144 at each stage of 0, 4, 8, 12, 18 DPA were fixed as described above. After a brief wash in distilled water, a piece of tissue was dissected from carpels/fruits at each developmental stage and immersed in a drop of HCl (1 M) on a glass slide, and then heated on an alcohol flame for several seconds. After being washed with distilled water and blotted with Kimwipes (Kimberly-Clarke, Roswell, GA), the samples were incubated in a drop of 4, 6-diamino-2-phenylindole (5 μg⋅ml−1, Sigma) for 10–15 min, then washed again in distilled water and squashed. Stained tissue was visualized under a fluorescence microscope with 20× and 40× objectives. The number of interphase and mitotic (from prometaphase to telophase) cells was determined in 10 separate carpels/fruits collected from 10 individual plants at each stage. The mitotic index was determined by calculating the ratio of mitotic cells to the total number of cells counted.
Preparation of RNA and Reverse Transcription Reaction.
Fresh tissues were collected at various stages from TA1143 and TA1144, frozen in liquid nitrogen, and ground to a fine powder. Total RNAs were isolated with Trizol reagent (Invitrogen). One microgram of total RNAs from each sample was treated with RNase free DNaseI (amplification grade, Invitrogen). First-strand cDNA was synthesized by reverse transcription with random hexamer. All reagents were from the Taqman Reverse Transcription Reagent kit (Applied Biosystems). The cycling parameters 10 min at 25°C, 30 min at 48°C, 5 min at 95°C were followed.
Quantitative Transcript Analysis.
fw2.2 mRNA was detected and quantified by using ABI Prism 7700 Sequence Detection System (Applied Biosystems). Three microliters of first-strand cDNA (1:1 diluted with distilled water) solutions was used as templates for the subsequent PCR amplifications. Concentrations of MgCl2, the primers, and the probe were optimized to yield the maximum fluorescent intensity (Δ_R_n). The sequences of forward primer, reverse primer, and probe specific to the fw2.2 gene are CAACCTTATGTTCCTCCTCACTATGTAT, GGGTCATCAAAACAATGACAAAGA, 6FAM-TGCCCCCGGCACCACCA-TRMRA, respectively. PCR amplification was carried out in a 28-μl reaction volume containing 1× Taqman Buffer A, 5.5 mM MgCL2, 900 nM each primer, 200 nM probe, 200 μM each deoxynucleoside triphosphate (dATP, dCTP, dGTP), 400 μM dUTP, 0.7 unit of AmpliTaq Gold (0.025 unit/μl), and 0.28 unit of AmpErase uracil-_N_-glycosylase (0.01 unit/μl).
An endogenous control for PCR quantification studies was provided by measuring 18S RNA levels by using the Taqman Ribosomal RNA Control Reagent kit (Applied Biosystems). The sequences of the primers and the probe contained in this kit were confirmed to perfectly match the tomato 18S RNA gene (data not shown). To monitor for potential carryover contamination through RT-PCR of the test samples, tubes with water and products of the reverse transcription reactions without Reverse Transcriptase were applied as negative controls. PCR reactions were performed in conditions recommended by the manufacturer (2 min at 50°C, 10 min at 95°C, and 40 cycles with 15 s at 95°C and 1 min at 60°C).
The CT (threshold cycle) value of a real-time PCR reaction is positively correlated with the amount of target RNAs (22). The relative amount of target (fw2.2), normalized to an endogenous reference (the 18S RNA levels) and relative to a calibrator (any one of the samples), is given by: 2−ΔΔCT (23), where ΔΔCT = [CT(fw2.2) − CT(18S)] − [CT(calibrator) − CT(18S)]. Each set of experiments was repeated three times, from extracting RNA to performing RT-PCR. The final relative quantification was the average value of the three repeats at each stage.
In Situ Hybridization.
To generate probes for hybridization, a 256-bp DNA fragment from the 3′ untranslated region of the fw2.2 cDNA was cloned into the pBluescript KS(+) vector (Stratagene). The resultant linearized plasmids by _Bam_HI and _Hin_dIII served as templates for synthesizing the antisense and sense strand RNA probes by T3 and T7 RNA polymerase, respectively, with the Digoxigenin (DIG) RNA Labeling Kit (Roche). Probe concentrations were estimated by dot blot comparisons with serial dilutions of standards in the DIG-RNA Kit (Roche).
Tomato fruit from TA1143 and TA1144 was harvested at 6 and 12 DPA, and the pericarp and the placenta tissues were fixed, dehydrated, and embedded in paraplast (Sigma) according to Jackson (24). Detailed procedures of hybridization and signal detection followed the protocol described by Coen et al. (25). After hybridization and washing, DIG-labeled RNA was detected by using anti-DIG antibody conjugated to alkaline phosphatase (Roche). Alkaline phosphatase activity was measured by using nitro blue tetrazolium (Roche) and bromo-chloro-indolyl phosphate (Roche). Chromogen production was then recorded with a digital camera (Polaroid DMC) mounted on a fluorescence and light microscope (Olympus).
Results
Fruit Development in TA1143 and TA1144.
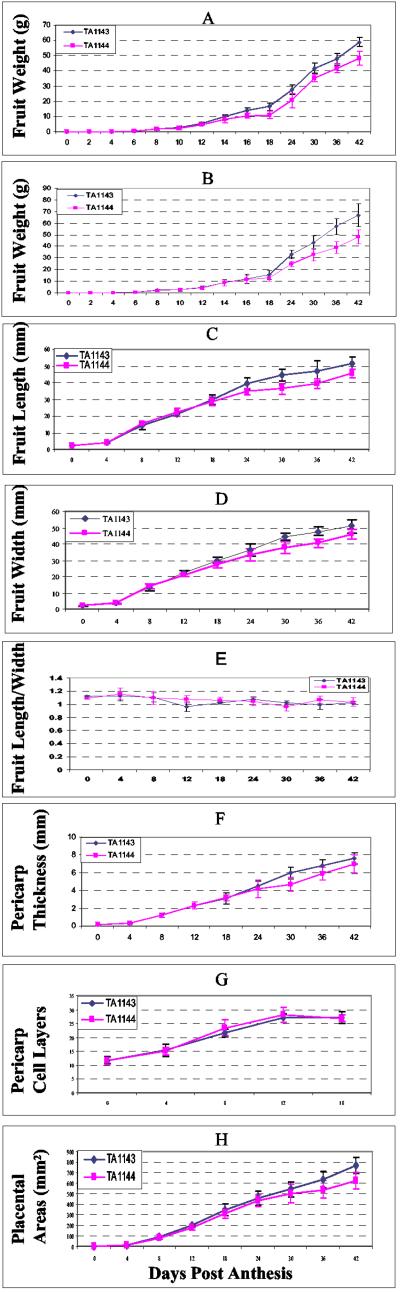
TA1143 and TA1144 are nearly isogenic lines containing the large- and small-fruited alleles for fw2.2, a major QTL controlling fruit size in cultivated and wild tomatoes (16, 18). Fig. 1 A and B depict the change in fruit weight from anthesis through maturity for TA1143 and TA1144, grown both in the greenhouse and the field. The plots were similar for both NILs in both locations, with TA1143 manifesting larger fruit mass than TA1144, even during the early stages of fruit development. The larger size of TA1143 fruit was reflected in both increased fruit length and width (Fig. 1 C and D), with no difference detected in fruit shape (length/width ratio) (Fig. 1E). These results are consistent with the findings of Nesbitt and Tanksley (19).
Figure 1.
Profiles for developing fruit of TA1143 and TA1144 NILs. The following traits were monitored: (A) fruit weight, greenhouse; (B) fruit weight, field; (C) fruit length, greenhouse; (D) fruit width (diameter), greenhouse; (E) fruit shape index (fruit length/width), greenhouse; (F) fruit wall (pericarp) thickness, greenhouse; (G) number of cell layers in pericarp, greenhouse; (H) area of placental tissue, greenhouse.
Pericarp wall thickness, as well as the number of cell layers and the size of the placental cortical areas of developing fruit of TA1143 and TA1144, was also recorded (Fig. 1 F–H). Pericarp thickness (Fig. 1F) and cell layer number (Fig. 1G) were indistinguishable in the two NILs early in fruit development. Although a difference in pericarp thickness became visible during the later stages of fruit development, at maturity the difference diminished (Fig. 1F). The placental cortical areas, however, were significantly larger in TA1143 than TA1144, with the increase being visible in early stages and persistent throughout fruit development (Fig. 1H).
Previous studies have demonstrated that the number of fertilized ovules and developing seeds in a fruit correlate with both the initial cell division rate and the final fruit size (1). For this reason, the number of seeds from 10 fruits of each of the two NILs was analyzed. No significant difference (P = 0.37) in seed number was found, suggesting that the fruit-weight difference in the two lines is regulated directly by fw2.2 and is not due to hormonal effects associated with fertility and seed formation.
Cell Size in Developing Fruit of TA1143 and TA1144.
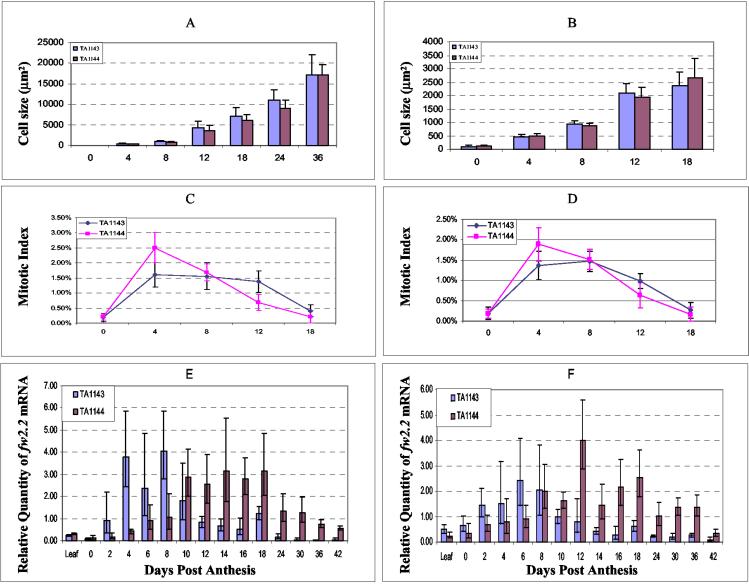
The large fruit of TA1143 could be due to the presence of more cells, larger cells, or a combination of both. Microscopic examination revealed that cell size in both the pericarp and placenta was not significantly different at any stage of development (Fig. 2 A and B). This result implies that _fw2.2_-induced fruit size variation is caused by a change in the rate of cell division rather than cell enlargement, consistent with the report of Frary et al. (18).
Figure 2.
Profiles for developing fruit of TA1143 and TA1144 NILs. The following traits were measured: (A) cell size in pericarp, greenhouse; (B) cell size in placental tissue, greenhouse; (C) mitotic index in pericarp, greenhouse; (D) mitotic index in placental tissue, greenhouse; (E) fw2.2 transcript levels, greenhouse; (F) fw2.2 transcript levels, field.
Mitotic Index in Developing Fruit of TA1143 and TA1144.
Mitotic indices for pericarp and placental tissues were determined at various developmental stages for both TA1143 (large-fruit NIL) and TA1144 (small-fruit NIL). The developmental profiles in both tissues were significantly different between the two NILs (Fig. 2 C and D; Table 1). Mitotic activity was very low at anthesis and indistinguishable between the NILs. However, by 4 DPA, the mitotic index was significantly higher in both the pericarp and placental tissues of TA1144 compared with TA1143 fruit. However, this mitotic activity rapidly declined in TA1144, and by 12 DPA, the mitotic index was significantly lower in both tissues compared with TA1143.
Table 1.
Mitotic indices for pericarp and placental tissues in developing fruit
| TA1143 | TA1144 | t test | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pericarp | Placenta | Pericarp | Placenta | Pericarp¶ | Placenta‖ | |||||||||||
| DPA | n* | Cell no.† | Mitosis no.‡ | Mitotic index | Cell no. | Mitosis no. | Mitotic index§ | n | Cell no. | Mitosis no. | Mitotic index | Cell no. | Mitosis no. | Mitotic index | P value | P value |
| 0 | 9 | 15675 | 27 | 0.18% (0.14%) | 19774 | 32 | 0.19% (0.16%) | 10 | 16538 | 31 | 0.2% (0.11%) | 19500 | 34 | 0.18% (0.11%) | 0.728 | 0.879 |
| 4 | 12 | 12894 | 207 | 1.6% (0.40%) | 12103 | 167 | 1.37% (0.34%) | 11 | 9644 | 229 | 2.5% (0.5%) | 9340 | 184 | 1.89% (0.41%) | 0.000 | 0.003 |
| 8 | 10 | 15502 | 243 | 1.56% (0.44%) | 15584 | 227 | 1.47% (0.24%) | 8 | 12477 | 208 | 1.68% (0.28%) | 10425 | 158 | 1.52% (0.25%) | 0.520 | 0.666 |
| 12 | 10 | 5000 | 69 | 1.38% (0.36%) | 5000 | 49 | 0.98% (0.18%) | 10 | 5624 | 38 | 0.68% (0.27%) | 4000 | 25 | 0.63% (0.31%) | 0.000 | 0.007 |
| 18 | 10 | 8565 | 36 | 0.4% (0.2%) | 12627 | 41 | 0.27% (0.19%) | 10 | 10186 | 25 | 0.21% (0.21%) | 11100 | 26 | 0.17% (0.17%) | 0.160 | 0.689 |
In summary, the small-fruit allele of fw2.2, found in TA1144, is associated with a rapid but brief rise in mitotic index immediately after fertilization. In contrast, the large-fruit allele, found in TA1143, is associated with a more gradual rise in mitotic activity, but this activity is sustained over a significantly longer period of fruit development (up to 12 DPA). The extended period of cell division found in TA1143 is most likely causally related to the larger fruit size ultimately attained by this NIL.
Developmental Expression of fw2.2: Quantitative Transcript Analysis.
Quantitative PCR revealed that developmental transcript levels of fw2.2 are significantly different for TA1143 vs. TA1144 (Fig. 2 E and F). In both TA1143 and TA1144, transcript levels were very low at anthesis, but in TA1143, transcript levels increased dramatically immediately after fertilization and peaked at around 6–8 DPA. This was followed by a very rapid decrease in transcript accumulation, such that by 12 DPA the levels were comparable to those seen at anthesis (Fig. 2 E and F). In contrast, postfertilization transcript levels rose more slowly in TA1144, peaked at 12–14 DPA and were sustained over a much longer period of fruit development. By 12 DPA, the levels of fw2.2 transcript in TA1144 were more than double those seen in TA1143. Not only was the timing of transcription delayed and extended in TA1144, but the total transcript level, over the entire period of fruit development, was nearly twice that seen in TA1143 (Fig. 2 E and F).
Thus it appears that the differences in transcript levels of fw2.2 between the small- and large-fruit alleles are both qualitative (a shift in timing of expression) and quantitative (a change in the overall amount of transcripts). The large-fruit allele (found in TA1143) is transcribed more rapidly, and transcript levels of fw2.2 peak earlier (6–8 DPA) after fertilization compared with the small-fruit allele (found in TA1144), for which transcript accumulation increases more slowly and does not peak until 12–14 DPA. The quantitative difference is manifested by the fact that, compared with the large-fruit allele, the small-fruit allele directs nearly twice as much total fw2.2 transcript over the entire course of fruit development (Fig. 2 E and F). The differences in fw2.2 expression seen for TA1143 and TA1144 were observed in both greenhouse- (Fig. 2E) and field-grown (Fig. 2F) plants, indicating that these differences are developmentally controlled and not greatly influenced by environmental changes. Finally, it is also worth noting that the timing of expression of the fw2.2 alleles (both small and large fruit) are inversely correlated with the mitotic indices of both pericarp and placental tissues reinforcing the idea that fw2.2 functions as a negative regulator of cell division (18) (Fig. 2 C and D).
Cellular Distribution of fw2.2 mRNA.
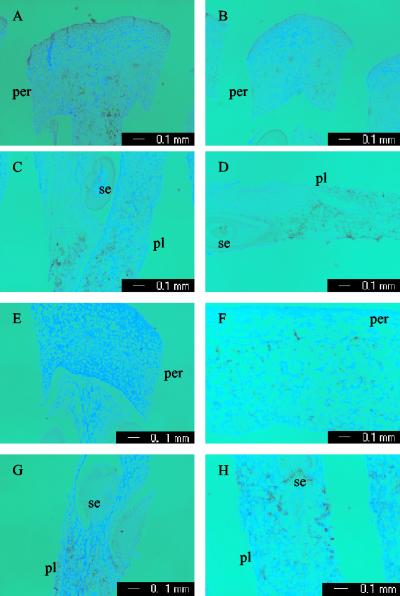
Despite its major effect on fruit size, fw2.2 is expressed at relatively low levels compared with other genes. Millions of cDNA clones have been screened, and yet not a single fw2.2 cDNA clone has been identified (18). The low expression of fw2.2 makes in situ detection difficult. Nonetheless, it was possible to obtain weak but reproducible signals for fw2.2 expression by using in situ techniques. The expression of fw2.2 mRNA within individual cells was determined by in situ hybridization by using a DIG-labeled antisense probe containing the 3′ untranslated region and a partial sequence in the coding region. The fruit tissues used for this purpose were collected from 6- and 12-DPA fruit, because fw2.2 mRNA peaked around these two stages in TA1143 and TA1144, respectively (Fig. 2 E and F). At 6 DPA, the fw2.2 transcripts were restricted to the inner pericarp and the septum tissues in the large fruit (Fig. 3A), whereas the expression of fw2.2 was detectable around vascular bundle regions in the small fruit (Fig. 3B). The strongest signals of fw2.2 were found in the regions proximal to seeds in the placenta of both NILs (Fig. 3 C and D). At 12 DPA, in the placental tissues, the spatial expression patterns of fw2.2 were similar in both NILs (Fig. 3 G and H). However, weak signals were observed in the inner pericarp of TA1144 (Fig. 3F) but not in the pericarp of TA1143 (Fig. 3E). This latter result may reflect some spatial difference in expression of the two alleles.
Figure 3.
In situ hybridization of fw2.2 in developing fruit. (A and C) Cross section of pericarp and placenta from 6-DPA fruit of TA1143. (B and D) Cross section of pericarp and placenta from 6-DPA fruit of TA1144. (E and G) Cross section of pericarp and placenta from 12-DPA fruit of TA1143. (F and H) Cross section of pericarp and placenta from 12-DPA fruit of TA1144. per, pericarp. pl, placenta. se, seed. Hybridization with sense probe was not shown.
Discussion
Natural Variation in Tomato Fruit Size Is Associated with Heterochronic Regulatory Mutation in fw2.2 Alleles.
It has long been hypothesized that heterochronic mutations that modulate the developmental timing of gene expression are a major cause of evolutionary change (26, 27). Presumably, even subtle changes in the timing of gene expression could result in major changes in phenotype (28). Although the intellectual arguments for this hypothesis are compelling, currently there are very few examples where the genes and mutations underlying natural variation are known—a prerequisite for evaluating the merits of the hypothesis.
In plants, a number of heterochronic mutations with significant effects on plant development have been identified (29–33); however, the relationship of these induced mutations to natural genetic variation and evolution is largely unknown. In the past few years, several genes underlying natural variation in QTL have been isolated in plants, allowing opportunities to determine the relationship between natural variation and heterochronic developmental mutations (18, 34–39). In two of these cases, the variation underlying the QTLs has been shown to be due to mutations in the coding region of the underlying gene (35, 36). However, for the other examples, the mutational causes of phenotypic variation are either uncertain (34, 39) or associated with changes in the regulatory region of the underlying genes (18, 20, 37). In these latter cases, the exact nature of the regulatory changes is unknown.
The fw2.2 QTL from tomato is an example where natural quantitative variation is clearly due to regulatory mutations (20). We show here that the 5′ regulatory differences between two naturally occurring fw2.2 alleles result in a change in both the timing of fw2.2 transcription (heterochronic changes) and the overall quantity of fw2.2 transcripts (Fig. 2 E and F). The two alleles differ in peak transcript levels by ≈1 wk (Fig. 2 E and F). This difference in timing of expression is associated with concomitant changes in mitotic activity in the early stages of fruit development. The change in timing of expression of the two alleles, combined with overall differences in total transcript levels, is sufficient to cause a major change in final fruit mass (Fig. 1 A and B). This study thus provides some of the first molecular evidence that heterochronic regulatory changes in gene expression constitute a natural mechanism of evolutionary change in plants. On the basis of the in situ hybridization experiments, these heterochronic regulatory changes may also be accompanied by subtle changes in spatial expression in the developing pericarp (Fig. 3 E and F).
fw2.2 Exercises Direct Control Over Cell Division and Not Indirectly Through Modulation of Fertility.
Cell division inside fruit can be stimulated by increases in the number of developing seeds in the fruit (1). Hence, the possibility exists that fw2.2 modulates cell division indirectly through changes in seed set/fertility. However, the fact that the seed yield per fruit was not significantly different between the fw2.2 NILs indicates that this is not the case, and that fw2.2 is not affecting fertility/seed set. This result, combined with experiments showing that fw2.2 does not modulate fruit size through changes in sink source relationships, gives strong support to the hypothesis that fw2.2 is a direct regulator of cell division in developing fruit (19).
fw2.2 Acts Directly as a Negative Regulator of Cell Division Early in Fruit Development.
The remarkable increase in mass during fruit development is accompanied by a coordinated increase in cell division (cell number) and cell growth (cell expansion) (1). Previous experiments have demonstrated there are four mechanisms by which cell division and cell growth may be connected: (i) control of cell growth by cell division (40); (ii) control of cell division by cell growth (41); (iii) coordinate control of cell growth and cell division (42); and (iv) independent regulation of cell growth and cell division (43). Results from the developmental studies of fw2.2 NILs are consistent with the fourth mechanism. Developmental studies show that fw2.2 controls cell division but not cell growth during fruit development (Fig. 2 A–D). Although both cell numbers and mitotic division rates differed significantly between the NILs during fruit development, the patterns of cell enlargement were indistinguishable between the NILs. Thus, fw2.2 modulates the pattern of cell division without affecting the pattern of cell expansion, suggesting that these processes are under different molecular control during fruit development.
For several plant genes, such as NtKIS1a (14), ICK1 (44), TSO1 (45), and KRP2/KRP3 (46), higher levels of transcript have been correlated with reduced cell division and smaller organ size. However, the manner in which these genes influence cell division (whether directly or indirectly) has not been well elucidated. In mammals and Xenopus, where more detailed studies have been conducted, specific genes/proteins have been shown to act directly as negative regulators of cell divisions (47, 48).
Previous work has shown that higher levels of fw2.2 transcription are correlated with reduced cell division (18) and, as discussed earlier, fw2.2 appears to exert its effects directly on the tissues of developing fruit rather than indirectly through changes in fertility or sink source relationships. The current work shows that fw2.2 transcript levels are inversely correlated (quantitatively and temporally) with the mitotic activity of pericarp and placental cells in the early stages of fruit development. Thus, the combined evidence supports the hypothesis that fw2.2 exerts a direct negative control over cell division in developing fruit. However, the mechanism by which fw2.2 controls cell division remains to be determined.
Acknowledgments
We thank Dr. Reinhard K. Strauginger for excellent technical assistance for performing real-time RT-PCR and Drs. Anne Frary, Amy Frary, Trudy Mackay, Jian Hua, and John Doebley for critical comments on the manuscript. This project was supported by grants from the National Science Foundation (DBI-0116076), the U.S. Department of Agriculture Plant Genome Program (97-35300-4384), and the U.S.–Israel Binational Agriculture Research and Development Fund (IS-3009-98C).
Abbreviations
DPA
days postanthesis
QTL
quantitative trait loci
NIL
nearly isogenic line
DIG
digoxigenin
References
- 1.Gillaspy G, Ben-David H, Gruissem W. Plant Cell. 1993;5:1439–1451. doi: 10.1105/tpc.5.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho L-C, Hewitt J D. In: The Tomato Crop. Atherton J-G, Rudich J, editors. New York: Chapman & Hall; 1986. pp. 201–239. [Google Scholar]
- 3.Bohner J, Bangerth F. Plant Growth Regul. 1988;7:141–155. [Google Scholar]
- 4.Joubes J, Phan T-H, Just D, Rothan C, Bergounioux C, Raymond P, Chevalier C. Plant Physiol. 1999;121:857–869. doi: 10.1104/pp.121.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallant P, Shiio Y, Cheng P-F, Parkhurst S M, Eisenman R N. Science. 1996;274:1523–1527. doi: 10.1126/science.274.5292.1523. [DOI] [PubMed] [Google Scholar]
- 6.Datar S A, Jacobs H W, de la Cruz A F, Lehner C F, Edgar B A. EMBO J. 2000;19:4543–4554. doi: 10.1093/emboj/19.17.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prober D A, Edgar B A. Cell. 2000;100:435–446. doi: 10.1016/s0092-8674(00)80679-0. [DOI] [PubMed] [Google Scholar]
- 8.Potter C J, Huang H, Xu T. Cell. 2001;105:357–368. doi: 10.1016/s0092-8674(01)00333-6. [DOI] [PubMed] [Google Scholar]
- 9.Mizukami Y, Fischer R L. Proc Natl Acad Sci USA. 2000;97:942–947. doi: 10.1073/pnas.97.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krizek B A. Dev Genet. 1999;25:224–236. doi: 10.1002/(SICI)1520-6408(1999)25:3<224::AID-DVG5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 11.Tsuge T, Tsukaya H, Uchimiya H. Development (Cambridge, UK) 1996;122:1589–1600. doi: 10.1242/dev.122.5.1589. [DOI] [PubMed] [Google Scholar]
- 12.Jones A M, Im K H, Savka M A, Wu M-J, DeWitt N G, Shillito R, Binns A N. Science. 1998;282:1114–1117. doi: 10.1126/science.282.5391.1114. [DOI] [PubMed] [Google Scholar]
- 13.Schoof H, Lenhard M, Haecker A, Mayer K F X, Jurgens G, Laux T. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 14.Jasinski S, Riou-khamlichi C, Roche O, Perennes C, Bergounioux C, Glab N. J Cell Sci. 2002;115:973–982. doi: 10.1242/jcs.115.5.973. [DOI] [PubMed] [Google Scholar]
- 15.Doerner P, Jorgensen J E, You R, Steppuhn J, Lamb C. Nature. 1996;380:520–523. doi: 10.1038/380520a0. [DOI] [PubMed] [Google Scholar]
- 16.Alpert K B, Grandillo S, Tanksely S D. Theor Appl Genet. 1995;91:994–1000. doi: 10.1007/BF00223911. [DOI] [PubMed] [Google Scholar]
- 17.Grandillo S H, Ku H M, Tanksley S D. Theor Appl Genet. 1999;99:978–987. [Google Scholar]
- 18.Frary A, Nesbitt T C, Frary A, Grandillo S, van der Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert K B, et al. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- 19.Nesbitt T C, Tanksley S D. Plant Physiol. 2001;127:575–583. [PMC free article] [PubMed] [Google Scholar]
- 20.Nesbitt T C, Tanksley S D. Genetics. 2002;162:365–379. doi: 10.1093/genetics/162.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alpert K B, Tanksley S D. Proc Natl Acad Sci USA. 1996;93:15503–15507. doi: 10.1073/pnas.93.26.15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassam B J, Allen T, Flood S, Stevens J, Wyatt P, Livak K J. Aust Biotechnol. 1996;6:285–294. [Google Scholar]
- 23.PE Applied Biosystems. User Bulletin No. 2: ABI PRISM 7700 Sequence Detection System. Foster City, CA: PE Applied Biosystems; 1997. [Google Scholar]
- 24.Jackson D. In: Molecular Plant Pathology: A Practical Approach. Gurr S J, McPherson M, Bowles D J, editors. London: IRL; 1992. pp. 163–174. [Google Scholar]
- 25.Coen E S, Romero J M, Doyle S, Elliott R, Murphy G, Carpenter R. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- 26.Gould S J. In: Evolution and Development. Bonner J-T, editor. New York: Springer; 1982. pp. 333–346. [Google Scholar]
- 27.Lord E M, Hill J P. In: Development as an Evolutionary Process. Raff R A, Raff E C, editors. New York: Liss; 1987. pp. 47–70. [Google Scholar]
- 28.Johnson N A, Porter A H. J Theor Biol. 2000;205:527–542. doi: 10.1006/jtbi.2000.2070. [DOI] [PubMed] [Google Scholar]
- 29.Asai K, Satoh N, Sasaki H, Satoh H, Nagato Y. Development (Cambridge, UK) 2002;129:265–273. doi: 10.1242/dev.129.1.265. [DOI] [PubMed] [Google Scholar]
- 30.Itoh J-L, Haseyawa H, Kitano H, Nagato Y. Plant Cell. 1998;10:1511–1521. doi: 10.1105/tpc.10.9.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dudley M, Poethig R. Development (Cambridge, UK) 1991;111:733–739. doi: 10.1242/dev.111.3.733. [DOI] [PubMed] [Google Scholar]
- 32.Evans M, Passas H, Poethig R. Development (Cambridge, UK) 1994;120:1971–1981. doi: 10.1242/dev.120.7.1971. [DOI] [PubMed] [Google Scholar]
- 33.Freeling M, Bertrand-Garcia R, Sinha N. BioEssays. 1992;14:227–236. doi: 10.1002/bies.950140405. [DOI] [PubMed] [Google Scholar]
- 34.Fridman E, Pleban T, Zamir D. Proc Natl Acad Sci USA. 2000;97:4718–4723. doi: 10.1073/pnas.97.9.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi Y, Shomura A, Sasaki T, Yano M. Proc Natl Acad Sci USA. 2001;98:7922–7927. doi: 10.1073/pnas.111136798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Din El-Assal S, Alonso-Blanco C, Peeters A J, Raz V, Koornneef M. Nat Genet. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- 37.Wang R-L, Stec A, Hey J, Lukens L, Doebley J. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- 38.White S E, Doebley J F. Genetics. 1999;153:1455–1462. doi: 10.1093/genetics/153.3.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thornsberry J M, Goodman M M, Doebley J, Kresovich S, Nielsen D, Buckler E S, IV. Nat Genet. 2001;28:286–289. doi: 10.1038/90135. [DOI] [PubMed] [Google Scholar]
- 40.Killander D, Zetterberg A. Exp Cell Res. 1965;38:272–284. doi: 10.1016/0014-4827(65)90403-9. [DOI] [PubMed] [Google Scholar]
- 41.Dirick L, Bohm T, Nasmyth K. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall C J. Curr Opin Cell Biol. 1996;8:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- 43.Edgar B A, Lehner C F. Science. 1996;274:1646–1652. doi: 10.1126/science.274.5293.1646. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke L C. Plant J. 2000;24:613–623. doi: 10.1046/j.1365-313x.2000.00899.x. [DOI] [PubMed] [Google Scholar]
- 45.Song J-Y, Leung T, Ehler L K, Wang C, Liu Z. Development (Cambridge, UK) 2000;127:2207–2217. doi: 10.1242/dev.127.10.2207. [DOI] [PubMed] [Google Scholar]
- 46.De Veylder L, Beeckman T, Beemster G T, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inze D. Plant Cell. 2001;13:1653–1668. doi: 10.1105/TPC.010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fero M L, Rivkin M, Tasch M, Porter P, Carow C E, Firpo E, Polyak K, Tsai L-H, Broudy V, Perlmutter R M, et al. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 48.Peunova N, Scheinker V, Cline H, Enikolopov G. J Neuroscience. 2001;21:8809–8818. doi: 10.1523/JNEUROSCI.21-22-08809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]