A dominant block to HIV-1 replication at reverse transcription in simian cells (original) (raw)
Abstract
Although nonhuman primates are genetically close to humans, their T cells do not support productive replication of HIV-1. In contrast, HIV-1 replicates in activated human CD4+ T cells, monocytes, and metabolically active human cells of a variety of cell types become permissive for HIV-1 replication when transduced to express CD4 and CCR5 or CXCR4. The molecular basis of this species restriction to HIV-1 replication was investigated by using African green monkey and rhesus macaque cell lines that were stably transduced to express human CD4 and CCR5. The cells supported replication of cognate viruses [simian immunodeficiency virus from African green monkeys (SIV-AGM) and macaques (SIVmac239)] but did not support replication of an R5-tropic cytopathic HIV-1. A β-lactamase-based HIV-1 entry assay was used to show that the virus efficiently entered the nonhuman primate cells. Provirus formation was reduced 50-fold compared with similarly infected human cells. Real-time PCR quantitation demonstrated that reverse transcription failed to initiate efficiently in the simian cells. The block to reverse transcription was overridden at multiplicity of infection >1 or by preincubation of the nonhuman primate cells with virus, a feature reminiscent of the Friend virus resistance gene-1 (FV-1), restriction to murine leukemia virus replication in mouse cells. Heterokaryon analysis in which human and simian cells were fused demonstrated that the block was dominant. These findings suggested that the primate cells contain a dominant inhibitor that prevents HIV-1 reverse transcription.
Although HIV-1 was introduced into the human population from a nonhuman primate reservoir in the chimpanzee fairly recently (1), it does not infect nonhuman primates and replicates poorly or not at all on activated nonhuman primate lymphocytes in vitro (2, 3). HIV-1 can replicate at detectable levels in experimentally infected chimpanzees (4–7), but virus loads remain low, and chimpanzee T cells in culture are largely refractory to infection (8). Studies in the pig-tailed macaque have similarly demonstrated detectable but inefficient virus replication in vivo and in vitro (9, 10). The inability of the virus to replicate in simian cells, which are genetically very similar to human, is presumably the result of adaptive alterations in HIV-1 that are incompatible with replication in nonhuman primates. The restricted host range of HIV-1 limits its use in primate models.
Retroviral host range is determined in part by the interaction of virus with target cell factors. For example, species-specific interactions between the surface component (SU) of the retroviral envelope glycoprotein and its cell surface receptor are required for virus entry. HIV-1 entry is mediated by the binding of gp120 to CD4 and then to a chemokine receptor, either CCR5 or CXCR4. HIV-1 does not enter mouse cells, because gp120 does not bind murine CD4 or CCR5 (11, 12). Retroviral host range can also be restricted postentry. Mouse cells engineered to express hCD4 and hCCR5 support HIV-1 entry, yet the virus fails to productively replicate due to blocks in provirus transcription and virus assembly. The virus genomic RNA is reverse transcribed and integrated, but due to inefficient Tat-mediated transactivation, viral mRNA transcripts fail to accumulate. Transactivation fails because murine cyclin T1 is altered at amino acid 160, preventing its interaction with HIV-1 Tat (13, 14). Expression of human cyclin-T1 largely restores Tat function, causing viral mRNAs to accumulate and virion proteins to be synthesized. The proteins, however, fail to efficiently assemble and release (15, 16).
Several studies have suggested that the block to HIV-1 replication in simian cells is postentry (17–19). Rhesus CD4 and CCR5 are very similar to their human homologues and are competent to mediate HIV-1 entry when expressed on human cells (20). Infection of rhesus peripheral blood mononuclear cells with HIV-1 results in only small amounts of reverse transcripts (18). Hofmann et al. (21) studied a large number of cell lines and found that titers of an HIV-1 reporter virus were lower than those of simian immunodeficiency virus SIVmac239 or murine leukemia virus-based vectors in Old World monkey cells than in New World monkey-derived cell lines.
Here we further analyzed the basis of the block to HIV-1 replication in nonhuman primate cells. The African green monkey cell line CV-1 and two cell lines from rhesus macaque were transduced to stably express the human entry cofactors hCD4 and hCCR5. Nevertheless, they failed to support productive replication of a highly cytopathic R5-tropic HIV-1. The cells were permissive for replication of cognate SIV. The block to HIV-1 replication was postentry and appeared to result from a failure to initiate reverse transcription. The block was overridden by high multiplicity of infection (moi) with glycoprotein of vesicular stomatitis virus (VSV-G) pseudotyped virus. Heterokaryon analysis suggested that the block was caused by a dominant inhibitor of reverse transcription or uncoating.
Materials and Methods
Cells.
Adherent cell lines HOS, 293T, CV-1 (African green monkey kidney cells), FrHL-2 [rhesus macaque lung cells (22)], and HeLa.CD4 were cultured in DMEM, 10% FBS. hCD4 and hCCR5 were introduced stably as described (15). OM10.1 and 5.25 were cultured in RPMI 1640/10% FBS/10 mM Hepes; MM221–92 [rhesus macaque T cells (21)] were cultured in RPMI 1640/20% FBS/25 mM Hepes/3.8 μM 2-mercaptoethanol/100 units of IL-2/ml. GHOST.CCR5 are HOS cells that contain an HIV-2 LTR-enhanced green fluorescent protein (EGFP) cassette and express hCD4 and hCCR5 (23). 5.25 cells are CEMx174 cells that contain LTR-EGFP and LTR-luciferase cassettes and express CD4 CCR5/CXCR4.
Viruses and Infections.
Replication-competent NL4.3, NL.BaL (NL4.3 with the env of BaL) (24), SIVmac239, and SIVagmSAB-1 [pSAB-1 (25)] virus stocks were prepared by harvesting the supernatant of transfected 293T cells. To generate VSV-G-pseudotyped HIV-1 or SIV, 293T cells were cotransfected with equal amounts of HIV-1 or SIV DNA [pNL4.3, pNL4.3R− pNL4.3R−E−, pNL4.3pol−R−E− (26), pNL-Bal, pSIVmac239, pSAB-1] and pCMV-VSV-G (27). HIV-1 and SIVmac single-cycle luciferase reporter viruses were produced by cotransfecting 293T cells with pNL-LucR−E−, pNL-RLucR−E− (15), or SIV-LucR−E− and JR.FL or VSV-G Env expression vector (28). Viruses were quantitated by p24_gag_ or p27_gag_ ELISA (Cellular Products). Infectious titer of replication-competent viruses was measured by end-point dilution in quadruplicate on 5.25 cells or GHOST.R5. Single-cycle viruses were titered on GHOST.R5 cells by flow cytometry. For reporter virus infections, the cells were seeded at 2.0 × 103 cells/well in 96-well plates and then infected with titered reporter virus stock. Luciferase activity was measured 3 days later with Luc-Lite (Packard) or Stop and Glow reagents (Promega) according to the manufacturer's directions on a Packard TopCount luminometer.
Preincubation Experiments.
Cells were preincubated for 6 h with virus at moi 10, washed five times with culture medium, and then infected with NL-LucR−E−(VSV-G) for 6 h at moi 0.1. Alternatively, the two viruses were mixed, and then the cells were infected for 6 h or with NL-Luc(VSV-G) moi 0.1 and then 6 h later exposed to virus at moi 10.
Entry Assay.
β-Lactamase-containing virions were produced by cotransfection of 293T cells with equal amounts of pNL-Bal and pMM310, a vector that encodes a β-lactamase-Vpr fusion protein (provided by Michael Miller, Merck). Cells (2.0 × 105) in six-well dishes were infected with β-lactamase-loaded virus at moi 0.1 and 1.0 in 1 ml. After 4–5 h, the cells were washed twice with PBS and then incubated with 2 μM CCF2/AM (GeneBLAzer Loading Kit, Aurora Biosciences, La Jolla, CA)/1% probenecid in 1.0 ml of serum-free DMEM/1 mM Hepes for 16–18 h at 25°C 5% CO2. The cells were then trypsinized, fixed in 1% paraformaldehyde/PBS for 10 min, and analyzed on a BD LSR 3 analytical flow cytometer (Becton Dickinson) with a UV laser and 470-nm long-pass dichroic filter exciting at 325 nm. Cleaved substrate was detected as blue fluorescence with a 424/44-nm bandpass filter, and uncleaved substrate was detected with a 516/20-nm bandpass filter. The results were plotted as the ratio of the blue/green to minimize differential dye loading.
Quantitative-Competitive PCR.
HOS and CV-1 cells (1.0 × 106) were infected with NL4.3(VSV-G) moi 1.0, and 4 days later genomic DNA was prepared. Increasing amounts of a competitor consisting of pNL4.3 deleted in env from _Nhe_I-_Hpa_I sites (nucleotides 6,628–8,028) was added to 500 ng of genomic DNA. Proviruses were amplified by using primers RC-9 and RC-12 (29) that flank env. PCR was for 5 min at 95°C, followed by 25 cycles of 2 min at 92°C, 30 s at 55°C, and 5 min at 72°C. Amplicons were visualized on 1% agarose gels stained with ethidium bromide and quantitated by densitometry.
Real-Time PCR Quantitation of Reverse Transcripts.
Virus stocks derived by transfection were treated with 10–20 units/ml of DNase-I (Roche) for 60 min at room temperature to remove plasmid and viral DNA. Cells (1.0 × 106) were infected in 100-mm dishes, and genomic DNA was prepared at the indicated time points. Early HIV-1 reverse transcripts were quantitated with primers ert2f (5′-GTG CCC GTC TGT TGT GTG AC-3′) and ert2r (5′-GGC GCC ACT GCT AGA GAT TT-3′) and the probe ERT2 probe [5′-(FAM)-CTA GAG ATC CCT CAG ACC CTT TTA GTC AGT GTG G-(TAMRA)-3′; late HIV-1 reverse transcripts were quantitated with primers MH531, MH532, and the probe LRT-P; HIV-1 2-LTR circles were quantitated with primers MH535, MH536, and the probe MH603; proviruses were quantitated with primers containing an Alu repeat SB704 and MH535 and the probe MH603 (30). Late reverse transcripts of SIVagmSAB-1 were quantitated with primer SMB14 (5′-ACA GCA GGC CGG AGG AA-3′), SMB15 (5′-GGA TCG TCT GGC CCT GGT-3′), and probe SMB16 [5′-(FAM)-ATT GGT ATG CAG ACA GAC CTC CGA CCA-(TAMRA)-3′]; and of SIVmac239 with primer SMB1 (5′-TTG GGA AAC CGA AGC AGG-3′), SMB2 (5′-TCT CTC ACT CTC CTT CAA GTC CCT-3′) with the probe SMB3 [5′-(FAM)-AAA TCC CTA GCA GAT TGG CGC CTG AA-(TAMRA)-3′]. Reaction mixtures contained Taqman universal master mix (Perkin–Elmer), 300 nM primers, 100 nM probe, and 500 ng of genomic DNA. PCR was for 2 min at 50°C, 10 min at 95°C followed by 40 cycles of 15 s at 95°C, 1 min at 60°C (1 min 30 s for Alu primers) on an ABI Prism 7700 (Applied Biosystems). Nevirapine was added to controls to 10 μm.
Heterokaryon Formation.
hCD4+ cells (4 × 104) were seeded in triplicate in a 24-well dish with an equal number of hCCR5+ cells. The cells were fused the next day by adding 200 μl of 50% (wt/vol) polyethylene glycol (PEG) 3350. After 5 min at room temperature, the PEG was removed by repeated washing with DMEM. The heterokaryons were cultured another 24 h and then infected with luciferase reporter virus. To monitor heterokaryon formation, one cell was labeled with CellTracker Green 5-chloromethylfluorescein diacetate (CMFDA) and the other with CellTracker Orange 5-(and -6)-(((4-chloromethyl)benzoyl)amino)tetramethylrhodamine (CMTMR) (Molecular Probes), and the next day the cells were analyzed by flow cytometry.
Western Blotting.
Virions and lysates were prepared 3 days after infection with NL4.3(VSV-G) as described (15). Lysates containing 10 μg of protein were separated by SDS/PAGE and transferred to polyvinylidene difluoride filters. Filters were probed with AIDS patient serum at 1:2,500 dilution (provided by D. Richman, University of California, San Diego) or rabbit anti-gp120 serum followed by horseradish peroxidase-conjugated rabbit anti-human or donkey anti-rabbit antibody and developed with chemiluminescence reagents.
Results
HIV-1 Does Not Replicate in Simian Cell Lines Expressing Human Entry Cofactors.
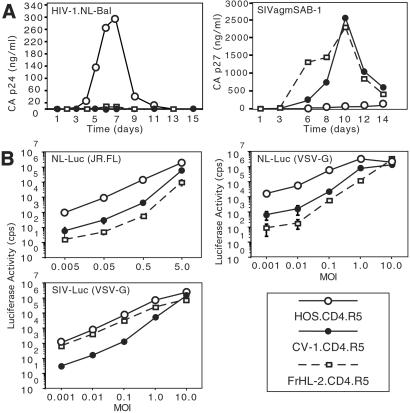
Human cell lines that express endogenous or transfected CD4 and CCR5 generally support productive HIV-1 infection. We were interested in determining whether nonhuman primate cells that express the human entry cofactors would become permissive for HIV-1 replication. Introducing the human entry cofactors into mouse cells does not restore permissiveness (15, 16), but nonhuman primates are genetically much closer to humans. The African green monkey kidney cell line, CV-1, and the rhesus macaque fibroblast cell line, FrHL-2, were transduced by retroviral vectors to express hCD4 and hCCR5. Similarly transduced human osteosarcoma cells, HOS.CD4.R5, were used for comparison. Expression of the entry cofactors was confirmed by flow cytometry (Fig. 8, which is published as supporting information on the PNAS web site, www.pnas.org). The simian cells expressed CD4 at a level similar to HOS.CD4.R5. CCR5 was about 5-fold lower than HOS.CD4.R5, but these levels are sufficient to mediate efficient virus entry (31). The simian and human cell lines were infected with the highly cytopathic R5-tropic HIV-1, NL-BaL (24). The virus replicated with rapid kinetics on HOS.CD4.CCR5 but failed to replicate detectably in either of the simian cell lines (Fig. 1A). In contrast, SIVagmSAB-1 replicated efficiently in rhesus and AGM cells but to a lesser extent in the human cell line (Fig. 1A). These findings suggest that there is a species-specific block to HIV-1 replication in both of the simian cell lines. SIVagm was less restricted in its host range, being able to infect both simian and human cells.
Figure 1.
HIV-1 does not infect simian cell lines. (A) HOS.CD4.R5, CV-1.CD4.R5, FrHL-2.CD4 were infected at moi 0.05 with HIV-1 and SIVagmSAB. Supernatant virions were quantitated by p24 or p27 ELISA at the indicated days. (B) Cells were infected with increasing amounts of NL-LucR−E−(JR.FL), NL-LucR−E−(VSV-G), or SIV-LucR−E−(VSV-G). Luciferase activity was determined 3 days postinfection.
A Postentry Block to HIV-1 Replication in Primate Cells.
To localize the block to an early or late step in virus replication, the cells were infected with single-cycle luciferase reporter virus pseudotyped by the R5-tropic HIV-1 glycoprotein JR.FL. At low moi (0.005–0.5), luciferase activity in the simian cells was 30- to 200-fold less than that of the human cells (Fig. 1B). The difference between simian and human became smaller as the moi approached 5. This was not specific to the envelope glycoprotein, because cells infected with VSV-G pseudotyped virus yielded similar results (Fig. 1B). The difference between human and simian infection narrowed as more virus was used until, at moi 10, simian cell activity approached that of human cells. The plateau in luciferase activity achieved at the highest moi was not due to saturation of the luciferase assay, because dilution of the cell lysate before measurement did not affect the results (data not shown). SIVmac239 luciferase reporter virus infected the rhesus and the human cells but was about 50-fold less infectious on the AGM cells (Fig. 1B). Thus, the block to HIV-1 infection of the simian cells is species-specific but is overridden at high moi.
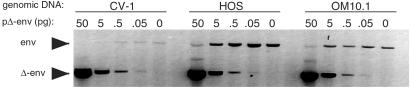
To determine whether the reduced luciferase activity in the infected cells was caused by a prior postintegration block, we quantitated the proviral load in the infected cells. Cells lacking entry cofactors were infected with NL4.3(VSV-G) at moi 1. Four days postinfection, genomic DNA was prepared, and provirus copy number was quantitated by quantitative-competitive PCR. This analysis showed that the CV-1 cells contained 50-fold fewer proviruses than HOS (Fig. 2).
Figure 2.
Infected simian cells contain low provirus loads. CV-1 and HOS were infected with NL4.3(VSV-G) at moi 1. Four days later, proviruses were quantitated by QC-PCR. Competitor HIV-1 DNA containing a deletion in env was added in the indicated amounts. DNA from OM10.1 cells, which contain a single provirus per cell, was used for calibration. The proviral load per cell calculated from band intensities normalized to OM10.1 was as follows: CV-1 = 0.1, HOS = 5.0.
Efficient Entry of HIV-1 into Simian Cells.
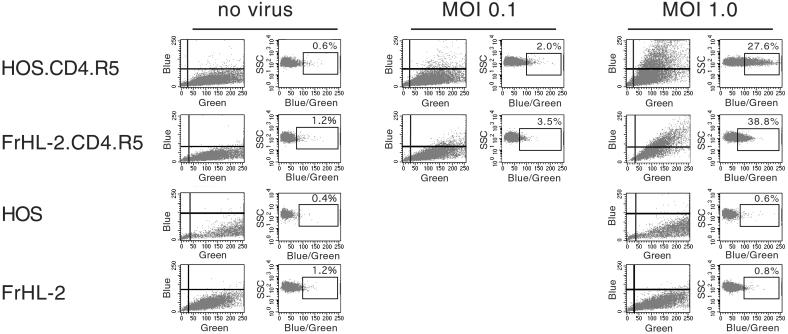
The transduced simian cell lines expressed hCD4 and hCCR5 at levels that on human cells are sufficient to meditate efficient HIV-1 entry. Whether the entry cofactors work as well on the simian cells was not clear. To determine this, we used an assay that specifically measures HIV-1 entry. HIV-1 virions loaded with an enzymatically active β-lactamase-Vpr fusion protein (BlaM-Vpr) were prepared by cotransfection. Target cells were infected with the virus and then exposed to CCF2/AM, a membrane-permanent BlaM substrate. Entry of the virus introduces into the cytoplasm active BlaM that cleaves the substrate. The ratio of cleaved to uncleaved substrate was measured by flow cytometry as the blue/green fluorescence ratio.
HOS.CD4.R5 and FrHL-2.CD4.R5 cells were infected with BlaM-Vpr-loaded virus at mois 0.1 and 1 and then incubated with the substrate. CV-1 cells were not used, because these were found not to take up the substrate. Blue/green fluorescence increased significantly for HOS.CD4.R5 and FrHL-2.CD4.R5 (Fig. 3). For the simian cells, the ratio was not as high as for the human cells, suggesting that fewer virions had entered per cell. This is likely due to the 5-fold lower abundance of CCR5 on FrHL-2.CD4.R5. Infection of the cells with VSV-G pseudotyped BLaM virus resulted in similar blue/green ratios in the two cell lines (Fig. 9, which is published as supporting information on the PNAS web site). Infection of HOS.CD4 and FrHL-2.CD4 cells that lack CCR5 resulted in no significant increase in fluorescence ratio, demonstrating that the assay detected only productive entry and not nonspecific virus uptake (data not shown). The results demonstrate that simian cells expressing hCD4 and hCCR5 support efficient HIV-1 entry, and thus that the block to replication is postentry.
Figure 3.
HIV-1 enters the simian cell lines. HOS.CD4.R5, FrHL-2.CD4.R5, HOS, and FrHL-2 were infected with NL-Bal(BlaM-Vpr). After 4 h, the virus was removed, and the cells were loaded with the BlaM substrate CC2F/AM for 16–18 h at 25°C. The blue/green ratio was quantitated by flow cytometry. Cleavage of the dye by BlaM upon virus entry causes an increase in blue/green. The data are presented as the intensity of blue and green and as side scatter vs. blue/green ratio. The blue/green measurement controls for cell-type differences in dye loading.
HIV-1 Reverse Transcription Is Inefficient in Simian Cells.
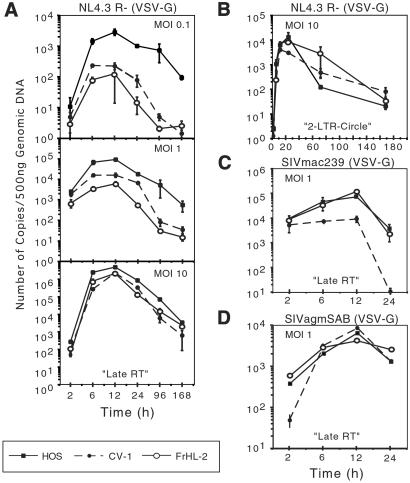
To more precisely pinpoint the block to replication in simian cells, we used real-time PCR to quantitate the synthesis of HIV-1 cDNA. Early reverse transcripts, late reverse transcripts, 2-LTR circles, and integrated provirus were quantitated by using specific primer and probe combinations. To limit infection to a single round, cells lacking entry cofactors were infected with VSV-G pseudotypes. The results showed that early and late transcripts accumulated with similar kinetics in the simian and human cells, but that the absolute number of transcripts was decreased in the simian cells (Fig. 4A and Fig. 10, which is published as supporting information on the PNAS web site). The difference between human and simian cells was more pronounced for the late than the early reverse transcripts. At moi 1, the difference in viral DNA abundance between HOS and CV-1 was less pronounced, and at moi 10, there was no significant difference.
Figure 4.
Real-time PCR analysis of HIV-1 and SIV reverse transcripts synthesized shortly after infection. Human and simian cells were infected with NL4.3R−(VSV-G) at moi 0.1, 1.0, and 10. Genomic DNA was prepared at the indicated time points, and reverse transcripts were quantitated by real-time PCR by using primers that detected (A) late reverse transcripts, (B) 2-LTR circles (2-LTR circles were measured at moi 10). Infection with SIVmac239(VSV-G) (C) and SIVagmSAB-1(VSV-G) (D). The data are plotted as copy numbers/500 ng of DNA. Cultures pretreated with nevirapine had 40 copies/500 ng of DNA and did not increase over time (not shown).
2-LTR circles, which are a marker of nuclear import of the preintegration complex, were present at similarly high levels in the human and simian cells at moi 10 (Fig. 4B). Quantitation of the proviral copy number using an Alu probe/primer combination 72 h postinfection demonstrated similar efficiencies of integration (3.3 × 105 for HOS, 7.38 × 104 for CV-1, and 7.95 × 104 for FrHL-2 proviruses/500 ng DNA). Normalizing for the total number of full-length viral cDNAs at 12 h postinfection (4.8 × 106, 2.1 × 106, and 2.3 × 106 copies/500 ng DNA), the relative efficiency of provirus formation was 6.86% for HOS, 3.51% for in CV-1, and 3.46% for FrHL-2. Thus, the findings suggested that nuclear import of the preintegration complex and integration is comparable in human and simian cells. Similar results on HOS.CD4.R5, CV1.CD4.R5, and FrHL2.CD4.R5 cells were also obtained by using nonpseudotyped replication-competent viruses produced by infected cells (data not shown). Real-time PCR analysis of lymphoid cell lines also yielded similar results. The rhesus T cell line MM221-89 contained 100-fold fewer HIV-1 late reverse transcripts compared with the human T cell line CEMx174 (Fig. 10). SIV-AGM reverse transcripts were present at similar amounts in human and primate cells (Fig. 4D). SIVmac239 reverse transcripts were not detectable in CV-1 cells, consistent with the inability of this virus to replicate in the cell line (Fig. 4C).
Simian Cells Contain an Inhibitory Activity That Acts Postentry.
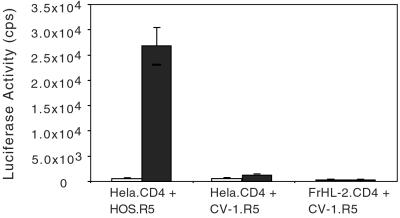
The block to reverse transcription in the simian cells could be due to the absence of a required factor or the presence of an inhibitory factor. To distinguish between these possibilities, we generated human–simian cell heterokaryons and tested them for the ability to be infected with luciferase reporter virus. To ensure that only the interspecies heterokaryons were measured, CD4 was expressed in one cell and CCR5 in the other. The heterokaryons were then infected with NL-LucR−E−(JR.FL). Heterokaryons formed between HeLa.CD4 and CV-1.R5 resulted in low luciferase activity, as did control fusions of FrHL-2.CD4 to CV-1.CCR5 (Fig. 5). In contrast, human/human heterokaryons formed by fusion of HeLa.CD4 to HOS.R5 had high luciferase activity. The infection required cell/cell fusion, as omission of the polyethylene glycol prevented luciferase activity. The findings suggested that the simian cells contain a cellular cofactor that interferes with HIV-1 infection.
Figure 5.
Infection of human/simian heterokaryons. Human and simian cell lines expressing hCD4 or hCCR5 were mixed (1:1) and then fused by exposure to polyethylene glycol for 5 min (filled bars) or left untreated as a control (open bars). The heterokaryons were infected with NL-LucR−E−(JR.FL), and luciferase activity was analyzed 3 days postinfection.
Efficient Virus Assembly and Release in Simian Cells.
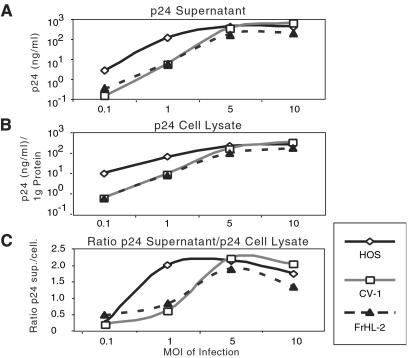
COS cells, which are derived from CV-1, have been widely used to produce HIV-1 by transfection. To confirm that the infected simian cells synthesized and processed HIV-1 proteins, the CV-1 and FrHL-2 were infected at different mois, and cell lysates and supernatants were then analyzed on immunoblots probed with AIDS patient and anti-gp120 serum (Fig. 11, which is published as supporting information on the PNAS web site). No differences in the pattern of viral proteins produced and released by the human and simian cells were noted. Consistent with the results described above, the simian cells produced relatively little virus at low moi but reached levels comparable to the human cells at high moi (Fig. 6 A and B). In addition, the ratio of extracellular to intracellular p24 was similar in the human and simian cells, demonstrating that the simian cells efficiently released virions (Fig. 6C).
Figure 6.
Simian cells efficiently release HIV-1 virions. HOS, CV-1, and FrHL-2 cells were infected with NL4.3(VSV-G) at the indicated moi. Input virus was thoroughly removed after 6 h and 3 days later, supernatant (A) and intracellular (B) p24 was measured. (C) The ratio of supernatant/lysate p24.
The Block to HIV-1 Infection in Simian Cells Is Saturable.
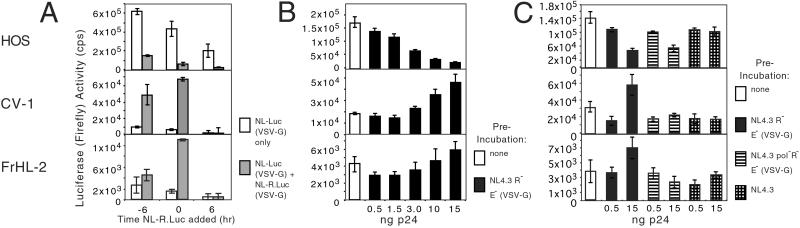
Studies in the murine system have shown that preincubation of cells at high moi relieves the block to murine leukemia virus infection in nonpermissive cells (32). We determined whether the block to HIV-1 infection could be overridden by preincubating the target cells with virus at high moi. The cells were exposed first at moi 10 to a reporter virus encoding Renilla luciferase, NL-R.LucR−E−(VSV-G). The cells were infected 6 h later at low moi with the firefly luciferase reporter virus NL-LucR−E−(VSV-G) (Fig. 7A). Alternatively, the two viruses were mixed and added to the cells. Preincubation of HOS cells with the Renilla virus caused a 5- to 8-fold decrease in infection with the firefly reporter virus. The mixture of the two viruses gave a similar result. This decrease was unexpected and may have been due to depletion of the pool of free nucleotide triphosphates in the cytoplasm as a result of the large number of initiated reverse transcription reactions. In contrast, preincubation of the FrHL-2 and CV-1 cells with the Renilla luciferase reporter virus had the opposite effect, causing a 2- to 6-fold increase in firefly luciferase activity. Similarly, coinfection caused a 7- to 12-fold increase in firefly reporter virus. To control for nonspecific effects, the Renilla virus was added 6 h postinfection with the firefly luciferase virus. This had no effect on firefly luciferase activity. Renilla luciferase activity was also quantitated in each of the experiments to ensure that the preinfection was not toxic. This activity was similar in each of the experiments (data not shown). The effect of preincubation on the subsequent infection was proportional to the amount of virus used (Fig. 7B). Taken together, the data suggested that an inhibitory factor in the simian cells was saturated by preincubating the target cells with virus.
Figure 7.
Preincubation with virus abrogates the block to HIV-1 infection. (A) Cells (HOS, CV-1, FrHL-2) were infected with Renilla reporter virus, NL-R.LucR−E−(VSV-G), at moi 10 6 h before, at the same time as, or 6 h after infection with firefly reporter virus, NL-LucR−E−(VSV-G). Firefly and Renilla luciferase activity was determined 3 days postinfection. The cells were infected with both viruses or with the firefly reporter virus alone. (B) The cells were preincubated with increasing amounts of the NL4.3R−E−(VSV-G) and infected 6 h later with the firefly reporter virus at moi 0.1. (C) The cells were preincubated for 6 h with 0.5 and 15 ng p24 of NL4.3R−E−(VSV-G), NL4.3pol−R−E−(VSV-G), and NL4.3 not pseudotyped followed by an infection of NL-LucR−E−(VSV-G).
The mechanism by which preincubation relieved the block to infection of the simian cells was investigated by exposing the target cells to variants of the virus used for preincubation. Preincubation of the cells with NL4.3 had no effect on the subsequent infection. This virus could not enter the target cells, which lacked CD4, thus the effect appeared to require virus entry. Virus that lacked pol also failed to mediate the effect, suggesting a requirement for reverse transcription (Fig. 7C).
Discussion
AGM and rhesus macaque cells lines that expressed hCD4 and hCCR5 readily supported replication of their cognate viruses, SIVagm and SIVmac239, but failed to support replication of a highly cytopathic R5 HIV-1. The failure of HIV-1 to replicate on the simian cells appeared to be due to inefficient initiation of reverse transcription after entry of the virus into the target cell cytoplasm. The block was relieved by infection of the target cells at high moi. The block was also relieved by preexposing the cells to virus. The block to infection was dominant, as human/simian heterokaryons could not be efficiently infected by HIV-1 reporter virus. These findings were most consistent with the presence of an inhibitory factor in the simian cells that reduced the frequency with which HIV-1 was reverse transcribed after entry.
These findings were most likely due to the species difference between the human and simian cells and not to cell-type differences. Tissue-specific factors are not generally required for HIV-1 replication. Actively dividing human cells of most any cell type will support HIV-1 replication once transduced to express the hCD4 and CCR5 or CXCR4. Cellular activation is required for HIV-1 replication, because resting cells generally do not support HIV-1 replication. The simian cells used here grow rapidly and are metabolically active. The ability of the cells to support SIV replication further pointed to the species specificity of the phenotype.
The block to HIV-1 replication in the simian cells is reminiscent of FV-1 in the murine system (32). FV-1 encodes an endogenous Gag protein that imposes a dominant block restricting the replication of N and B tropic murine leukemia virus (MuLV). The restriction maps to a small region of CA. The block to HIV-1 infection of simian cells is similar in that it is overridden at high moi and is due to a dominant inhibitory factor. FV-1 restriction differs in that it occurs after reverse transcription, perhaps at nuclear import. The block in simian cells occurs earlier, acting at or before reverse transcription. Towers et al. (33) described an FV-1-like restriction, termed Ref1, that prevents the replication of N-tropic MuLV in human cells. Ref-1 restriction is determined by amino acid 110 of CA and, like the restriction to HIV-1 replication in simian cells, is at reverse transcription. Moreover, the restriction is also dominant and overridden by high moi (34). A similar gene may act in the simian cells to block HIV-1 reverse transcription. The failure to reverse transcribe could be due to inhibition of reverse transcription itself or could be secondary to a failure to uncoat.
The BlaM entry assay that was used here is likely to be useful for other related applications. It is currently the only method for directly detecting HIV-1 entry. Syncytium formation assays are indirect and can be misleading. Initial attempts to measure virus entry by quantitating trypsin-resistant p24 postinfection were unreliable. The BlaM assay was accurate, quantitative, and specifically detected productive virus entry and not nonspecific uptake of virus.
Further understanding of the basis of this species specificity will require mapping the phenotype to a virus component. The ability of SIV-HIV chimers, SHIV, to replicate in simian cells rules out env, tat, rev, vpu, and nef (35, 36). Vif is not likely to have been responsible, because adherent cell lines are generally Vif permissive, which leaves Gag/Pol as potential targets of the inhibitory activity. Additionally, there could be a role for the viral RNA. Attempts to adapt HIV-1 to replicate in simian cell lines have been unsuccessful. Generation of an HIV-1 that replicates in simian cells will require identification of the component of the virus that restricts its replication.
Supplementary Material
Supporting Figures
Acknowledgments
We thank Susan Hedrick (Salk Institute) for technical assistance; and Michael Miller (Merck), Henrietta Nymark-McMahon (Salk Institute), Douglas Richman [University of California, San Diego (UCSD)], Carolyn Wilson (U.S. Food and Drug Administration), and Ronald Desrosiers (New England Regional Primate Research Center) for cells and reagents. pSAB-1 was obtained through the AIDS Research and Reference Reagent Program from Mojun Jin and Beatrice Hahn (University of Alabama). The studies were funded by National Institutes of Health Grants AI43252, CA7214, AI42397, and DA14497; and by a UCSD Center for AIDS Research (CFAR) development grant and Deutsche Forschungsgemeinschaft Grant MU 1608 (to C.M.). N.R.L. is an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation.
Abbreviations
SIV
simian immunodeficiency virus
VSV-G
glycoprotein of vesicular stomatitis virus
moi
multiplicity of infection
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, et al. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 2.Levy J A, Shimabukuro J, McHugh T, Casavant C, Stites D, Oshiro L. Virology. 1985;147:441–448. doi: 10.1016/0042-6822(85)90146-1. [DOI] [PubMed] [Google Scholar]
- 3.McClure M O, Sattentau Q J, Beverley P C, Hearn J P, Fitzgerald A K, Zuckerman A J, Weiss R A. Nature. 1987;330:487–489. doi: 10.1038/330487a0. [DOI] [PubMed] [Google Scholar]
- 4.Fultz P N, McClure H M, Swenson R B, Anderson D C. Intervirology. 1989;30:51–58. doi: 10.1159/000150124. [DOI] [PubMed] [Google Scholar]
- 5.Morrow W J, Homsy J, Eichberg J W, Krowka J, Pan L Z, Gaston I, Legg H, Lerche N, Thomas J, Levy J A. AIDS Res Hum Retroviruses. 1989;5:233–245. doi: 10.1089/aid.1989.5.233. [DOI] [PubMed] [Google Scholar]
- 6.Nara P, Hatch W, Kessler J, Kelliher J, Carter S. J Med Primatol. 1989;18:343–355. [PubMed] [Google Scholar]
- 7.ten Haaft P, Murthy K, Salas M, McClure H, Dubbes R, Koornstra W, Niphuis H, Davis D, van der Groen G, Heeney J. AIDS. 2001;15:2085–2092. doi: 10.1097/00002030-200111090-00003. [DOI] [PubMed] [Google Scholar]
- 8.Benton P A, Timanus D K, Shearer M H, White G L, Lee D R, Kennedy R C. Dev Comp Immunol. 1999;23:97–105. doi: 10.1016/s0145-305x(98)00045-7. [DOI] [PubMed] [Google Scholar]
- 9.Agy M B, Frumkin L R, Corey L, Coombs R W, Wolinsky S M, Koehler J, Morton W R, Katze M G. Science. 1992;257:103–106. doi: 10.1126/science.1621083. [DOI] [PubMed] [Google Scholar]
- 10.Otten R A, Brown B G, Simon M, Lupo L D, Parekh B S, Lairmore M D, Schable C A, Schochetman G, Rayfield M A. AIDS. 1994;8:297–306. doi: 10.1097/00002030-199403000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Landau N R, Warton M, Littman D R. Nature. 1988;334:159–162. doi: 10.1038/334159a0. [DOI] [PubMed] [Google Scholar]
- 12.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 13.Garber M E, Wei P, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mariani R, Rutter G, Harris M E, Hope T J, Krausslich H G, Landau N R. J Virol. 2000;74:3859–3870. doi: 10.1128/jvi.74.8.3859-3870.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bieniasz P D, Cullen B R. J Virol. 2000;74:9868–9877. doi: 10.1128/jvi.74.21.9868-9877.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibata R, Sakai H, Kawamura M, Tokunaga K, Adachi A. J Gen Virol. 1995;76:2723–2730. doi: 10.1099/0022-1317-76-11-2723. [DOI] [PubMed] [Google Scholar]
- 18.Himathongkham S, Luciw P A. Virology. 1996;219:485–488. doi: 10.1006/viro.1996.0276. [DOI] [PubMed] [Google Scholar]
- 19.Kimball L E, Bosch M L. J Med Primatol. 1998;27:99–103. doi: 10.1111/j.1600-0684.1998.tb00233.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petricciani F C, Huang C C, Rubin B A, Yang D P, Minecci L C, Kadanka Z, Earley E M. J Biol Stand. 1976;4:43–49. doi: 10.1016/0092-1157(76)90038-x. [DOI] [PubMed] [Google Scholar]
- 23.Cecilia D, KewalRamani V N, O'Leary J, Volsky B, Nyambi P, Burda S, Xu S, Littman D R, Zolla-Pazner S. J Virol. 1998;72:6988–6996. doi: 10.1128/jvi.72.9.6988-6996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mariani R, Rasala B A, Rutter G, Wiegers K, Brandt S M, Krausslich H G, Landau N R. J Virol. 2001;75:3141–3151. doi: 10.1128/JVI.75.7.3141-3151.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin M J, Hui H, Robertson D L, Muller M C, Barre-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paxton W, Connor R I, Landau N R. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emi N, Friedmann T, Yee J K. J Virol. 1991;65:1202–1207. doi: 10.1128/jvi.65.3.1202-1207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, et al. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 29.Connor R I, Sheridan K E, Lai C, Zhang L, Ho D D. J Virol. 1996;70:5306–5311. doi: 10.1128/jvi.70.8.5306-5311.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butler S L, Hansen M S, Bushman F D. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 31.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goff S P. Cell. 1996;86:691–693. doi: 10.1016/s0092-8674(00)80141-5. [DOI] [PubMed] [Google Scholar]
- 33.Towers G, Bock M, Martin S, Takeuchi Y, Stoye J P, Danos O. Proc Natl Acad Sci USA. 2000;97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Towers G, Collins M, Takeuchi Y. J Virol. 2002;76:2548–2550. doi: 10.1128/jvi.76.5.2548-2550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibata R, Kawamura M, Sakai H, Hayami M, Ishimoto A, Adachi A. J Virol. 1991;65:3514–3520. doi: 10.1128/jvi.65.7.3514-3520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexander L, Du Z, Howe A Y, Czajak S, Desrosiers R C. J Virol. 1999;73:5814–5825. doi: 10.1128/jvi.73.7.5814-5825.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figures